Abstract
Intratumoral heterogeneity of human epidermal growth factor receptor 2 (HER2) gene amplification has been reported to occur with variable frequencies in breast cancers. However, there have been few studies of its clinicopathological significance. We used tissue microarrays to evaluate two aspects of intratumoral heterogeneity of HER2 gene amplification: regional heterogeneity and genetic heterogeneity. We examined 96 invasive breast cancers in which HER2 amplification had been diagnosed in whole sections, and determined the clincopathological characteristics of those tumors. HER2 regional heterogeneity, defined as the existence of amplification/negative or amplification/equivocal patterns in different tissue microarray cores of a tumor, was present in 17 (18%) of the 96 cases. HER2 genetic heterogeneity, defined as the presence of tumor cells with a HER2/chromosome enumeration probe 17 ratio higher than 2.2 in 5–50% of the tumor cells, was found in 11 cases (11%), all of which showed HER2 regional heterogeneity. The cases with intratumoral heterogeneity of HER2 gene amplification were characterized by low grade or equivocal HER2 amplification and equivocal (2+) HER2 expression in whole sections. The patients with intratumoral heterogeneity of HER2 gene amplification had significantly shorter disease-free survival times than those with homogeneous HER2 gene amplification, and this effect was also evident in subgroup analysis by hormone receptor status. In multivariate analysis, intratumoral HER2 heterogeneity retained its status as an independent prognostic factor for disease-free survival. In conclusion, intratumoral heterogeneity of HER2 gene amplification is present in a subset of HER2-amplified breast cancers, especially in cases with low-grade HER2 amplification and equivocal HER2 expression, indicating a need for HER2 testing on more representative, larger tumor samples for accurate assessment of HER2 status in such cases. The patients with this heterogeneity have decreased disease-free survival, suggesting that genetic instability, and hence aberrant HER2 amplification in subclones of such tumors, may be associated with breast cancer progression.
Similar content being viewed by others
Main
The HER2 (human epidermal growth factor receptor-2) gene, located at chromosome 17q12–21, is the gene most frequently amplified in breast cancer. Overexpression of HER2, usually attributable to HER2 gene amplification, has been identified in 15–20% of breast cancers and is associated with poor prognosis.1, 2, 3 After the introduction of trastuzumab, a humanized monoclonal antibody against the extracellular portion of the HER2 protein, evaluation of HER2 status in breast cancer by immunohistochemistry and/or fluorescence in-situ hybridization (FISH) has been standard practice, in the metastatic setting and, more recently, in adjuvant and neoadjuvant settings, to identify the patients most likely to benefit from trastuzumab.4, 5, 6 Therefore, accurate assessment of HER2 status is essential for clinical decision-making in the treatment of patients with breast cancer. The current guidelines proposed by the American Society of Clinical Oncology/College of American Pathologist (ASCO/CAP) in 2007 states that HER2 status should be initially assessed by immunohistochemistry, using a semiquantitative scoring system, and confirmed by FISH, in all immunohistochemical score 2+ equivocal cases.7
HER2 status has been thought to be fairly homogeneous within primary tumors and to remain constant during breast cancer progression. Many studies have shown a high level of concordance, although incomplete, between the HER2 status of a primary tumor and that of its metastases.8, 9, 10 However, several studies have reported significant discordance.11, 12, 13, 14 Some studies have suggested that HER2 gene amplification can be acquired during tumor progression and that this could be a cause of this discordance.12, 13 Others have demonstrated primary intratumoral heterogeneity of both HER2 expression and HER2 gene amplification.8, 13, 14
Intratumoral heterogeneity of HER2 gene amplification has been demonstrated in a subset of breast cancers.15, 16, 17, 18, 19, 20, 21, 22, 23 However, as the previous studies targeted different sets of samples, the frequency of intratumoral HER2 heterogeneity was quite variable and was not comparable between the studies. Andersson et al15 found HER2 heterogeneity within the invasive components of breast cancers in only 1 of 78 tumors. However, their tumors included only 13 cases with HER2 gene amplification. Glöckner et al17 examined the amplification status of growth regulatory genes including HER2, and found HER2 heterogeneity in 36% of the amplified cases. The high frequency of heterogeneity in their study might be related to their use of laser-assisted microdissection and quantitative polymerase chain reaction rather than FISH. Shin et al23 demonstrated intratumoral heterogeneity for HER2 gene amplification in 5 (16%) of 31 invasive ductal carcinomas with grade 3 architecture, using tissue microarrays, which are an efficient approach for studying intratumoral heterogeneity. Brunelli et al16 also used tissue microarrays and reported that intratumoral HER2 heterogeneity was present in 4 (13%) of 30 breast cancers with HER2 amplification and 3+ immunoexpression.
Intratumoral heterogeneity of HER2 gene amplification may have clinical implications, as it may contribute to inaccurate assessment of HER2 status and may affect treatment responses to HER2-targeted therapy including trastuzumab, as a result of selection of subclones lacking HER2 gene amplification.24 However, studies of the heterogeneity of HER2 gene amplification in primary tumors have been mostly confined to small samples, and its clinical significance has been rarely determined. Recently, the CAP addressed this issue and published a separate recommendation.19 They defined HER2 genetic heterogeneity as the presence of tumor cells with HER2/chromosome enumeration probe (CEP) 17 signal ratios >2.2 in 5–50% of the tumor cells tested. However, there have been no studies using this definition to evaluate intratumoral heterogeneity of HER2 gene amplification, except a recent study by Bartlett et al.25
In the present study, we compared HER2 amplification status in three cores of tissue microarrays chosen from different areas of each tumor in 96 invasive breast cancers that were considered to have HER2 gene amplification in whole sections, using the ASCO/CAP criteria for HER2 amplification7 and the CAP definition for HER2 genetic heterogeneity.19 In addition, we determined the clincopathological characteristics of tumors with intratumoral heterogeneity of HER2 gene amplification.
Materials and methods
Patients and Tissue Samples
We retrospectively searched the FISH data for surgically resected primary invasive breast cancers in the files of the Department of Pathology, Seoul National University Bundang Hospital from 2003 to 2009. After excluding cases with initial metastases, we selected 96 invasive breast cancers with HER2 gene amplification in whole tissue sections. About 20–40 cells were randomly evaluated to get HER2/CEP17 ratio in the whole section. They comprised 3 cases with ‘equivocal’ amplification (HER2/CEP17 ratio 1.8–2.2), 36 with low-grade amplification (ratio >2.2–<4.0) and 57 with high-grade amplification (ratio≥4.0). Of the 96 cases, 69 had 3+ HER2 immunohistochemical scores, 26 had 2+ scores and one scored 1+. Baseline characteristics of the patients are summarized in Table 1. Hematoxylin and eosin-stained slides were reviewed for every case, and the following histopathological variables were determined: histological subtype, T stage, nodal status, Nottingham combined histological grade, venous invasion, lymphatic invasion, tumor border and presence or absence of ductal carcinoma in situ component. Expression of standard biomarkers including estrogen receptor, progesterone receptor, HER2, p53 and Ki-67 were recorded in the following ways. Expression of HER2 was scored according to the ASCO/CAP guidelines:7 0, no staining; 1+, weak and incomplete membranous staining in ≥10% of the tumor cells; 2+, weak-to-moderate, complete membranous staining in ≥10% of the tumor cells and 3+, strong, complete membranous staining in ≥30% of the tumor cells. Estrogen and progesterone receptors were regarded as positive if there were at least 1% positive tumor nuclei.26 For p53, cases with 10% or more positive staining were grouped as positive. For Ki-67 proliferation index, cases with ≥20% positive tumor cells were regarded as having a high proliferation index. All cases were independently reviewed by two breast pathologists (SYP and HS). The study was approved by the institutional review board of Seoul National University Bundang Hospital, waiving the requirement for informed consent.
Tissue Microarray Construction
We used tissue microarrays to evaluate whether there were regional differences in HER2 gene amplification. All slides including slides immunohistochemically stained for standard biomarkers were reviewed, and the most representative tumor section was selected for each case. If the tumor showed regional differences in HER2 immunoexpression, differentially stained areas were selected. Three representative core tissue sections (2 mm in diameter) were taken from different areas of the invasive tumors and arranged in new tissue microarray blocks using a trephine apparatus (Superbiochips Laboratories, Seoul, Korea). Thus, three cores were included in the tissue microarray block for each case and analyzed separately.
FISH Assays for HER2 Gene Amplification
Fluorescence in-situ hybridization was performed using the PathVysion assay (Abbott Molecular, Downers Grove, IL), as previously described.27 Briefly, 4 μm deparaffinized tissue microarray sections were incubated in pretreatment solution (Abbott Molecular) at 80°C for 30 min, then in protease solution (Abbott Molecular) for 20 min at 37°C. Co-denaturation of the probe and the DNA in the tissue sections was achieved by incubation at 73°C for 5 min using a HYBrite (Abbott Molecular), and this was followed by 16-h hybridization at 37°C. Post-hybridization washes were performed according to the protocol supplied. Slides were then counterstained with 4′,6-diamidino-2-phenylindole in an antifade solution, and viewed with a fluorescence microscope.
A total of 50 cells were evaluated for each core and the genetic variables were reported as: HER2 gene copy number, chromosome 17 copy number and average HER2 gene-to-chromosome 17 ratio. HER2 gene amplification was determined on the basis of the overall ratio defined by the ASCO/CAP criteria.7 The HER2 gene was considered to be amplified if the ratio of HER2 to CEP17 signals was >2.2. A ratio of ≥4.0 was defined as high-grade amplification; a ratio >2.2 and <4.0 as low level amplification. Cases showing a ratio of ≥1.8 and ≤2.2 were considered to be equivocal for amplification. The cut-off values for chromosome 17 copy number changes were adopted from Ma et al28 with modifications. Specimens with signals in the range of 1.25–2.25 were defined as having disomy 17. The other cases were considered to have aneusomy 17; either monosomy 17 (<1.25 signals per cells), low polysomy 17 (>2.25 but ≤3.75 signals per cell) or high polysomy 17 (>3.75 signals per cell).
Intratumoral heterogeneity of HER2 gene amplification was evaluated by two different methods. First, if three cores from a case gave discordant results for HER2 gene amplification, that is, if a case had an amplification/negative or amplification/equivocal pattern in the different cores, the case was considered to have HER2 regional heterogeneity. Second, HER2 genetic heterogeneity was defined, according to the CAP guidelines,19 as the existence of tumor cells with a HER2/CEP17 ratio >2.2 in 5–50% of all the tumor cells in the three cores.
Statistical Analysis
The statistical significance of the data was assessed using Statistical Package, SPSS version 15.0 for Windows (SPSS, Chicago, IL). The associations of HER2 intratumoral heterogeneity with clinicopathological characteristics of the tumors were analyzed using Fisher's exact test or the χ2 test, depending on the test conditions. Survival curves were estimated using the Kaplan–Meier product-limit method, and the significance of differences between survival curves was determined using the log-rank test. Covariates that were statistically significant in the univariate analysis were then included in the multivariate analysis using Cox proportional hazards regression model, and the hazard ratio and its 95% confidence interval were assessed for each factor. P values <0.05 were considered statistically significant. All P values reported are two-sided.
Results
Frequency of Intratumoral Heterogeneity of HER2 Gene Amplification
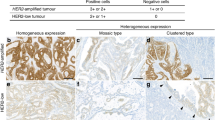
We determined the HER2/CEP17 ratio in the three cores of each of the 96 invasive breast cancers (Table 2). Of the 57 cases that were seen to have high-grade amplification in the whole sections, 44 cases were scored as high-grade amplification, 3 were scored as low-grade amplification, 8 were scored as high- /low-grade amplification and the remaining two cases were scored as high-grade amplification/negative for amplification in the three tissue microarray cores. Of the 36 cases with low-grade amplification in the whole sections, 20 were scored as low-grade amplification, 3 were scored as high- /low-grade amplification, another 3 cases were scored as high-grade amplification/negative for amplification, 6 were scored as low-grade amplification/equivocal for amplification and the remaining 4 were scored as low-grade amplification/negative for amplification. Three cases that were equivocal in the whole sections were scored as low-grade amplification in all three cores in one case and low-grade amplification/equivocal for amplification in the other two cases. If cases scored as amplification/negative or equivocal for amplification were considered to have HER2 regional heterogeneity, 17 (18%) of the 96 cases had HER2 regional heterogeneity: 5 of these were high-grade amplification/negative for amplification, 8 low-grade amplification/equivocal for amplification and 4 low-grade amplification/negative for amplification (Table 3; Figures 1 and 2). We also compared HER2 protein expression with FISH results in each case. A total of 84 cases yielded the same immunohistochemical findings for HER2 in all three cores; 69 were 3+, 14 were 2+ and one was 1+. HER2 expression differed in the three cores in 12 cases; the cores were 3+ or 2+ in 5 cases, 3+/2+/1+ in 3 cases, 2+ or 1+ in 2 cases and 2+ or 0 in the remaining cases. Of the 12 cases in which the HER2 immunohistochemical scores in the three cores differed, 8 were among the 17 cases with HER2 regional heterogeneity and the remaining 4 did not show HER2 regional heterogeneity (P<0.001).
Box plots depicting HER2/CEP17 ratios of three representative examples of breast cancers in the three tissue microarray cores. The box shows the 25th–75th percentile, the horizontal line inside the box represents the median, the whiskers extend to the 10th and 90th percentiles, and the outlying circles are individual data points outside the 10th and 90th percentiles. (a) A case with low-level amplification in all three cores. (b) A case with high-level amplification in all three cores. (c) A case showing low-level amplification/high-level amplification/negative for amplification, respectively, in the three cores.
Two representative examples of HER2 regional heterogeneity. (a) An invasive ductal carcinoma (case 74) showing equivocal (2+) HER2 expression in core 2. HER2 amplification is seen only in core 2. (b) An invasive ductal carcinoma (case 96) showing heterogeneous HER2 expression in the three cores (3+, core 1; 2+, core 2 and 1+, core 3). HER2 amplification is confined to core 1. Abbreviations: H&E, hematoxylin and eosin stain; IHC, immunohistochemisty; FISH, fluorescence in-situ hybridization.
We counted 50 nuclei per core; so a total of 150 nuclei were evaluated for each case. When HER2 genetic heterogeneity was defined as the existence of tumor cells with a HER2/CEP17 ratio >2.2 in 5–50% of all tumor cells (8–75 tumor cells in our cases), according to the CAP guidelines, 11 cases (11%) were found to have HER2 genetic heterogeneity, all of which were among the 17 cases with HER2 regional heterogeneity (Table 3). In contrast to HER2 regional heterogeneity, HER2 genetic heterogeneity was not associated with heterogeneity of HER2 protein expression in the cores.
Association of Intratumoral Heterogeneity of HER2 Gene Amplification with the Clinicopathological Characteristics of the Tumors
To characterize the cases with intratumoral heterogeneity of HER2 gene amplification, we analyzed the relationship between this heterogeneity and various clinicopathological parameters of the tumors (Table 4). HER2 regional heterogeneity was more frequently found in the cases with low grade or equivocal amplification in whole sections than in those with high-grade amplification (P<0.001). Moreover, it was more frequent in the cases with 2+ or 1+ HER2 immunohistochemical socres than in those with 3+ (P<0.001). The cases with HER2 regional heterogeneity tended to be hormone receptor-positive (P=0.069). Other clinicopathological parameters including T stage, N stage, chromosome 17 polysomy, Ki-67 proliferation index and p53 status were not associated with HER2 regional heterogeneity.
HER2 genetic heterogeneity defined by the CAP guidelines was also more frequent in the cases with low grade or equivocal amplification than in those with high-grade amplification in whole sections (P=0.006). It was also more frequent in the cases with 2+ or 1+ HER2 immunoexpression (P<0.001). In addition, the cases with HER2 genetic heterogeneity tended to be hormone receptor-positive (P=0.059).
Intratumoral Heterogeneity of HER2 Gene Amplification as a Poor Prognostic Factor
We also investigated the prognostic effect of HER2 intratumoral heterogeneity in the patients with HER2-amplified breast cancer. At the time of the analysis, the median follow-up was 4 years (range, 1–7 years). There were four (4%) loco-regional recurrences and nine (9%) distant metastases as first events. In Kaplan–Meier survival analyses, the patients with HER2 genetic heterogeneity had shorter disease-free survival times than those without it (P=0.004; Figure 3a). Similarly, the patients with HER2 regional heterogeneity had shorter disease-free survival times than the remaining patients (P=0.018; Figure 3b). In addition to HER2 intratumoral heterogeneity, high T stage (T1–2 vs T3–4; P<0.001) and nodal metastasis (N0 vs N1–3; P=0.024) were associated with poor disease-free survival. The patients with hormone receptor-negative tumor tended to have shorter disease-free survival (P=0.110). Other clinicopathological variables including adjuvant trastuzumab therapy, neoadjuvant/adjuvant chemotherapy, radiotherapy, histologic grade, Ki-67 proliferation index and p53 status were not associated with disease-free survival.
Disease-free survivals according to HER2 genetic or HER2 regional heterogeneity. The cases with HER2 genetic heterogeneity (a) and HER2 regional heterogeneity (b) show significantly poorer disease-free survival than the other cases. These findings are consistently observed in the subgroup analyses by hormone receptor status (c–f). Abbreviation: HR, hormone receptor.
Subgroup analyses by hormone receptor status also revealed survival differences between the patients with HER2 genetic heterogeneity and those without it in both subgroups (P=0.012 for the hormone receptor-negative group; P=0.010 for the hormone receptor-positive group; Figures 3c and e). With regard to HER2 regional heterogeneity, the patients with heterogeneity had shorter disease-free survival times than the remaining patients in the hormone receptor-negative subgroup (P=0.034; Figure 3d) and tended to have shorter disease-free survival times in the hormone receptor-positive subgroup (P=0.053; Figure 3f).
In multivariate analysis including T stage, N stage and HER2 genetic heterogeneity, T stage (pT1–2 vs pT3–4; hazard ratio, 11.659; 95% confidence interval, 3.107–43.748; P<0.001) and HER2 genetic heterogeneity (hazard ratio, 6.160; 95% confidence interval, 1.740–21.806; P=0.005) were independent prognostic factors for disease-free survival. Again, in multivariate analysis including T stage, N stage and HER2 regional heterogeneity, T stage (pT1–2 vs pT3–4; hazard ratio, 13.035; 95% confidence interval, 3.379–50.280; P<0.001) and HER2 regional heterogeneity (hazard ratio, 5.839; 95% confidence interval, 1.714–19.888; P=0.005) were independent prognostic factors.
Discussion
Genetically unstable tumor clones continue to mutate at a rapid rate,29 and thus intratumoral genetic heterogeneity may arise from random genetic alterations during clonal evolution, resulting in genetic subclones of cells within the primary tumor. In a previous study, we showed that there is a high degree of genetic heterogeneity both within and between distinct breast cancer cell populations based on the expression of CD44 and CD24, thus highlighting the value of analyzing tumors as ecosystems.30 Likewise, heterogeneity of HER2 expression and/or amplification exists in some HER2-positive breast cancers, as shown in this study. Supporting this, Szollosi et al.31 demonstrated marked cell to cell heterogeneity in both HER2 protein expression and HER2 gene copy number in HER2-amplified breast cancer cell lines and primary breast cancer samples. However, there have been no previous studies addressing the clinical significance of intratumoral heterogeneity in HER2 gene amplification, except a recent study by Bartlett et al.25 They evaluated HER2 genetic heterogeneity according to CAP guidelines in 6461 cases from two pathology laboratories and TEAM (Tamoxifen vs Exmestane Adjuvant Multicenter) pathology study, and reported that no prognostic impact was found when <30% of cells exhibited ratios of >2.2. As they included heterogeneous groups of patients and used FISH reports for the evaluation of heterogeneity, it is hard to compare their results with those of our study that included only HER2-amplified cases and evaluated three distinct areas in a tumor. However, interestingly, most of our cases with HER2 genetic heterogeneity have 30–50% of cells with HER2/CEP17 ratios >2.2.
In the present study, we showed that intratumoral heterogeneity of HER2 gene amplification is associated with short disease-free survival of the patients. It is not clear whether the cases with intratumoral heterogeneity of HER2 amplification arise from HER2 amplification in HER2-negative tumors or the loss of HER2 amplification in HER2-positive tumors. However, acquisition of HER2 amplification in genetically unstable HER2-negative tumors is more likely. In particular, hormone receptor negative tumors with intratumoral heterogeneity of HER2 amplification are probably genetically unstable triple negative tumors in which subclones amplified HER2 during tumor progression. Intratumoral genetic heterogeneity may indicate tumor evolution, adaptation to environmental stress and differential response to treatment. Also, intratumoral genetic heterogeneity is commonly caused by chromosomal instability, cell–cell variation in chromosome structure or number across a tumor population, which is associated with poor prognosis in solid tumors, including breast cancer.32, 33 Therefore, intratumoral HER2 heterogeneity may be associated with poor prognosis of the patients.
Intratumoral heterogeneity of HER2 gene amplification has been connected with breast cancers with low-grade HER2 amplification or equivocal (2+) HER2 expression in previous studies.16, 21, 22 Our results confirmed these findings. The frequency of HER2 regional heterogeneity or HER2 genetic heterogeneity was significantly higher in the cases with low grade or equivocal amplification in whole sections than in those with high-grade amplification. Moreover, HER2 regional heterogeneity or HER2 genetic heterogeneity was also more frequent in the cases with 2+ or 1+ HER2 immunohistochemical scores than in those with 3+ scores. We found that HER2 expression in the three tissue microarray cores was more heterogeneous in the cases with 2+ or 1+ HER2 expression in the whole sections than in those with strong 3+ HER2 expression (33 vs 4%; P<0.001, data not shown). In accord with our findings, Lewis et al21 showed that HER2 staining heterogeneity on a slide, or in different blocks of a tumor, was a frequent feature of HER2 2+ equivocal cases. We also found that heterogeneous HER2 protein expression was associated with regional heterogeneity of HER2 gene amplification. We believe that the variability of HER2 protein expression within a tumor is not simply a technical problem attributable to poor fixation, inadequate antigen retrieval or suboptimal immunohistochemical procedure, but represents real biological heterogeneity. We suggest that when a tumor displays heterogenous HER2 immunoreactivity, one should include differently stained areas in the FISH analysis to compare the immunohistochemical data with FISH results. The CAP recommended that HER2 FISH should be scanned on entire tumor slides, and that at least two (and up to four) representative fields of the invasive carcinoma should be analyzed to determine whether HER2 genetic heterogeneity is present.19
It has been suggested that intratumoral genetic heterogeneity underlies therapeutic resistance.34, 35 However, the implications of intratumoral heterogeneity of HER2 gene amplification for therapeutic resistance to HER2-targeted therapy has been little studied. It has been reported that patients with HER2 protein 2+ metastatic breast cancer do not respond to single agent trastuzumab therapy.5 Moreover, it was shown in a neoadjuvant setting that the level of HER2 amplification assessed by FISH was positively correlated with the rate of pathological response to trastuzumab-based neoadjuvant therapy.36 Intratumoral HER2 heterogeneity has generally been reported in breast cancers with low grade or equivocal HER2 amplification or protein expression, as shown in this study. Thus, it is possible that intraumoral HER2 heterogeneity has a role in the therapeutic resistance to trastuzumab in breast cancer patients with low grade or equivocal HER2 gene amplification by selecting subclones lacking HER2 gene amplification. Further studies will be needed to address the clinical relevance of intratumoral HER2 heterogeneity for therapeutic resistance to HER2-targeted therapy including trastuzumab.
Regional heterogeneity of HER2 gene amplification, which was present in 17 (18%) of the 96 cases we studied, raises a question concerning the accuracy of HER status evaluated by core biopsy. HER2 status is usually determined on resected specimens, which allow identification of intratumoral heterogeneity of HER2 gene amplification and HER2 protein expression. However, with the increasing use of neoadjuvant chemotherapy and inclusion of trastuzumab as a part of neoadjuvant chemotherapy regimens for patients with HER2-positive breast cancer,6 HER2 status is now more often determined on core biopsy specimens. D’Alfonso et al37 reported that the concordance between HER2 amplification status determined by FISH on core biopsies and on subsequent excision specimens of the same tumor was excellent (86% for all cases; 95% when equivocal FISH cases were excluded), and they suggested that intratumoral heterogeneity of HER2 assessed by FISH is not a significant confounding factor when analyzing core biopsy specimens. However, their study included only 21 HER2 FISH-positive or equivocal cases among the 100 cases examined. When analyzing only 21 HER2 FISH-positive or equivocal cases on excision specimen in their study, 7 cases (33%) are revealed to be HER2 FISH-negative on core biopsy. Thus, intratumoral heterogeneity of HER2 gene amplification may contribute to inaccurate assessment of HER2 status on small biopsy samples. Striebel et al22 supported this idea by showing that the HER2 status of 10 (59%) of 17 breast cancers with equivocal HER2 amplification on core biopsy changed in the resection specimens, and they stressed the heterogeneity of gene amplification and protein expression in breast cancers with low HER2 gene copy numbers. Recently, Wu et al38 reported a case of intratumoral heterogeneity of HER2 gene amplification, which showed HER2 gene amplification on core biopsy but not in metastases. In that case, only focal HER2 amplification was found in the primary tumor. Thus, the cases with low grade or equivocal for HER2 amplification and those with 2+ HER2 expression on breast core biopsy may need to be confirmed on subsequent resection specimens, not to lose the chance to be eligible for HER2-targeted therapy.
In conclusion, HER2 regional heterogeneity and HER2 genetic heterogeneity defined by the CAP guidelines was present in 18 and 11% of invasive breast cancers in our study. The cases with intratumoral HER2 heterogeneity were characterized by low grade or equivocal HER2 amplification and equivocal HER2 expression. Our findings emphasize the need for HER2 testing on more representative and larger tumor samples in such cases for accurate assessment of HER2 status. More importantly, our study shows, for the first time, the patients with heterogeneous HER2 amplification have significantly shorter disease-free survival times than patients with homogeneous HER2 gene amplification, suggesting that intratumoral heterogeneity of HER2 gene amplification may be associated with breast cancer progression. The impact of intratumoral heterogeneity of HER2 gene amplification on the treatment outcomes of HER2-targeted therapy requires further investigation.
References
Press MF, Pike MC, Chazin VR, et al. Her-2/neu expression in node-negative breast cancer: direct tissue quantitation by computerized image analysis and association of overexpression with increased risk of recurrent disease. Cancer Res 1993;53:4960–4970.
Slamon DJ, Clark GM, Wong SG, et al. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science 1987;235:177–182.
Tandon AK, Clark GM, Chamness GC, et al. HER-2/neu oncogene protein and prognosis in breast cancer. J Clin Oncol 1989;7:1120–1128.
Romond EH, Perez EA, Bryant J, et al. Trastuzumab plus adjuvant chemotherapy for operable HER2-positive breast cancer. N Engl J Med 2005;353:1673–1684.
Vogel CL, Cobleigh MA, Tripathy D, et al. Efficacy and safety of trastuzumab as a single agent in first-line treatment of HER2-overexpressing metastatic breast cancer. J Clin Oncol 2002;20:719–726.
Madarnas Y, Trudeau M, Franek JA, et al. Adjuvant/neoadjuvant trastuzumab therapy in women with HER-2/neu-overexpressing breast cancer: a systematic review. Cancer Treat Rev 2008;34:539–557.
Wolff AC, Hammond ME, Schwartz JN, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. J Clin Oncol 2007;25:118–145.
Gong Y, Booser DJ, Sneige N . Comparison of HER-2 status determined by fluorescence in situ hybridization in primary and metastatic breast carcinoma. Cancer 2005;103:1763–1769.
Tanner M, Jarvinen P, Isola J . Amplification of HER-2/neu and topoisomerase II alpha in primary and metastatic breast cancer. Cancer Res 2001;61:5345–5348.
Gancberg D, Di Leo A, Cardoso F, et al. Comparison of HER-2 status between primary breast cancer and corresponding distant metastatic sites. Ann Oncol 2002;13:1036–1043.
Fabi A, Di Benedetto A, Metro G, et al. HER2 protein and gene variation between primary and metastatic breast cancer: significance and impact on patient care. Clin Cancer Res 2011;17:2055–2064.
Meng S, Tripathy D, Shete S, et al. HER-2 gene amplification can be acquired as breast cancer progresses. Proc Natl Acad Sci U S A 2004;101:9393–9398.
Regitnig P, Schippinger W, Lindbauer M, et al. Change of HER-2/neu status in a subset of distant metastases from breast carcinomas. J Pathol 2004;203:918–926.
Lower EE, Glass E, Blau R, et al. HER-2/neu expression in primary and metastatic breast cancer. Breast Cancer Res Treat 2009;113:301–306.
Andersson J, Linderholm B, Bergh J, et al. HER-2/neu (c-erbB-2) evaluation in primary breast carcinoma by fluorescent in situ hybridization and immunohistochemistry with special focus on intratumor heterogeneity and comparison of invasive and in situ components. Appl Immunohistochem Mol Morphol 2004;12:14–20.
Brunelli M, Manfrin E, Martignoni G, et al. Genotypic intratumoral heterogeneity in breast carcinoma with HER2/neu amplification: evaluation according to ASCO/CAP criteria. Am J Clin Pathol 2009;131:678–682.
Glockner S, Buurman H, Kleeberger W, et al. Marked intratumoral heterogeneity of c-myc and cyclin D1 but not of c-erbB2 amplification in breast cancer. Lab Invest 2002;82:1419–1426.
Hanna W, Nofech-Mozes S, Kahn HJ . Intratumoral heterogeneity of HER2/neu in breast cancer—a rare event. Breast J 2007;13:122–129.
Vance GH, Barry TS, Bloom KJ, et al. Genetic heterogeneity in HER2 testing in breast cancer: panel summary and guidelines. Arch Pathol Lab Med 2009;133:611–612.
Cottu PH, Asselah J, Lae M, et al. Intratumoral heterogeneity of HER2/neu expression and its consequences for the management of advanced breast cancer. Ann Oncol 2008;19:595–597.
Lewis JT, Ketterling RP, Halling KC, et al. Analysis of intratumoral heterogeneity and amplification status in breast carcinomas with equivocal (2+) HER-2 immunostaining. Am J Clin Pathol 2005;124:273–281.
Striebel JM, Bhargava R, Horbinski C, et al. The equivocally amplified HER2 FISH result on breast core biopsy: indications for further sampling do affect patient management. Am J Clin Pathol 2008;129:383–390.
Shin SJ, Hyjek E, Early E, et al. Intratumoral heterogeneity of her-2/neu in invasive mammary carcinomas using fluorescence in-situ hybridization and tissue microarray. Int J Surg Pathol 2006;14:279–284.
Moeder CB, Giltnane JM, Harigopal M, et al. Quantitative justification of the change from 10 to 30% for human epidermal growth factor receptor 2 scoring in the American Society of Clinical Oncology/College of American Pathologists guidelines: tumor heterogeneity in breast cancer and its implications for tissue microarray based assessment of outcome. J Clin Oncol 2007;25:5418–5425.
Bartlett AI, Starcyznski J, Robson T, et al. Heterogeneous HER2 gene amplification: impact on patient outcome and a clinically relevant definition. Am J Clin Pathol 2011;136:266–274.
Hammond ME, Hayes DF, Dowsett M, et al. American Society of Clinical Oncology/College Of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J Clin Oncol 2010;28:2784–2795.
Hyun CL, Lee HE, Kim KS, et al. The effect of chromosome 17 polysomy on HER-2/neu status in breast cancer. J Clin Pathol 2008;61:317–321.
Ma Y, Lespagnard L, Durbecq V, et al. Polysomy 17 in HER-2/neu status elaboration in breast cancer: effect on daily practice. Clin Cancer Res 2005;11:4393–4399.
Jackson AL, Loeb LA . The mutation rate and cancer. Genetics 1998;148:1483–1490.
Park SY, Gonen M, Kim HJ, et al. Cellular and genetic diversity in the progression of in situ human breast carcinomas to an invasive phenotype. J Clin Invest 2010;120:636–644.
Szollosi J, Balazs M, Feuerstein BG, et al. ERBB-2 (HER2/neu) gene copy number, p185HER-2 overexpression, and intratumor heterogeneity in human breast cancer. Cancer Res 1995;55:5400–5407.
Carter SL, Eklund AC, Kohane IS, et al. A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat Genet 2006;38:1043–1048.
Burrell RA, Juul N, Johnston SR, et al. Targeting chromosomal instability and tumour heterogeneity in HER2-positive breast cancer. J Cell Biochem 2010;111:782–790.
Heppner GH, Miller BE . Tumor heterogeneity: biological implications and therapeutic consequences. Cancer Metastasis Rev 1983;2:5–23.
Merlo LM, Pepper JW, Reid BJ, et al. Cancer as an evolutionary and ecological process. Nat Rev Cancer 2006;6:924–935.
Arnould L, Arveux P, Couturier J, et al. Pathologic complete response to trastuzumab-based neoadjuvant therapy is related to the level of HER-2 amplification. Clin Cancer Res 2007;13:6404–6409.
D’Alfonso T, Liu YF, Monni S, et al. Accurately assessing her-2/neu status in needle core biopsies of breast cancer patients in the era of neoadjuvant therapy: emerging questions and considerations addressed. Am J Surg Pathol 2010;34:575–581.
Wu JM, Halushka MK, Argani P . Intratumoral heterogeneity of HER-2 gene amplification and protein overexpression in breast cancer. Hum Pathol 2010;41:914–917.
Acknowledgements
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (Grant No. 2010-0004822).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Seol, H., Lee, H., Choi, Y. et al. Intratumoral heterogeneity of HER2 gene amplification in breast cancer: its clinicopathological significance. Mod Pathol 25, 938–948 (2012). https://doi.org/10.1038/modpathol.2012.36
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2012.36
Keywords
This article is cited by
-
Impact of the OncotypeDX score and HER2 RNA PCR levels on HER2-low IHC levels in primary and metastasized tumors
BMC Cancer (2023)
-
HER2-low breast cancer: evolution of HER2 expression from primary tumor to distant metastases
BMC Cancer (2023)
-
Radiomic tumor phenotypes augment molecular profiling in predicting recurrence free survival after breast neoadjuvant chemotherapy
Communications Medicine (2023)
-
Reproducibility and intratumoral heterogeneity of the PAM50 breast cancer assay
Breast Cancer Research and Treatment (2023)
-
Potential of antibody–drug conjugates (ADCs) for cancer therapy
Cancer Cell International (2022)