Abstract
The MYC onco-protein is a transcription factor that regulates cell proliferation, metabolism, protein synthesis, mitochondrial function and stem cell renewal. A region on chromosome 8q24 encompassing the MYC locus is amplified in prostate cancer, but this occurs mostly in advanced disease suggesting that MYC alterations occur late in prostate cancer. In contrast, MYC mRNA is elevated in most prostate cancers, even those of relatively low stage and grade (eg Gleason score 6) suggesting that MYC plays a role in initiation. However, since MYC protein levels are tightly regulated, elevated MYC mRNA does not necessarily imply elevated MYC protein. Thus, it is critical to determine whether MYC protein is elevated in human prostate cancer, and if so, at what stage of the disease this elevation occurs. Prior studies of MYC protein localization have been hampered by lack of suitable antibodies and controls. We utilized a new anti-MYC antibody coupled with genetically defined control experiments to localize MYC protein within human tissue microarrays consisting of normal, atrophy, PIN, primary adenocarcinoma, and metastatic adenocarcinoma. Nuclear overexpression of MYC protein occurred frequently in luminal cells of PIN, as well as in most primary carcinomas and metastatic disease. MYC protein did not correlate with gain of 8q24, suggesting alternative mechanisms for MYC overexpression. These results provide evidence that upregulation of nuclear MYC protein expression is a highly prevalent and early change in prostate cancer and suggest that increased nuclear MYC may be a critical oncogenic event driving human prostate cancer initiation and progression.
Similar content being viewed by others
Main
A central problem in cancer biology is the identification of the molecular changes that cause neoplastic transformation. Studies of prostate cancer have revealed a large number of genetic and epigenetic alterations in oncogenes, tumor suppressor genes, caretaker genes, and telomeres;1, 2, 3 yet the elucidation of what changes are required for the initiation and maintenance of neoplastic transformation, the ordering of events of these changes, and the phenotype of the cell types involved remain poorly characterized. One such proto-oncogene implicated in prostate cancer development and progression is MYC (Although most authors have referred to this gene and protein as C-MYC, the official gene name is the V-MYC avian myelocytomatosis viral oncogene homolog, and the official gene symbol is MYC.).
MYC is present on human chromosome 8q24 and encodes the MYC protein which is a transcription factor that plays a key role in regulating a number of cellular processes including cell-cycle progression, metabolism, ribosome biogenesis, protein synthesis, mitochondrial function, and stem cell self renewal.4, 5 MYC is overexpressed in a large variety of tumor types, which in many cases is associated with somatic genetic alterations such as translocations and gene amplification.6 In prostate cancer, there is evidence that MYC is involved in disease progression since a region encompassing the MYC locus (8q24) is somatically amplified at low levels in a subset of patients6, 7, 8, 9 and the presence of amplification correlates with both high histological grade and a worse prognosis.7, 8 Whether there is amplification of MYC in the likely precursor to many prostate adenocarcinomas, high-grade prostatic intraepithelial neoplasia (HGPIN), is controversial because MYC amplification has been reported in up to 50% of HGPIN lesions,9 but more recent experiments revealed a lack of MYC amplification in HGPIN.10 Other genes, such as TRPS1, EIF3S3, RAD21, KIAA0916, and PSCA are within or near the 8q24 region and, at times, are also amplified in prostate carcinoma11, 12, 13, 14, 15 complicating the data implicating MYC as the key target of amplification in this chromosomal region.
It has been long known that a subset of prostate cancer lesions express elevated levels of MYC mRNA16, 17, 18 relative to benign matched prostate tissues, and recent transcription profiling studies have confirmed and extended these findings indicating that a large fraction of prostate cancer cases overexpress MYC mRNA as compared to matched normal appearing tissues (see results section). Further, targeted overexpression of the human MYC gene in the mouse prostate results in PIN,19, 20 early invasive prostate adenocarcinoma20 and rare metastatic adenocarcinoma,20 providing definitive evidence that MYC overexpression can drive neoplastic transformation in the mouse prostate, and supporting a model whereby MYC may play a role in initiation of human prostate cancer development. Additionally, retrovirally induced overexpression of MYC can transform primary cultures of benign prostate epithelial cells.21 Nevertheless, because of a lack of suitable antibodies that can be readily applied for cellular and subcellular localization in archival tissues, the phase of prostate cancer development in which MYC protein is expressed in humans is still unclear. It is critical to directly ascertain MYC protein levels because MYC protein levels are tightly regulated by post-transcriptional and post-translational mechanisms, and the presence of MYC mRNA does not necessarily imply the presence of MYC protein.22, 23 Therefore, the precise role of MYC protein in the early phases of human prostate cancer development, if any, remains undefined. Clearly, an improved assessment of the role of MYC in all stages of prostate cancer progression would be provided by an ability to use human archival tissue specimens to readily localize the MYC protein.
While there have been prior studies that describe MYC protein expression as detected by immunohistochemistry in prostate cancer7, 24, 25 and even one prior study in high-grade PIN,7 the results of these studies are difficult to interpret. For example, in two of these studies MYC staining was localized either exclusively7 or nearly exclusively24 to the cytoplasm. This lack of nuclear staining is surprising because all of the known functions of MYC in cellular transformation have been ascribed to actions in the nucleus; endogenous MYC has been localized to the nucleus,26 and in cells genetically modified to express exogenous MYC, the protein localizes predominantly to the nucleus.27, 28 In the third paper that examined MYC staining in prostate cancer, staining was localized predominantly to the nucleus and was positive in 33 of 45 cases.25 Surprisingly, however, there was very little difference in MYC staining between benign and malignant epithelial cells.25
Perhaps the most compelling reason to revisit this question of MYC protein expression and localization in prostate cancer is the fact that no prior studies of MYC immunohistochemical staining in prostate cancer reported on the performance of positive control experiments beyond staining of presumed positive tumor tissues, nor did they perform negative control experiments beyond simply leaving out the primary antibody. We conclude that MYC protein expression and localization in prostate cancer remains poorly defined and that the determination of the localization of immunoreactivity in normal, pre-neoplastic and neoplastic prostate tissues is still a critical unanswered question in prostate carcinogenesis. In this paper, we employ for the first time a newly developed rabbit monoclonal antibody and a highly sensitive immunohistochemical protocol, developed in conjunction with genetically defined control experiments, to obtain strong nuclear staining for MYC in human clinical prostate cancer.
Materials and methods
Antibodies
The anti-MYC rabbit monoclonal antibody used for both western blotting and immunohistochemistry was obtained from Epitomics (Burlingame, CA, USA, Clone Y69).
Tissues and Tissue Microarray Construction
This study was approved by our institutional internal review board. Tissue specimens were obtained from radical retropubic prostatectomies performed at the Johns Hopkins Hospital. Patient ages ranged from 37 to 89 years (mean=59), the final Gleason sums varied from five to nine (mean=6.6), and the pathological stages ranged from T2N0Mx to T3BN1Mx (see Table 1). For metastatic tissue samples, tissues were obtained from pelvic lymph node dissections that were or were not accompanied by radical prostatectomy, or surgical excisions of distant metastases. All tissues were fixed in 10% neutral-buffered formalin and processed into paraffin blocks.
Nine high-density tissue microarrays were employed for MYC immunostaining. Matched cancer and normal tissue microarrays (N=6) were constructed with matched samples from 40 patients per array as described previously.29 Additional tissue microarrays were designed to compare matched normal and atrophy and matched PIN and normal. A final tissue microarray was constructed of tissues from hormone naive metastatic prostate cancer obtained from surgical specimens from pelvic lymph nodes, soft tissue, and bone.29
Images of each tissue microarray core were captured by scanning of the tissue microarray slides using the BLISS scanner (Bacus laboratories, Chicago, IL, USA) and were imported into the TMAJ images application (http://tmaj.pathology.jhmi.edu) as described.29 Histologic diagnoses were applied to all images and these diagnoses are listed in Table 2. All images and image data are available for viewing and download at http://demarzolab.pathology.jhmi.edu/Pubs.html.
Immunohistochemistry
Immunohistochemistry was performed with the Dako catalyzed signal amplification kit (DAKO Cytomation, Carpentaria, CA, USA). Slides were steamed for 40 min in EDTA solution (Zymed, South San Francisco, CA, USA) for antigen retrieval. Slides were incubated with the rabbit monoclonal anti-MYC antibody overnight at 4°C. Biotinylated anti-rabbit IgG (Vector, Burlingame, CA, USA) was used as secondary antibody. Staining was visualized using 3,3′-Diamino-benzidine (DAB) (Sigma, Saint Louis, MO, USA, FAST 3,3′-Diamino-benzidine Tablets) and slides were counterstained with hematoxylin.
Western blotting
P493-6 cells are Epstein–Barr virus transformed human B lymphocytes30 that express a tetracycline-regulated (Tet-off) conditional MYC construct. The cells were cultured in RPMI+10% fetal calf serum with penicillin/streptomycin. They were grown to ∼3 × 105 cells/ml and treated with 0.1 μg/ml of tetracycline for 24 h. To prepare the cell lysates, the cells were collected by centrifugation at 2000 g for 5 min. The pellet was washed once with Hank's Balanced Salt Solution (HBSS) and the supernatant discarded. Ice-cold RIPA buffer was added and the cells were incubated on ice for 15 min. The mixture was then centrifuged at 14 000 g for 20 min at 4°C and the supernatant was collected. Protein concentrations were determined using the BCA protein assay kit (Pierce Biotechnology, Rockford, IL, USA). Samples were prepared in Laemmli sample buffer with β-mercaptoethanol and run on 5–20% gradient SDS-polyacrylamide gels (Bio-Rad). Proteins were transferred to PVDF membranes and the membranes were blocked using 5% non-fat dry milk in Tris-buffered saline with 1% Tween 20 (TBST) for 3 h at room temperature. The membrane was then incubated overnight at 4°C with the rabbit monoclonal anti-MYC antibody (1:1000 dilution), followed by 1 h of incubation at room temperature with the secondary horseradish peroxidase-conjugated Donkey polyclonal anti-rabbit antibody (1:5000 dilution, Amersham, Piscataway, NJ, USA). Proteins were visualized using an ECL chemiluminescence detection system, following the manufacturer's protocol and Hyperfilm ECL (Amersham). As a protein loading and integrity control, immunoblots were also probed with an anti-actin antibody (1:5000 dilution in 5% NFDM for 1 h, followed by 1:5000 of secondary antibody [Mouse IgG, HRP-Linked Whole Ab from sheep, GE Healthcare NA931] in 1% NFDM for 1 h at room temperature). Prostate cancer cells (LnCaP, CWR22Rv1, LAPC-4, C42B, PC3 and DU145) lines were obtained from the American Type Culture Collection and grown to 80% confluence according to the ATCC recommendations. All cell lines were fixed in formalin and paraffin-embedded for use in immunostaining for MYC as above. LnCaP and CWR22Rv1 cells were lysed and analyzed for western blotting as above.
Lo-MYC Mice
Lo-MYC mice, which are transgenic for the human MYC gene under the control of the rat probasin promoter,20 were obtained from the Mouse Repository of the National Cancer Institute Mouse Models of Human Cancer Consortium at NCI Fredrick, MD, USA. Animals were housed in an animal facility maintained on a 12-h light/dark cycle, at a constant temperature (22±2°C) and relative humidity (55±15%). Tap water and food were available ad libitum.
Analysis of Immunohistochemical Staining
The tissue microarray scanned images were assigned a diagnosis by both BG and AMD. For image analysis we used FrIDA (FRamework for Image Dataset Analysis),31 a custom open source image analysis software package (available at http://sourceforge.net/projects/fridajhu/) for the analysis of RGB color image datasets, including those generated from scanning of tissue microarray slides. Hue Saturation and Brightness (HSB) segmentation ranges for DAB brown staining and hematoxylin alone (nuclei not staining brown) were defined from the tissue microarray image set, and a region of interest (ROI) was created from each tissue microarray image. The ‘MYC ratio’ was defined as the DAB area in the ROI divided by the sum of the DAB area and the hematoxylin area in the ROI. The ROI mask was created by including epithelial structures while excluding the stroma and glands that did not correspond with the primary diagnosis in the tissue microarray core. Thresholds for automated image analysis were set such that any brown staining within nuclei detected visually was included.
We estimate that on average between 50 and 1000 cells on each tissue microarray core were circled. Many of the samples contained heterogeneous staining for MYC. However, because our currently employed image analysis technology only allows us to quantify overall areas and intensity values, and does not allow us to count individual cells, we simply provided the area fraction of positively stained nuclei and did not attempt to quantify heterogeneity. In a previous study from our laboratory using image analysis on tissue microarrays we used similar methods after scanning slides using the Automated Cellular Imaging System (ACIS® from DAKO Inc.) in which we validated the strong correlation between the number of cells staining positive by visual estimation and the area of positive staining obtained by image analysis.29 In the current study, therefore, the area fraction of staining is roughly equivalent to the fraction of nuclei staining positively. We have also recently found that the quantitative image analysis data obtained using FrIDA is highly correlated with that obtained using the ACIS (AM De Marzo, unpublished observations). The ‘MYC Score’ was defined as the DAB integrated intensity divided by the sum of the DAB area and the hematoxylin area. Statistical analyses were performed using SAS. To compare the distribution of MYC staining for each histologic type to normal, MYC values were first transformed (to the power of 1/4) to approximate a normal distribution. P-values were then calculated by linear regression with robust variance estimation to account for the correlation between observations. In addition to providing overall values for area of nuclear staining (MYC ratio) and the MYC score, we also compared the values among tissue types such that positive staining was defined when the overall MYC ratio was greater than the median MYC ratio for all samples.
Results
Overexpression of MYC mRNA in Prostate Cancer
Figure 1 shows a box-plot of relative MYC expression at the mRNA expression level in which we compared matched cases of prostate cancer and normal appearing prostate tissues. The data was extracted from a dataset that we recently published.32 The majority of cases contained markedly elevated MYC mRNA expression in tumor tissue, as compared with matched normal appearing prostate tissue or benign prostatic hyperplasia tissue. These findings were further validated by querying the Oncomine database33 in which five separate publications deposited mRNA profiling data using prostate cancer tissues.34, 35, 36, 37, 38 In each experiment evaluating tumor tissue compared with normal or BPH tissue in non-pretreated patients (n=7 separate experiments from the five different papers), MYC mRNA was found to be markedly elevated in the cancer tissue (Figure 2). These results showing highly consistent upregulation of MYC at the mRNA level in prostate cancer led us to determine whether MYC protein was also elevated in human prostate cancer clinical specimens.
Box plots of MYC mRNA expression values in BPH (n=9), normal (n=25), and cancer tissues (n=25). The expression values were normalized to the common BPH reference denominator as described in detail.32 Each box is lined at lower quartile, median, and upper quartile score values for each group, and extended to 95 percentiles by the whiskers. The ‘+’ symbols mark data values beyond the ends of the whiskers.
Differential expression of MYC mRNA in the prostate. The results from seven experiments from five different studies34, 35, 36, 37, 38 obtained through the Oncomine database are represented as box plots as output from the Oncomine website built in features. NP: normal prostate, PCa: primary prostatic carcinoma, NAP: normal adult prostate, NPP: normal pre-pubertal prostate, HRM: hormone-refractory metastatic prostate carcinoma, AT: atrophic epithelium, PIN: prostatic intraepithelial neoplasia, MET: metastatic prostate carcinoma, CaLN: metastatic prostatic carcinoma in lymph node. a–g represent the different experiments with investigators as indicated.
Specificity of MYC Immunohistochemical Staining
Initial attempts at immunohistochemistry employed the well-known mouse monoclonal antibody, clone 9E10, which recognizes the epitope from MYC that is often used to ‘tag’ recombinant proteins for use in immuno-precipitation and western blotting of the tagged proteins. As a positive control we used prostate tissues from transgenic mice that overexpress human MYC (8-week-old ‘Lo-MYC’ mice) in the mouse prostate.20 Despite several attempts using different immunohistochemistry protocols and kits, we were unable to obtain specific nuclear staining with the 9E10 antibody (data not shown).
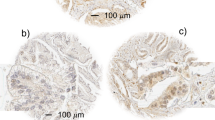
Next we tried a recently introduced rabbit monoclonal antibody raised against N terminus of the human MYC protein that has been reported by the manufacturer to perform well in immunohistochemistry (see Materials and methods). The specificity of the rabbit monoclonal antibody is shown at Figure 3. As an initial control, we used the antibody for western blotting using a cell line that expresses MYC under the control of a tetracycline-regulated promoter (‘Tet-off’ system).30 As seen in Figure 3a, with tetracycline, there were no bands present. After tetracycline withdrawal, a strong band appeared at a predicted molecular weight of ∼64 kDa, which is near the apparent molecular weight at which the MYC protein is known to migrate.39 In addition, prostate carcinoma cell lines (LnCaP and CWR22Rv1) were subjected to western blotting using this same antibody and each showed a single band at the same apparent molecular weight (Figure 3a). To test the specificity of this antibody for immunohistochemistry we stained prostate tissue sections from the Lo-MYC20 mouse. As shown in Figure 3, although there was very little or no staining in the prostates of the wild type mouse litter mates (FVB), there was intense staining in the nuclei of the epithelial cells of the MYC-expressing transgenic animals in their ventral prostate (Figure 3c). As another control we stained liver tissue from an additional transgenic mouse model that expresses MYC specifically in the liver in a doxycycline repressible fashion,40 and we found strong nuclear staining in hepatocytes for MYC in the mouse that was not treated with doxycycline but no expression was seen in hepatocytes from mice that were pretreated with doxycycline (data not shown). Taken together, the western blot and the immunostaining data establish the specificity of this antibody for MYC for immunohistochemistry on archival tissue specimens. Interestingly, all prostate cancer cell lines tested that were subjected to formalin fixation and paraffin embedding, including LnCaP, CWR22rv1, LAPC-4, C42B, PC3, and DU145 showed strong staining for MYC in the nuclei of the majority of tumor cells (not shown).
Verification of the specificity of the anti-MYC antibody. (a) Western blot of LnCaP, CWR22Rv1 and p493 cell lines, the latter without (−Tet) or with (+Tet) application of tetracycline, stained with anti-MYC antibody. (b) Wild type FVB mouse prostate tissue stained with anti-MYC antibody showing negative staining. (c) Lo-MYC mouse prostate tissue, expressing the human MYC protein, stained with anti-MYC antibody showing strong staining in epithelial cells, primarily within nuclei. The Lo-MYC mice are on the FVB background.
Qualitative and Quantitative Analyses of MYC Immunohistochemical Staining in Human Prostate Tissues
MYC staining in normal epithelium was often completely absent (Figure 4a). In other cases, there were low levels of MYC staining with a median MYC ratio of 2.2% and a median MYC score of 2.6 (Table 2). When staining was present, it was restricted to nuclei and was generally more commonly seen in basal epithelial cells rather than luminal epithelial cells (Figure 5a and b). There was also strong staining of the endothelial cell nuclei in some of the tissue microarray cores (Figure 5a and b), although this finding was quite variable. Non-endothelial stromal cells were negative. When using the overall median MYC ratio of MYC staining for the entire set of tissue microarray spots as a cutoff value (7.55%) for scoring as positive, 23% of the tissue microarray spots containing normal-appearing epithelium scored as positive. In prostate atrophy lesions, there also was variable staining of nuclei with the median MYC ratio being similar to normal prostate epithelium (Table 2) (21% were above the 7.55% median cutoff value). However, in contrast to the normal epithelium, there was a shift in the compartmentalization of staining from predominantly basal cell staining in normal epithelium to predominantly luminal cell staining in atrophy (Figure 4). To quantify the relative number of luminal and basal cells staining positively in atrophy as compared with normal, we manually counted the number of cells staining for MYC in each compartment in a subset of normal regions and atrophy lesions on one of the tissue microarrays (n=63 normal and 68 atrophy tissue microarray spots were counted) in which there was some MYC staining. The median number of cells positive in the luminal compartment in normal prostate was 0.25% and in atrophy was 0.75% (P<0.0001 sign rank test), and the median number of cells positive in the basal compartment of normal was 0.75% and in atrophy was 0.25% (P<0.0001 sign rank test).
Human prostate tissues stained with anti-MYC antibody. (a) Normal prostate tissue. (b) Focal prostate atrophy (simple atrophy). Note the nuclear staining in luminal epithelial cells (arrows). (c) High-grade PIN. (d) PIN, higher power view. Widespread nuclear staining in the luminal epithelial cells can be seen (arrows).
MYC staining in primary and metastatic prostatic adenocarcinoma. (a) Low-power view of tissue microarray spot containing Gleason pattern 3 adenocarcinoma and mixed stroma and benign glands. Note the pronounced difference in staining between benign and malignant glands (arrows). (b) Higher power view. MYC staining in the benign prostate glands is weaker compared with the malignant glands, and is confined to the basal epithelial cell layer (arrows). A few weakly staining endothelial cells can be seen. (c) Primary prostatic carcinoma, Gleason pattern 4. (d) Metastatic prostate carcinoma in a lymph node.
In prostatic intraepithelial neoplasia (PIN), MYC staining was elevated in both low- and high-grade lesions as compared with normal epithelium and atrophic epithelium (Figure 4, Table 2). The median MYC ratio of low-grade PIN was 8.4% and the median MYC score was 8.6, and both of these values were significantly higher than normal epithelium. High-grade PIN showed a stepwise increase in staining with a median MYC ratio of 23.5% and a median MYC score of 25.8. Lesions with both low- and high-grade PIN mixed together had values for MYC staining intermediate between those considered pure low and high grade (Table 2). When using the overall median ratio of MYC staining for the entire set of tissue microarray spots as a cutoff value, 76% of PIN lesions were considered positive. Like atrophic epithelium, the vast majority of staining in PIN was restricted to the nuclei of the luminal component (Figure 4).
In carcinoma, the majority of cases showed overexpression compared with matched normal epithelium such that in many of the cases most of the tumor cell nuclei were strongly positive for MYC staining (Figure 5, Table 2). Prostate carcinoma typically invades between and around benign normal appearing prostate acini and quite often the nuclei in the cancerous glands stained strongly while the adjacent normal appearing glands were negative. When using the cutoff value for the MYC ratio described above, 81.6% of carcinoma lesions were considered positive. We averaged all tissue microarray spots for a given histology (ie normal, atrophy, PIN, carcinoma) for a given patient and the distribution of staining for these is shown in Figure 6.
We next analyzed the scoring data for the tissue microarray spots containing carcinoma stratified by Gleason pattern. As seen in Table 2, Gleason pattern 4 lesions showed less MYC staining than Gleason pattern 3 lesions, although the staining in Gleason pattern 4 lesions remained significantly elevated compared with normal epithelium. Interestingly, although the number of cases was small, Gleason pattern 5 lesions showed higher staining values than Gleason pattern 4, similar to but not quite as high as Gleason pattern 3 (Table 2). When comparing MYC staining to pathological stage at radical prostatectomy, no relation was found (P=0.134, Kruskal–Wallis).
We also stained a tissue microarray containing non-pretreated metastatic prostate cancer from either pelvic lymph nodes or soft tissue or bone metastases. While the overall MYC staining was not as high as primary Gleason pattern 3, it was generally much higher than normal prostate, with 68% of cases being above the median MYC ratio.
On two tissue microarrays we also had data on the presence from FISH analysis of chromosome 8p22 loss (LPL locus), 8 centromere gain, and 8q24.12-24.13 gain (encompassing the MYC locus).10 As previously noted in this dataset there was a strong relation between gain of 8q24 and Gleason pattern,10 and this was also true for the smaller subset of cases that we also have MYC protein staining data on in the present study (Table 3). Interestingly, there was no relation between 8q24 copy number and MYC protein staining (Table 4), yet there was an inverse relation between chromosome 8p loss and MYC protein staining.
Discussion
In this study, we show that overexpression of MYC protein in prostate cancer is a remarkably frequent event. Unlike previous studies that reported that MYC protein is overexpressed nearly exclusively in the cytoplasm in prostate cancer cells,7, 24 we found MYC protein primarily in the nuclei of all expressing cells tested. The specificity of the staining was supported by genetically defined control experiments. Although it has been known for many years that MYC mRNA is elevated in human prostate cancer, as a number of mechanisms can regulate the stability of the MYC protein itself,22, 23 it was important to determine whether MYC protein is also overexpressed in prostate cancer.
As MYC staining was significantly higher in the majority of cases of prostate cancer, when compared with matched normal tissues, these results raise the possibility that MYC might be a useful biomarker in tissues or body fluids. For example, immunohistochemistry analysis of MYC may prove useful in combination with other markers such as basal-specific keratins, p63, and alpha methyl acyl CO-A racemase, to enhance diagnostic accuracy if applied to prostate needle biopsies in difficult cases in which the diagnosis of cancer is uncertain. Further, MYC levels either assayed by immunohistochemistry in intact cells in the urine, or by enzyme-linked immunosorbent assay (ELISA) or other protein quantification methods in the urine may add value as an early detection and disease-monitoring biomarker.
As MYC is so commonly overexpressed at the protein level in prostate cancer, the current findings raise the possibility that MYC might be an excellent therapeutic target in this disease. New approaches being developed to target MYC or MYC-dependent tumors in vivo are being developed (eg Goga et al41) and if these, or similar approaches, reach the clinic it will be important to test their therapeutic efficacy in prostate cancer patients.
Although MYC protein was increased in most cases of primary and metastatic prostate cancer that we examined, it was significantly lower in higher grade lesions (Gleason patterns 4–5) and in metastatic lesions from hormone naive patients as compared with intermediate grade lesions (primarily Gleason pattern 3). Yet, MYC expression in the higher grade and metastatic lesions was still much higher in most cases than normal appearing prostate epithelium, indicating that MYC overexpression may still be important in these less differentiated lesions. These results do raise the possibility that there may be some pressure to keep the levels from becoming too high in the higher grade lesions. This might, for example, occur since high levels of MYC are known to induce apoptosis in a number of cellular contexts.23 Interestingly, while MYC protein levels were somewhat lower in high-grade cancers, results from a number of previous studies and from specimens used in the current study indicate that high-grade prostate cancers are much more likely to harbor increases in chromosome 8q24. Our current finding that these increases do not correlate with MYC protein could support a number of different possibilities, including one whereby other genes are the main target for 8q24 amplification in prostate cancer, or that higher grade tumors are under pressure to amplify the MYC locus because for some reason they are ‘programmed’ to keep the levels somewhat lower than more low-grade tumors. Studies in different data sets with long term follow-up will be useful to determine whether MYC protein levels correlate with a poor clinical outcome, as has already been shown for 8q24 gain.8 Interestingly, in the present study, although no clear mechanism can currently explain this finding, MYC protein levels were inversely related to loss of chromosome 8p22 (LPL locus).
Although it has been widely assumed that relatively undifferentiated tissue stem cells may be the target progenitor cell of neoplastic transformation in solid organ tumors, it is also likely that cells partially differentiated along a particular lineage (committed progenitor cells or transit-amplifying cells) can become transformed by aberrant reactivation of self renewal genes normally active only in the stem cells.42 Prostate epithelium consists of two distinct cell populations, basal and luminal. Although definitive evidence is still lacking, most evidence in both rodents and humans indicate that stem cells reside in the basal cell compartment and that luminal cells are derived from basal cells in a hierarchical system in which basal stem-like cells give rise to progeny that differentiate into transit/amplifying (TA) cells.43, 44, 45 These TA cells have a phenotype intermediate between basal and luminal cells and can differentiate into mature luminal cells. Although some prostate cancers tend to accumulate at least some cells with markers expressed predominantly in basal as opposed to luminal cells (eg keratin 5, bcl-2 and c-met), the vast majority of primary human prostate cancers consists of populations of cells expressing phenotypic differentiation markers closely matching prostatic luminal cells. This is also the case for the presumed precursor lesion of many prostate carcinomas, high-grade prostatic intraepithelial neoplasia. Therefore, if prostate stem cells do have a basal cell phenotype, then it would appear that the target progenitor cell for neoplastic transformation in the prostate is not the prostate epithelial stem cell. Rather, we and others have postulated that partially differentiated luminal cells are the target progenitor cells for neoplastic transformation in the prostate.45, 46, 47, 48, 49 Results from the present study showing nuclear expression of MYC primarily in the basal compartment of normal appearing epithelium, yet in the luminal cells of focal atrophy, low- and high-grade PIN, are supportive of this concept. In addition, as MYC is known to be expressed strongly in many tissues during embryogenesis, it is one of the four key genes that can be used to reprogram differentiated fibroblasts into cells that are characteristic of embryonic stem cells,50 and is known to activate an embryonic stem cell-like transcriptional program during the induction of cancer stem cells in vitro,51 it is intriguing to speculate that activation of MYC protein expression in prostate luminal cells may stimulate reprogramming of these cells into cancer stem-like cells. Along somewhat parallel lines, MYC inhibits ‘terminal’ differentiation of a number of cell types,23 and MYC protein elevation in prostate cancer may be responsible for preventing ‘terminal’ differentiation and, therefore, preventing permanent exit from the cell cycle in the luminal cell compartment in PIN and in the luminal-like cells in the carcinoma.
Although MYC is clearly associated with cell-cycle progression in many cell types, it appears that the overexpression of MYC in PIN and adenocarcinoma is not simply reflective of an increased proliferative fraction, as the number of cells positive for MYC were generally much more than that seen when one uses cellular markers of proliferation, such as Ki-67. Thus, although our data suggest that MYC is expressed as part of the normal proliferative physiology of prostate epithelial cells, the tumor cells somehow perturb this system to increase MYC beyond what is physiologically normal. The fact that MYC mRNA and protein are elevated in most prostate cancers raises the intriguing question as to how MYC overexpression is occurring. There are a number of molecular mechanisms for MYC overexpression in various cancers6 and the present findings that MYC protein is indeed overexpressed in a large number of prostate cancers should stimulate additional work to seek out these mechanisms.
To conclude, MYC protein overexpression occurred commonly in PIN, early clinically localized prostate cancers, and in hormone naive metastatic adenocarcinoma. Given the findings that MYC overexpression is the critical factor driving neoplastic transformation in a number of cancers,6 forced overexpression of MYC in the mouse prostate can drive the development of PIN and early prostate cancer lesions,19, 20 and retrovirally induced overexpression of MYC can transform primary cultures of benign prostate epithelial cells,21 the current results in human clinical specimens are consistent with a hypothesis in which deregulation of MYC expression is a critical, early and widely prevalent oncogenic event in prostate cancer.
References
Isaacs W, De Marzo A, Nelson W . Focus on prostate cancer. Cancer Cell 2002;2:113.
Abate-Shen C, Shen MM . Molecular genetics of prostate cancer. Genes Dev 2000;14:2410–2434.
Tomlins SA, Rhodes DR, Perner S, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science 2005;310:644–648.
Dang CV, O’Donnell KA, Zeller KI, et al. The c-Myc target gene network. Semin Cancer Biol 2006;16:253–264.
Knoepfler PS . Why Myc? An unexpected ingredient in the stem cell cocktail. Cell Stem Cell 2008;2:18–21.
Nesbit CE, Tersak JM, Prochownik EV . MYC oncogenes and human neoplastic disease. Oncogene 1999;18:3004–3016.
Jenkins RB, Qian J, Lieber MM, et al. Detection of c-myc oncogene amplification and chromosomal anomalies in metastatic prostatic carcinoma by fluorescence in situ hybridization. Cancer Res 1997;57:524–531.
Sato K, Qian J, Slezak JM, et al. Clinical significance of alterations of chromosome 8 in high-grade, advanced, nonmetastatic prostate carcinoma. J Natl Cancer Inst 1999;91:1574–1580.
Qian J, Jenkins RB, Bostwick DG . Detection of chromosomal anomalies and c-myc gene amplification in the cribriform pattern of prostatic intraepithelial neoplasia and carcinoma by fluorescence in situ hybridization. Mod Pathol 1997;10:1113–1119.
Bethel CR, Faith D, Li X, et al. Decreased NKX3.1 protein expression in focal prostatic atrophy, prostatic intraepithelial neoplasia and adenocarcinoma: association with Gleason score and chromosome 8p deletion. Cancer Res 2006;66:10683–10690.
Nupponen NN, Porkka K, Kakkola L, et al. Amplification and overexpression of p40 subunit of eukaryotic translation initiation factor 3 in breast and prostate cancer. Am J Pathol 1999;154:1777–1783.
Reiter RE, Sato I, Thomas G, et al. Coamplification of prostate stem cell antigen (PSCA) and MYC in locally advanced prostate cancer. Genes Chromosomes Cancer 2000;27:95–103.
Saramaki O, Willi N, Bratt O, et al. Amplification of EIF3S3 gene is associated with advanced stage in prostate cancer. Am J Pathol 2001;159:2089–2094.
Tsuchiya N, Kondo Y, Takahashi A, et al. Mapping and gene expression profile of the minimally overrepresented 8q24 region in prostate cancer. Am J Pathol 2002;160:1799–1806.
Porkka KP, Tammela TL, Vessella RL, et al. RAD21 and KIAA0196 at 8q24 are amplified and overexpressed in prostate cancer. Genes Chromosomes Cancer 2004;39:1–10.
Fleming WH, Hamel A, MacDonald R, et al. Expression of the c-myc protooncogene in human prostatic carcinoma and benign prostatic hyperplasia. Cancer Res 1986;46:1535–1538.
Buttyan R, Sawczuk IS, Benson MC, et al. Enhanced expression of the c-myc protooncogene in high-grade human prostate cancers. Prostate 1987;11:327–337.
Meng MV, Dahiya R . Molecular genetics of prostate cancer. In: Carroll PR, Grossfeld GD (eds). Prostate Cancer, Vol. BC Decker Inc., Hamilton, Ontario,2002, pp 42–59.
Zhang X, Lee C, Ng PY, et al. Prostatic neoplasia in transgenic mice with prostate-directed overexpression of the c-myc oncoprotein. Prostate 2000;43:278–285.
Ellwood-Yen K, Graeber TG, Wongvipat J, et al. Myc-driven murine prostate cancer shares molecular features with human prostate tumors. Cancer Cell 2003;4:223–238.
Williams K, Fernandez S, Stien X, et al. Unopposed c-MYC expression in benign prostatic epithelium causes a cancer phenotype. Prostate 2005;63:369–384.
Sears RC . The life cycle of C-myc: from synthesis to degradation. Cell Cycle 2004;3:1133–1137.
Adhikary S, Eilers M . Transcriptional regulation and transformation by Myc proteins. Nat Rev Mol Cell Biol 2005;6:635–645.
Yang G, Timme TL, Frolov A, et al. Combined c-Myc and caveolin-1 expression in human prostate carcinoma predicts prostate carcinoma progression. Cancer 2005;103:1186–1194.
Fox SB, Persad RA, Royds J, et al. p53 and c-myc expression in stage A1 prostatic adenocarcinoma: useful prognostic determinants? J Urol 1993;150:490–494.
Persson H, Leder P . Nuclear localization and DNA binding properties of a protein expressed by human c-myc oncogene. Science 1984;225:718–721.
Stone J, de Lange T, Ramsay G, et al. Definition of regions in human c-myc that are involved in transformation and nuclear localization. Mol Cell Biol 1987;7:1697–1709.
Smith KP, Byron M, O’Connell BC, et al. c-Myc localization within the nucleus: evidence for association with the PML nuclear body. J Cell Biochem 2004;93:1282–1296.
Faith D, Isaacs WB, Morgan JD, et al. Trefoil factor 3 overexpression in prostatic carcinoma: prognostic importance using tissue microarrays. The Prostate 2004;61:215–227.
Schuhmacher M, Staege MS, Pajic A, et al. Control of cell growth by c-Myc in the absence of cell division. Curr Biol 1999;9:1255–1258.
Cornish T, De Marzo AM, Gurel B, et al. FRIDA an Open Source Framework for Image Dataset Analysis. Advancing Practice, Instruction and Innovation Through Informatics: Pittsburgh, PA, 2007.
Dunn TA, Chen S, Faith DA, et al. A novel role of myosin VI in human prostate cancer. Am J Pathol 2006;169:1843–1854.
Rhodes DR, Kalyana-Sundaram S, Mahavisno V, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18 000 cancer gene expression profiles. Neoplasia 2007;9:166–180.
Lapointe J, Li C, Higgins JP, et al. Gene expression profiling identifies clinically relevant subtypes of prostate cancer. Proc Natl Acad Sci USA 2004;101:811–816.
Dhanasekaran SM, Dash A, Yu J, et al. Molecular profiling of human prostate tissues: insights into gene expression patterns of prostate development during puberty. FASEB J 2005;19:243–245.
Varambally S, Yu J, Laxman B, et al. Integrative genomic and proteomic analysis of prostate cancer reveals signatures of metastatic progression. Cancer Cell 2005;8:393–406.
Tomlins SA, Mehra R, Rhodes DR, et al. Integrative molecular concept modeling of prostate cancer progression. Nat Genet 2007;39:41–51.
Yu YP, Landsittel D, Jing L, et al. Gene expression alterations in prostate cancer predicting tumor aggression and preceding development of malignancy. J Clin Oncol 2004;22:2790–2799.
Alitalo K, Ramsay G, Bishop JM, et al. Identification of nuclear proteins encoded by viral and cellular myc oncogenes. Nature 1983;306:274–277.
Gao P, Zhang H, Dinavahi R, et al. HIF-dependent antitumorigenic effect of antioxidants in vivo. Cancer Cell 2007;12:230–238.
Goga A, Yang D, Tward AD, et al. Inhibition of CDK1 as a potential therapy for tumors over-expressing MYC. Nat Med 2007;13:820–827.
Krivtsov AV, Twomey D, Feng Z, et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature 2006;442:818–822.
van Leenders G, Dijkman H, Hulsbergen-van de Kaa C, et al. Demonstration of intermediate cells during human prostate epithelial differentiation in situ and in vitro using triple-staining confocal scanning microscopy. Lab Invest 2000;80:1251–1258.
Litvinov IV, De Marzo AM, Isaacs JT . Is the Achilles’ heel for prostate cancer therapy a gain of function in androgen receptor signaling? J Clin Endocrinol Metab 2003;88:2972–2982.
De Marzo AM, Nelson WG, Meeker AK, et al. Stem cell features of benign and malignant prostate epithelial cells. J Urol 1998;160:2381–2392.
Verhagen AP, Ramaekers FC, Aalders TW, et al. Colocalization of basal and luminal cell-type cytokeratins in human prostate cancer. Cancer Research 1992;52:6182–6187.
Meeker AK, Hicks JL, Platz EA, et al. Telomere shortening is an early somatic DNA alteration in human prostate tumorigenesis. Cancer Res 2002;62:6405–6409.
van Leenders GJ, Gage WR, Hicks JL, et al. Intermediate cells in human prostate epithelium are enriched in proliferative inflammatory atrophy. Am J Pathol 2003;162:1529–1537.
De Marzo AM, Platz EA, Sutcliffe S, et al. Inflammation in prostate carcinogenesis. Nat Rev Cancer 2007;7:256–269.
Meissner A, Wernig M, Jaenisch R . Direct reprogramming of genetically unmodified fibroblasts into pluripotent stem cells. Nat Biotechnol 2007;25:1177–1181.
Wong DJ, Liu H, Ridky TW, et al. Module map of stem cell genes guides creation of epithelial cancer stem cells. Cell Stem Cell 2008;2:333–344.
Acknowledgements
We thank Srinivasan Yegnasubramanian, William G Nelson and Alan K Meeker for helpful discussions regarding this paper. Also we thank Helen Fedor, Marcela Southerland and the Brady Urological Institute Prostate Specimen Repository supported by the Prostate SPORE grant for Tissue Microarrays. We also thank Kristen Lecksell for excellence in tissue microarray slide scanning and image handling. Grants were received from Public Health Services NIH/NCI Specialized Program in Research Excellence (SPORE) in Prostate Cancer #P50CA58236 (Johns Hopkins). AMD is the Beth W. and A. Ross Myers Scholar supported through the Patrick C. Walsh Prostate Cancer Research Fund.
Author information
Authors and Affiliations
Corresponding author
Additional information
Disclosure/conflict of interest
The authors have nothing to disclose.
Rights and permissions
About this article
Cite this article
Gurel, B., Iwata, T., M Koh, C. et al. Nuclear MYC protein overexpression is an early alteration in human prostate carcinogenesis. Mod Pathol 21, 1156–1167 (2008). https://doi.org/10.1038/modpathol.2008.111
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2008.111
Keywords
This article is cited by
-
Mechanism-centric regulatory network identifies NME2 and MYC programs as markers of Enzalutamide resistance in CRPC
Nature Communications (2024)
-
Taraxasterol suppresses the proliferation and tumor growth of androgen-independent prostate cancer cells through the FGFR2-PI3K/AKT signaling pathway
Scientific Reports (2023)
-
O-GlcNAcylation: an important post-translational modification and a potential therapeutic target for cancer therapy
Molecular Medicine (2022)
-
Prostate-specific oncogene OTUD6A promotes prostatic tumorigenesis via deubiquitinating and stabilizing c-Myc
Cell Death & Differentiation (2022)
-
Cardio-onco-metabolism: metabolic remodelling in cardiovascular disease and cancer
Nature Reviews Cardiology (2022)