Abstract
Uveal melanoma is a lethal cancer with a strong propensity to metastasize. Limited therapeutic options are available once the disease has disseminated. A strong predictor for metastasis is the loss of chromosome 3. Inactivating mutations in BAP1 encoding the BRCA1-associated protein 1 and located on chromosome 3p21.1, have been described in uveal melanoma and other types of cancer. In this study, we determined the prevalence of somatic BAP1 mutations and examined whether these mutations correlate with the functional expression of BAP1 in uveal melanoma tissue and with other clinical, histopathological and chromosomal parameters. We screened a cohort of 74 uveal melanomas for BAP1 mutations, using different deep sequencing methods. The frequency of BAP1 mutations in our study group was 47%. The expression of BAP1 protein was studied using immunohistochemistry. BAP1 staining was absent in 43% of the cases. BAP1 mutation status was strongly associated with BAP1 protein expression (P<0.001), loss of chromosome 3 (P<0.001), and other aggressive prognostic factors. Patients with a BAP1 mutation and absent BAP1 expression had an almost eightfold higher chance of developing metastases compared with those without these changes (P=0.002). We found a strong correlation between the immunohistochemical and sequencing data and therefore propose that, immunohistochemical screening for BAP1 should become routine in the histopathological work-up of uveal melanoma. Furthermore, our analysis indicates that loss of BAP1 may be particularly involved in the progression of uveal melanoma to an aggressive, metastatic phenotype.
Similar content being viewed by others
Main
With an incidence in the Western world of about 5 per million people per year, uveal melanoma is the most common primary malignancy in the eye.1 Approximately half of the individuals who were diagnosed with uveal melanoma will develop metastatic disease, with a 4–6-month median survival period when metastasized to the liver.2 Several prognostic parameters are available to identify the patients at risk of developing metastases including cytogenetic aberrations such as loss of chromosome 1p, loss of chromosome 3, gain of chromosome 8, and abnormalities on chromosome 6.3, 4, 5, 6, 7
Harbour et al,8 reported inactivating somatic mutations in BAP1, the gene encoding BRCA-associated protein 1 in the predominantly metastasizing (class 2) uveal melanoma. BAP1 is located on chromosome 3p21.1, which is frequently deleted in uveal melanoma. Monosomy 3 is considered to be a relatively early event in uveal melanoma pathogenesis, and several studies have shown that it strongly correlates with decreased survival.4, 5, 6, 7, 9 BAP1 is a nuclear deubiquitinase that catalysis the removal of single ubiquitin moieties from ubiquitin chains or cleavage of the isopeptide bond between ubiquitin and the substrate protein.10 It is involved in several biological processes, including chromatin dynamics, the DNA damage response, and regulation of the cell cycle and cell growth.11, 12, 13 Inactivating somatic and germline BAP1 mutations have been identified in a variety of cancers, including malignant pleural mesotheliomas, cutaneous melanoma, atypical cutaneous melanocytic tumors, meningioma, lung adenocarcinoma, and renal cell carcinoma.14, 15, 16, 17, 18, 19 The number of reported cancer-prone families with germline BAP1 mutations is rising and suggesting a BAP1 cancer syndrome. However, the prevalence of germline BAP1 mutations in uveal melanoma patients is low compared with BAP1 mutations of somatic origin.8, 16, 20 Although somatic mutations in BAP1 are highly prevalent in metastasizing primary uveal melanoma, the role of BAP1 in the progression of uveal melanoma towards metastatic disease requires further investigation.
The purpose of this study was to identify BAP1 mutations in uveal melanoma patients and examine whether these mutations coincide with the protein expression of BAP1 in uveal melanoma tissue. We also investigated whether BAP1 mutations in uveal melanoma were associated with additional clinical, histopathological, and chromosomal parameters.
Materials and methods
Tissue Samples
Uveal melanoma specimens were collected from patients who underwent enucleation between the period 1993 and 2012 at the Erasmus University Medical Center and the Rotterdam Eye Hospital (Rotterdam, The Netherlands). Clinical and histopathological features, such as tumor localization, tumor diameter and thickness, age at time of diagnosis, cell type, and the presence of extracellular matrix patterns were evaluated. Cell type was scored by hematoxylin and eosin (H&E) staining according to the modified Callendar classification system. The presence of extracellular matrix patterns were examined with periodic acid–Schiff staining without hematoxylin. The study was performed according to the tenets of the Declaration of Helsinki and an informed consent was obtained before the operation.
DNA Extraction
DNA was isolated from fresh tumor samples using the QIAamp DNA-mini kit (Qiagen, Venlo, The Netherlands) according to the manufacturer’s instructions. The DNA concentration was measured using the NanoDrop ND-1000 Spectrophotometer (NanoDrop technologies, Wilmington, DE, USA) and Picogreen assay (Molecular Probes, Eugene, OR, USA). DNA was stored at −20 °C.
Copy Number Analysis
The DNA copy number status of the tumor was examined with single nucleotide polymorphism (SNP) array and fluorescent in situ hybridization (FISH) analysis. Two hundred nanograms of fresh tumor DNA was used as input for whole-genome analysis by SNP array (Illumina 610Q BeadChip, Illumina, San Diego, CA, USA). The data were analyzed with version 6 of the Nexus software (Biodiscovery, El Segundo, CA, USA). Chromosomal abnormalities were validated with FISH on directly fixed tumor cells using centromeric or locus-specific probes for chromosome 1, 3, 6, and 8, as described previously.21
Sequence Data Analysis
A 10.2 kb region containing the entire BAP1 gene was amplified from primary choroidal and ciliary body melanomas by long-range polymerase chain reaction (PCR) kit (Takara Holdings, Kyoto, Japan) using the primers 5′-GGCGCCGCTGTACTGGAGCTTTAGT-3′ and 5′-CGGCAGAGGAGAGCGGGACAGAGG-3′. Details of the PCR protocol are available upon request. If no PCR product could be obtained, two additional primers (5′-GGCAGCCTCCCCACAAGCCAAGG-3′ and 5′-CGGCAGAGGAGAGCGGGACAGAGG-3′) were used to amplify the gene as two overlapping 6.6 kb and 4.2 kb fragments. The amplified DNA was then purified using the QIAquick PCR Purification Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Sample preparation was performed according to the Illumina TruSeq v3 protocol and the samples were sequenced on the Hiseq2000 with a v3 paired-end flow cell for a read length of two times 100 bp with an index of 7 bp. The reads were aligned against the human reference genome build 19 (hg19) using BWA22 and the NARWHAL pipeline.23 Genetic variants were called using tools from the genome analysis toolkit,24 Picard and samtools.25 A VCF and Mpileup file for each sample were generated and processed with an in-house variant annotation tool.
The BAP1 region was captured and a unique index code with a length of 7 bp was incorporated into the sample using a HaloPlex Target Enrichment Kit (Agilent Technologies, Santa Clara, CA, USA). Sample preparation was performed as above and the samples were sequenced on the MiSeq using v2 flow cell for a paired-end read a length of 150 bp. Adapter trimming, alignment, variant calling, and annotation were performed as above.
Whole exome sequencing was performed using the Agilent version 4 capture kit on at least 1 μg of genomic tumor DNA, followed by sample preparation, sequencing, alignment, variant calling, and annotating, as described above (Koopmans et al, manuscript in preparation).
Variants were validated by Sanger sequencing. Oligonucleotide primers were designed from intronic sequences to amplify all coding sequence of BAP1 with PCR. The primers are listed in the Supplementary Table S1, and PCR amplification and Sanger sequencing protocols are available upon request.
Immunohistochemical Staining
Immunohistochemistry was performed with an automated immunohistochemistry staining system (Ventana BenchMark ULTRA, Ventana Medical Systems, Tucson, AZ, USA) using the alkaline phosphatase method and a red chromogen. In brief, following deparaffinization and heat-induced antigen retrieval for 64 min, the tissue sections were incubated with a mouse monoclonal antibody raised against amino acids 430–729 of human BAP1 (clone sc-28383, 1:50 dilution, Santa Cruz Biotechnology, Dallas, TX, USA) for 1 h at 36 °C. A subsequent amplification step was followed by incubation with hematoxylin II counter stain for 8 min and then a blue-colouring reagent for 8 min according to the manufacturer’s instructions (Ventana). Liver, tonsil, breast tissue, and the retinal pigment epithelium were used as positive controls for BAP1 expression. An ophthalmic pathologist independently evaluated the histopathological characterization of the tissue sections and the immunohistochemistry stainings. In some cases with suspected clonal subpopulations, multiple staining and double staining of BAP1 and HMB-45 and/or CD45 was conducted using the 3,3'-diaminobenzidine method. The samples were scored positive or negative by masked screening.
Statistical Analysis
The co-occurrence of BAP1 mutations with absence of BAP1 expression and other clinical, histopathological, and genetical data were calculated using either the χ2-test or Fisher’s exact test (categorical variables) and the Mann–Whitney test (continuous variables). The influence of these variables on survival was determined with the Kaplan–Meier method for categorical variables and using the Cox regression method for continuous variables. Survival analysis was performed on the basis of disease-free survival, defined as the time period from enucleation until the development of metastasis or death due to metastasis. Death due to another cause or lost to follow-up was treated as censored. Subsequently, Cox multivariate proportional hazards regression (forward logistic regression method) was used to confirm that the variables were independent predictors of survival. All tests were two-sided. An effect was considered significant if the P-value was 0.05 or less. The statistical analyses were performed with the SPSS-20 software package.
Results
Tumor Samples
A total of 74 patients with histopathologically proven uveal melanoma were included in our study. There were 34 men and 40 women, and the mean age was 63 years (range 37–86). The mean disease-free survival was 52 months (5–209). Thirty-five patients were alive at the last follow-up, 29 patients developed metastatic disease of which 26 died, 9 patients died due to another cause and 1 patient was lost during follow-up (with a survival of 69 months). Detailed clinicopathological data are provided in Table 1 and the Supplementary Table S2. The majority of uveal melanoma were of choroidal origin (n=64), only 10 tumors (14%) originated from the ciliary body. From the 64 choroidal tumors, 8 invaded the ciliary body. Most cases displayed a spindle or mixed cell morphology (n=64) and only 10 cases (14%) revealed a pure epithelioid phenotype. The tumors ranged from 5 to 21 mm in diameter (a mean value of 13.6 mm) and from 1.5 to 15 mm thick (a mean value of 7.8 mm). According to the 7th edition of the American Joint Committee on Cancer TNM classification (TNM7) for uveal melanoma, we classified uveal melanoma on the basis of the anatomic extent of the primary tumor (T).26 Seven tumors (10%) were classified as T1, 26 (35%) as T2, 35 (47%) as T3, and 6 (8%) as T4. Extracellular matrix patterns were present in 36 uveal melanoma (49%).
Genetic and Histopathological Analyses
FISH and SNP array analysis
Cytogenetic analysis was performed using a SNP array (n=59) and FISH (n=66). The study group contained 46 tumors with loss of chromosome 3 (62%). Three tumors appeared to have a loss of heterozygosity (LOH) region ranging from 0.57 to 4.1 Mb around the BAP1 gene. Therefore, we coded them as LOH for chromosome 3p21.1. A SNP array of one case with LOH is shown in Figure 1a. Other chromosomal aberrations included loss of chromosome 1p (n=23, 31%), gain of chromosome 6p (n=37, 51%), loss of chromosome 6q (n=29, 40%), loss of chromosome 8p (n=16, 22%), gain of chromosome 8p (n=15, 20%), and gain of chromosome 8q (n=43, 58%). Eight tumors were polyploid with either a triploid (n=5, 7%) or tetraploid (n=3, 4%) status. Seven out of these eight tumors showed relative loss of chromosome 3 compared with their baseline chromosome status.
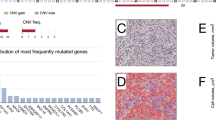
Histopathological and genetic features of two uveal melanoma cases. (a) In case S42, loss of heterozygosity (LOH) of 3p21.1 was observed on single nucleotide polymorphism (SNP) array. (b) The tumor shows mainly spindle cells in a hematoxylin and eosin (H&E) staining (400 × ). (c) Immunohistochemistry revealed no expression of BAP1 in the tumor cells and positive expression in endothelium and inflammatory cells (400 × ). (d) In case S4, the SNP array displays a disomy 3. (e) Spindle tumor cells in the uveal melanoma are shown in a H&E staining (200 × ). (f) Immunohistochemistry revealed a strong nuclear positivity for BAP1 (400 × ).
Mutation analysis
In all, 57 tumor samples were sequenced using the long-range PCR approach, generating 2 992 269–46 389 045 mapped reads per sample. Six samples were sequenced using the HaloPlex method. Of these, two samples were also sequenced with the long-range approach. Nineteen tumors were subjected to whole exome sequencing, in which the coding region of the entire genome was sequenced. Of these, seven were also sequenced with the long-range approach. Two groups of untypical uveal melanoma were selected for the exome sequencing: seven uveal melanomas with monosomy 3 and a follow-up of more than 60 months without any metastasis, and 10 disomy 3 tumors who did develop metastasis. Two samples were polyploid with relative chromosome 3 loss and metastatic disease. For all 19 samples, a mean coverage over 68 × was reached for the target regions (Koopmans et al, manuscript in preparation). For the current study, we only investigated the BAP1 gene in the samples that were subjected to exome sequencing.
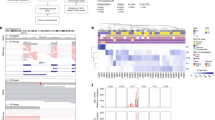
A BAP1 mutation was detected in 35 uveal melanoma samples (47%). These included 7 missense mutations, 3 nonsense mutations, 12 deletions, and 2 insertions leading to a frameshift, 1 in-frame deletion and 10 mutations located at a splice site. The mutations were located throughout the gene (Figure 2a). Thirty-three out of the 35 variants were validated using Sanger sequencing.
Schematic overview of sequence data and immunohistochemical analyses. (a) A schematic overview of the BAP1 gene and its functional domains. BAP1 is composed of an N-terminal UCH domain (orange; amino acid (aa) 1–250), an HCF1-binding domain (HBM)-like motif (blue; aa 363–366), an UCH37-like domain (ULD) (green; aa 634–693), and a nuclear localization signal (NLS) consisting of two parts (blue; aa 656–661 and aa 717–722). BAP1 has been reported to interact with BARD1 (aa 182–365), HCF1 (aa 365–385), BRCA1 (aa 596–721), and YY1 (aa 642–686).33 The binding site for BAP1 antibody is depicted with a dashed line (aa 430–729). The found mutations and indels are shown below, classified according to their type and position. (b) A multilevel doughnut chart was constructed for all samples (n=74) regarding the chromosome 3 status (outer ring), BAP1 mutations status (middle ring), and BAP1 expression (inner ring).
Immunohistochemistry
We assessed BAP1 expression by immunohistochemistry. In 31 of the 73 uveal melanoma investigated (43%), BAP1 expression was below the level of detection in the tumor. In these samples, the retinal pigment epithelium stained positive. One of the paraffin slides could not be examined due to insufficient material (sample S20). In the upper panel of Figure 1, an uveal melanoma is shown with LOH of the BAP1 gene and no detectable BAP1 staining (Figures 1a–c). In the lower panel, a uveal melanoma with a disomy 3 and positive BAP1 expression is shown (Figures 1d–f). In four tumors, a subpopulation of cells, ranging from 20 to 80% of the total, was observed, that were negative for BAP1 staining. The remaining tumor cells (corresponding 80–20% of the total), stained positive for BAP1 (Supplementary Table S2). In these four samples, a staining with HMB-45 was performed. We observed that both the BAP1-positive and -negative cells stained positive for HMB-45 (Figure 3).
Histopathological features of a uveal melanoma case with heterogeneous expression of BAP1. (a) In case S7, a heterogeneous distribution of BAP1 expression was observed throughout the tumor with immunohistochemistry (400 × ). (b) Staining with HMB-45 revealed strong positivity (3,3'-diaminobenzidine staining, brown color) concluding that the cells investigated were melanoma cells (400 × ).
The majority of the BAP1-negative tumors also harbored a BAP1 mutation (Figure 2b). More specifically, 30 out of the 31 uveal melanomas that did not show any BAP1 staining had a BAP1 mutation (SNPs, base insertions, or deletions). Four of the 42 BAP1-positive tumors harbored a BAP1 mutation (Table 2). As shown in Figure 2b, two of the 28 tumors possessing a normal chromosome 3 copy number had a BAP1 sequence variant (sample S11 and S71; Supplementary Table S2). In the group of uveal melanoma with loss of chromosome 3 (including the polyploid tumors with a relative chromosome 3 loss), 33 of the 46 tumors harbored a BAP1 mutation. We did not observe any BAP1 staining in 30 of the corresponding samples. Immunohistochemistry could not be conducted for sample S20. Two tumors, S23, and S43, stained positive for BAP1 despite their monosomy 3 and BAP1 mutation status. Tumor S23 contained a heterozygous deletion of 22 bp in exon 16 (p.R666fs) leading to a frameshift in 74% of the reads. In this sample, heterogeneous levels of BAP1 expression were observed. We estimated that BAP1 expression was absent in 20% of the tumor, whereas the remaining 80% of the tumor cells did stain positive for BAP1. Tumor S43 had a 7 bp frameshift deletion in exon 16 (p.E673X) in 54% of the reads. However, in this tumor 100% of the tumor tissue showed normal BAP1 expression. Lastly, there was one BAP1 wild-type uveal melanoma (sample S3) that did not stain positive for BAP1. In this case, BAP1 staining was absent in ∼50% of the tumor cells.
Statistical Analysis
BAP1 mutations strongly coincided with an absent BAP1 expression (P<0.001) and monosomy 3 (P<0.001) (Table 2). The presence of epithelioid cells (P=0.001) and extracellular matrix patterns (P=0.005) were also significantly overrepresented in uveal melanoma with BAP1 mutations. The sensitivity and specificity for the detection of BAP1 depletion by immunohistochemistry compared with mutation analysis were 88% and 97%, respectively. For survival analysis, we included only the unselected uveal melanoma samples (n=55) and excluded uveal melanoma specimens that were selected for exome sequencing. Univariate analyses showed that the disease-free survival was significantly shorter in patients with a BAP1 mutation (32 vs 133 months, P<0.001, Figure 4a). We examined whether BAP1 mutations influenced the prognosis of monosomy 3 patients by constructing Kaplan–Meier curves and performing the Log-rank test. Patients with monosomy 3 uveal melanoma and a BAP1 mutation seem to have a worse prognosis, although this was not statistically significant (P=0.122, Figure 4b). Patients with tumors with a negative BAP1 staining also had a significant shorter disease-free survival compared with tumors with positive BAP1 staining (31 vs 133 months, P<0.001). Other factors that affected the survival in uveal melanoma patients were: age at time of diagnosis (P=0.040), largest basal tumor diameter (P=0.005), the presence of epithelioid cells (P=0.003), the presence of extracellular matrix patterns (P=0.001), and chromosome 3 loss (P<0.001). Considering the strong interaction between BAP1 mutation status and BAP1 expression, we validated whether this concurrent inactivation of the gene and protein is an independent parameter for disease-free survival. The possible confounding variables were analyzed in a multivariate model. After correcting for these variables, we found that patients with a concurrent BAP1 mutation and a negative BAP1 expression have a 7.7 times greater chance of developing metastases compared with those without these aberrations (P=0.002) (Table 3). The largest basal diameter of the tumor was also an independent predictor for disease-free survival (P=0.016). The age at time of diagnosis, presence of epithelioid cells, extracellular matrix patterns, and loss of chromosome 3 did not reach significance and were rejected.
Kaplan–Meier estimate of disease-free survival in uveal melanoma patients. (a) Kaplan–Meier survival curves displaying melanoma-related mortality for 55 patients on the basis of BAP1 mutation status and (b) the survival curves for 34 patients with loss of chromosome 3. The table shows the number of events and cases at risk overtime at the respective time point. Log-rank tests were used to compare survival distributors across subgroups.
Discussion
In this study, we found that nearly half of the investigated uveal melanoma tumors harbored an inactivating BAP1 mutation and that this was strongly associated with the absence of BAP1 staining, monosomy 3, and other prognostic features of aggressive tumors, such as the presence of epithelioid cells and extracellular matrix patterns. Nonetheless, a few discrepancies were observed between BAP1 mutation status and BAP1 immunohistochemistry. For two samples (S11 and S71), the immunohistochemistry results can be explained by the fact that both tumors were disomic for chromosome 3 and harbored a heterozygous BAP1 mutation. Thus, presumably the remaining wild-type allele led to a normal positive staining. As mentioned previously, LOH of a small region containing the BAP1 gene was found in three tumors by SNP array analysis. These tumors (S21, S37, and S42) were classified as loss of chromosome 3p21.1 although chromosome 3 was not entirely deleted. In addition, S21 and S42 harbored a hemizygous BAP1 mutation (Figures 1a–c and Figure 2b). Our study confirms that biallelic inactivation of BAP1 in uveal melanoma tumor tissue is required to prevent BAP1 protein expression, through loss of a copy of chromosome 3 and a BAP1 mutation in the remaining copy. Two uveal melanomas (S23 and S43) with monosomy 3 had a positive BAP1 staining, despite harboring an out-of-frame deletion in exon 16. One possibility for the positive staining could be that the truncated proteins (p.R666fs and p.E673X) are still detected by the BAP1 antibody. Interestingly, both deletions were heterogeneous for hemizygous mutant suggesting that a normal population of cells is still present in the tumor, which could have led to a positive staining. However, this is not supported by the observation that, 80% and 100%, respectively, of the S23 and S43 tumor cells stained positive for BAP1. In two cases with evident heterogeneous subpopulations of cells with and without BAP1 expression within the same uveal melanoma (S5 and S7), the percentage of BAP1-negative uveal melanoma cells was equal to the percentage of chromosome 3 loss. Even though in our study, the percentage of BAP1 mutation does not always correlate with the percentage of absent expression, in most cases, these percentages are high enough to classify the tumors in the correct category. In one tumor sample (S3), 50% of the cells did not stain positive for BAP1 even though no BAP1 mutation was detected by exome sequencing. To be sure that the investigated clonal subpopulations were melanoma cells, we carried out a staining with HMB-45 and confirmed that this was the case. In this tumor S3, LOH of chromosome 3 was detected by FISH and SNP array analysis. Possibly, intronic variants that cannot be detected with exome sequencing prevent BAP1 expression in some of the cells in this tumor. Alternatively, the apparent LOH might reflect a more complex genetic rearrangement, where BAP1 is lost in a proportion of the cells comprising the tumor.
Somatic BAP1 mutations have been described in other cancers, such as malignant pleural mesotheliomas and cutaneous melanoma, and the absence of BAP1 expression in mesotheliomas has been demonstrated by immunohistochemistry.15 In contrast to our uveal melanoma cohort, 25% of the mesotheliomas without a BAP1 mutation did not display any immunohistochemistry staining for BAP1.
In two uveal melanomas, a mutation was detected by one of the NGS approaches but could not be validated with Sanger sequencing (S58 and S63). In both cases, the percentage of reads with the mutation was quite low (5% and 4%, respectively). A limitation of conventional Sanger sequencing is that low mosaicism variants are difficult to detect below a level of ∼20%27 and this is likely to be the reason why S58 and S63 could not be validated. Both tumor samples stained BAP1-negative.
A recent study suggested that BAP1 inactivation might be more characteristic of epithelioid mesotheliomas.28 We also found a correlation between BAP1 inactivation and uveal melanoma with an epithelioid cell type suggesting that BAP1 deficiency may be particularly involved in the pathogenesis of uveal melanoma with an aggressive phenotype.
Previous research has shown that BAP1 mutations are present in 47% (27/57) of the primary uveal melanoma and 84% (26/31) of class 2 uveal melanoma.8 Our findings support the hypothesis that somatic BAP1 mutations promote metastases. In the overall study group, we found a BAP1 mutation in 62% (18/29) of metastasizing uveal melanoma, and it is important to note that this could be an underestimation, since in a few patients, limited follow-up data was available. Selection bias could have occurred because untypical uveal melanomas were selected for exome sequencing. Therefore, we excluded these tumors from our survival analysis. Nonetheless, it would be interesting to enlarge the study group with random, nonselected uveal melanoma with a longer follow-up. After excluding the exome sequencing samples, there were six patients without metastasis after a follow-up of 4 years or longer. None of these individuals had a BAP1 mutation. Of the 18 patients who developed metastases, 15 uveal melanomas harbored a BAP1 mutation.
In the current study, we only investigated the BAP1 expression in tumors from enucleated eyes. Over the years, eye-sparing therapies have proved to be equally effective in terms of patient survival compared with radical treatment.29, 30 With eye-sparing therapies biopsies can be taken for prognostication and it is also possible to perform BAP1 immunohistochemistry on biopsy specimens in our institute. This technique has an additive value in determining the patients’ prognosis.
Recent work of Matatall and associates31 demonstrated that BAP1 depletion induces a primitive, stem-like phenotype and these findings implicate BAP1 in the maintenance of melanocyte identity in uveal melanoma cells. Therapeutic strategies that target these specific pathways in uveal melanoma are urgently needed. Currently, therapeutic agents targeting BAP1 deficiency are being investigated. Histone deacetylase inhibitors have shown to reverse the effects of BAP1 depletion in uveal melanoma cells.32 As therapeutic options emerge, it is important to be able to rapidly identify the patients, enucleated and conservatively treated patients, who would benefit from a specific intervention. Given the costs of BAP1 mutation analysis, immunohistochemistry offers an economical and fast alternative. In our study, we demonstrated that there is a strong association between BAP1 staining and BAP1 mutation status with a sensitivity of 88% and a specificity of 97%. We propose that, the BAP1 immunohistochemistry should be implemented in the routine histopathological examination of uveal melanoma.
References
Singh AD, Turell ME, Topham AK . Uveal melanoma: trends in incidence, treatment, and survival. Ophthalmology 2011;118:1881–1885.
Woodman SE . Metastatic uveal melanoma: biology and emerging treatments. Cancer J 2012;18:148–152.
Kilic E, Naus NC, van Gils W et al. Concurrent loss of chromosome arm 1p and chromosome 3 predicts a decreased disease-free survival in uveal melanoma patients. Invest Ophthalmol Vis Sci 2005;46:2253–2257.
Prescher G, Bornfeld N, Hirche H et al. Prognostic implications of monosomy 3 in uveal melanoma. Lancet 1996;347:1222–1225.
Sisley K, Parsons MA, Garnham J et al. Association of specific chromosome alterations with tumour phenotype in posterior uveal melanoma. Br J Cancer 2000;82:330–338.
Sisley K, Rennie IG, Parsons MA et al. Abnormalities of chromosomes 3 and 8 in posterior uveal melanoma correlate with prognosis. Genes Chromosomes Cancer 1997;19:22–28.
White VA, Chambers JD, Courtright PD et al. Correlation of cytogenetic abnormalities with the outcome of patients with uveal melanoma. Cancer 1998;83:354–359.
Harbour JW, Onken MD, Roberson ED et al. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science 2010;330:1410–1413.
Kilic E, van Gils W, Lodder E et al. Clinical and cytogenetic analyses in uveal melanoma. Invest Ophthalmol Vis Sci 2006;47:3703–3707.
Nishikawa H, Wu W, Koike A et al. BRCA1-associated protein 1 interferes with BRCA1/BARD1 RING heterodimer activity. Cancer Res 2009;69:111–119.
Farmer H, McCabe N, Lord CJ et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 2005;434:917–921.
Jensen DE, Proctor M, Marquis ST et al. BAP1: a novel ubiquitin hydrolase which binds to the BRCA1 RING finger and enhances BRCA1-mediated cell growth suppression. Oncogene 1998;16:1097–1112.
Ventii KH, Devi NS, Friedrich KL et al. BRCA1-associated protein-1 is a tumor suppressor that requires deubiquitinating activity and nuclear localization. Cancer Res 2008;68:6953–6962.
Abdel-Rahman MH, Pilarski R, Cebulla CM et al. Germline BAP1 mutation predisposes to uveal melanoma, lung adenocarcinoma, meningioma, and other cancers. J Med Genet 2011;48:856–859.
Bott M, Brevet M, Taylor BS et al. The nuclear deubiquitinase BAP1 is commonly inactivated by somatic mutations and 3p21.1 losses in malignant pleural mesothelioma. Nat Genet 2011;43:668–672.
Njauw CN, Kim I, Piris A et al. Germline BAP1 inactivation is preferentially associated with metastatic ocular melanoma and cutaneous-ocular melanoma families. PloS one 2012;7:e35295.
Pena-Llopis S, Vega-Rubin-de-Celis S, Liao A et al. BAP1 loss defines a new class of renal cell carcinoma. Nat Genet 2012;44:751–759.
Testa JR, Cheung M, Pei J et al. Germline BAP1 mutations predispose to malignant mesothelioma. Nat Genet 2011;43:1022–1025.
Wiesner T, Obenauf AC, Murali R et al. Germline mutations in BAP1 predispose to melanocytic tumors. Nat Genet 2011;43:1018–1021.
Aoude LG, Vajdic CM, Kricker A et al. Prevalence of germline BAP1 mutation in a population-based sample of uveal melanoma cases. Pigment Cell Melanoma Res 2013;26:278–279.
Naus NC, Verhoeven AC, van Drunen E et al. Detection of genetic prognostic markers in uveal melanoma biopsies using fluorescence in situ hybridization. Clin Cancer Res 2002;8:534–539.
Li H, Durbin R . Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 2009;25:1754–1760.
Brouwer RW, van den Hout MC, Grosveld FG et al. NARWHAL, a primary analysis pipeline for NGS data. Bioinformatics 2012;28:284–285.
McKenna A, Hanna M, Banks E et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20:1297–1303.
Li H, Handsaker B, Wysoker A et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009;25:2078–2079.
Malignant melanoma of the uvea In: Edge SD, Byrd DR, Compton CC, Fritz AG, Green FL, Trotti A, (eds). AJCC Cancer Staging Manual 7th edn. Springer: New York, 2010, pp 547–559.
Rohlin A, Wernersson J, Engwall Y et al. Parallel sequencing used in detection of mosaic mutations: comparison with four diagnostic DNA screening techniques. Hum Mutat 2009;30:1012–1020.
Yoshikawa Y, Sato A, Tsujimura T et al. Frequent inactivation of the BAP1 gene in epithelioid-type malignant mesothelioma. Cancer Sci 2012;103:868–874.
Seddon JM, Gragoudas ES, Albert DM et al. Comparison of survival rates for patients with uveal melanoma after treatment with proton beam irradiation or enucleation. Am J Ophthalmol 1985;99:282–290.
Seddon JM, Gragoudas ES, Egan KM et al. Relative survival rates after alternative therapies for uveal melanoma. Ophthalmology 1990;97:769–777.
Matatall KA, Agapova OA, Onken MD et al. BAP1 deficiency causes loss of melanocytic cell identity in uveal melanoma. BMC Cancer 2013;13:371.
Landreville S, Agapova OA, Matatall KA et al. Histone deacetylase inhibitors induce growth arrest and differentiation in uveal melanoma. Clin Cancer Res 2012;18:408–416.
Eletr ZM, Wilkinson KD . An emerging model for BAP1's role in regulating cell cycle progression. Cell Biochem Biophys 2011;60:3–11.
Acknowledgements
This study was supported by a grant of the Combined Ophthalmic Research Rotterdam (CORR) and Stichting Nederlands Oogheelkundig Onderzoek (SNOO).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Modern Pathology website
Supplementary information
Rights and permissions
About this article
Cite this article
Koopmans, A., Verdijk, R., Brouwer, R. et al. Clinical significance of immunohistochemistry for detection of BAP1 mutations in uveal melanoma. Mod Pathol 27, 1321–1330 (2014). https://doi.org/10.1038/modpathol.2014.43
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2014.43
Keywords
This article is cited by
-
5-Methylcytosine immunohistochemistry for predicting cutaneous melanoma prognosis
Scientific Reports (2024)
-
Genome-wide aberrant methylation in primary metastatic UM and their matched metastases
Scientific Reports (2022)
-
Loss of polycomb repressive complex 1 activity and chromosomal instability drive uveal melanoma progression
Nature Communications (2021)
-
Uveal melanoma
Nature Reviews Disease Primers (2020)
-
Loss of BAP1 in Pheochromocytomas and Paragangliomas Seems Unrelated to Genetic Mutations
Endocrine Pathology (2019)