Abstract
Despite recent breakthroughs in identifying mucosal-associated invariant T (MAIT) cell antigens (Ags), the precise requirements for in vivo MAIT cell responses to infection remain unclear. Using major histocompatibility complex–related protein 1 (MR1) tetramers, the MAIT cell response was investigated in a model of bacterial lung infection employing riboflavin gene-competent and -deficient bacteria. MAIT cells were rapidly enriched in the lungs of C57BL/6 mice infected with Salmonella Typhimurium, comprising up to 50% of αβ-T cells after 1 week. MAIT cell accumulation was MR1-dependent, required Ag derived from the microbial riboflavin synthesis pathway, and did not occur in response to synthetic Ag, unless accompanied by a Toll-like receptor agonist or by co-infection with riboflavin pathway-deficient S. Typhimurium. The MAIT cell response was associated with their long-term accumulation in the lungs, draining lymph nodes and spleen. Lung MAIT cells from infected mice displayed an activated/memory phenotype, and most expressed the transcription factor retinoic acid–related orphan receptor γt. T-bet expression increased following infection. The majority produced interleukin-17 while smaller subsets produced interferon-γ or tumor necrosis factor, detected directly ex vivo. Thus the activation and expansion of MAIT cells coupled with their pro-inflammatory cytokine production occurred in response to Ags derived from microbial riboflavin synthesis and was augmented by co-stimulatory signals.
Similar content being viewed by others
Introduction
Mucosal-associated invariant T (MAIT) cells are emerging as an important class of T cells with features of both innate and adaptive immunity. The function of MAIT cells is still being elucidated, but a key role is suggested by their conservation across many mammalian species and their high abundance in human blood and mucosal tissues. MAIT cells recognize conserved microbial metabolites generated during riboflavin synthesis, captured by the monomorphic MHC-related protein 1 (MR1).1, 2, 3, 4, 5, 6 MAIT cells express a conserved αβ-T-cell antigen receptor (TCR), consisting of a near-invariant TCRα-chain coupled with a restricted set of β-chains that are relatively conserved in humans and mice.1, 2, 3, 4, 5, 6 In mice, MAIT TCRs use TRAV1-TRAJ33 (Vα19-Jα33) generally assembled with TRBV19 (Vβ6) or TRBV13 (Vβ8).7, 8, 9 In humans, most MAIT cells express TRAV1-2-TRAJ33 (Vα7.2-Jα33), but TRAV1-2-TRAJ12 or TRAV1-2-TRAJ20 are also used.8 These TCRα-chains predominantly form receptors with TRBV20 (Vβ2) or TRBV6 (Vβ13) β-chains,9 although many other TRBV genes are also used at lower frequency.8
To date, characterization of MAIT cells in models of infectious diseases has largely depended on analysis of the CD4, CD8 double negative (DN) T-cell population and PCR detection of the Vα19Jα33 invariant TCR-α chain as there is no Vα19-specific monoclonal antibody (mAb) or the use of Vα19Jα33 TCR transgenic. Cα−/− mice.5, 10, 11, 12, 13 Although Vα19 is characteristic of MAIT cells, it is also expressed by non-MAIT T cells. In addition, most conventionally housed inbred mice, with the exception of the wild-derived inbred CAST/EiJ strain,14 have very low numbers of blood and lymphoid MAIT cells compared with humans,9 and while TCR transgenic models have been used to help circumvent this problem,15, 16 the relationship between MAIT cells generated in TCR transgenic vs. wild-type (WT) mice is unclear.
We recently identified by-products of microbial riboflavin synthesis as a major natural source of antigen (Ag) driving MAIT cell activation in vitro.2, 4, 17 It is unclear whether this pathway is the exclusive source of Ag in vivo, as Vβ chain and complementarity determining region 3β (CDR3β) diversity in MAIT TCRs and differential responses to diverse pathogens might reflect diversity in MAIT Ags.18, 19, 20 Indeed, a number of MAIT cell Ags were identified, including 5-(2-oxopropylideneamino)-6-D-ribitylaminouracil (5-OP-RU), 5-(2-oxoethylideneamino)-6-D-ribitylaminouracil (5-OE-RU), 6,7-dimethyl-8-D-ribityllumazine (RL-6,7-diMe), and 7-hydroxy-6-methyl-8-D-ribityllumazine (RL-6-Me-7-OH), which vary considerably in potency, as well as non-stimulatory MR1 ligands, including 6-formyl-pterin (6-FP), a natural breakdown product of folic acid.2, 4 5-OP-RU and 5-OE-RU, the most potent MAIT cell Ags, are created from the riboflavin precursor 5-amino-6-D-ribitylaminouracil (5-A-RU) by non-enzymatic condensation with small ubiquitous metabolites methylglyoxal and glyoxal, respectively.2 This finding has enabled the production of highly specific MR1-Ag tetramers that stain all MAIT cells in mice and humans.2 Here we have examined the behavior, functionality, and phenotype of mouse MAIT cells, detected using MR1-5-OP-RU tetramers, in a bacterial lung infection model in C57BL/6 mice. Our data reveal that MAIT cell activation and accumulation in vivo was dependent on the presence of a functional riboflavin synthesis pathway in the microbial pathogen and expression of MR1 in the host mice. The cell-surface phenotype and functional properties of MAIT cells indicate a responsiveness of MAIT cells to infection. These studies have substantially advanced our understanding of the functional capacity of MAIT cells in vivo.
Results
MAIT cells accumulate in the lungs of mice following infection with Salmonella Typhimurium
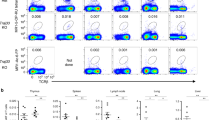
Using Salmonella enterica serovar Typhimurium (S. Typhimurium), we previously showed that the ability to stimulate MAIT cells in vitro depends on the presence of an intact microbial riboflavin pathway, specifically involving genes necessary for the production of riboflavin precursor compounds,4 including 5-A-RU.2 We showed how 5-A-RU recombines with small metabolites to form MAIT-activating ligands, including the most potent identified to date, 5-OP-RU.2 Tetramers of human or mouse MR1 loaded with 5-OP-RU specifically detect MAIT cells in the respective species.2, 8, 21, 22 Mouse MR1-5-OP-RU tetramers were used here to identify MAIT cells (defined as TCRβ+MR1-5-OP-RU tetramer+ cells) in single-cell suspensions derived from the lungs, reproductive tract, liver, spleen, thymus, blood, and mesenteric lymph nodes (LNs) of uninfected C57BL/6 mice. MAIT cells represented ∼2% of αβ-T cells in the reproductive tract, and about 0.5–2% of αβ-T cells in the lung, with lower proportions in blood and other organs (Figure 1a), consistent with recent findings.21 In contrast, we could not detect MAIT cells from C57BL/6.MR1−/− mice (Figure 1a,b), confirming earlier findings that MR1 is vital for MAIT cell development.6, 16
Mucosal-associated invariant T (MAIT) cells respond to Salmonella Typhimurium in vivo. (a) Representative plots and scatter plots showing MAIT cell percentage among TCRβ+ lymphocytes in a range of organs from five uninfected C57BL/6 mice. MAIT cells are defined by positive major histocompatibility complex–related protein 1 (MR1) tetramer and T-cell receptor-β (TCRβ) staining. Numbers represent MAIT cells as a percentage of TCRβ+ lymphocytes. (b) Representative plots showing TCRβ+ lymphocytes (with percentage of MAIT cells) harvested from the lungs of uninfected C57BL/6 or infected (d7 postinfection (p.i.) with 106 S. Typhimurium BRD509) C57BL/6 or C57BL/6 MR1−/− mice. (c) Treatment of mice with anti-MR1 mAb 26.5, but not an isotype control, blocked the accumulation of MAIT cells upon S. Typhimurium infection. Three mice per group were injected with 0.25 mg indicated antibodies or no Ab intraperitoneally 1 day prior to infection and three times p.i. (d1, d3, and d5). 106 BRD509 were inoculated intranasally (i.n.) at d0. At d7 p.i., mice were killed and lung cells were examined for MAIT cell accumulation. Statistics were performed using Student’s t-test (**P<0.01, error bars: s.e.m.). Uninfected MR1−/− mice were used as the negative control. The experiment was performed twice with similar results. (d) Dose (of S. Typhimurium BRD509) response of MAIT cells as a percentage of TCRβ+ cells at d7 p.i. Five mice per group were examined (mean±s.e.m.). The experiment was performed three times with similar results. (e) Absolute numbers of MAIT cells (circle), conventional non-MAIT cells (triangle), and total TCRβ+ cells (square) recovered from the lungs were expressed over a time course following i.n. infection with 106 S. Typhimurium BRD509. Five mice per group were examined (mean±s.e.m.). The experiment was performed twice with similar results. CFU, colony-forming units; mLN, mesenteric lymph node.
We next examined the impact of lung infection on MAIT cells in a model involving inoculation of mice with live-attenuated vaccine strain S. Typhimurium BRD509.23, 24 Although S. Typhimurium is not a natural pathogen in the lung, it is known to activate MAIT cells in vitro and genetic manipulation of this bacterium now allows us to probe the Ag requirements for MAIT cell activation in vivo. Following intranasal (i.n.) infection with S. Typhimurium BRD509, there was a substantial enrichment of MAIT cells in the lungs of C57BL/6 mice such that they comprised 25–50% of all infiltrating lung αβ-T cells after 1 week (Figure 1b,c, see Supplementary Figure S1 online). MAIT cell enrichment was absent in C57BL/6.MR1−/− mice (Figure 1b,c) and was blocked by treatment with MR1-specific mAbs, 26.5 or 8F2.F925, 26 (Figure 1c, see Supplementary Figure S1). Thus the MAIT cell accumulation and enrichment in this model was MR1 dependent. The accumulation of lung MAIT cells was also dependent on the size of the bacterial inoculum (Figure 1d) and peaked at day 7 postinfection (d7 p.i.) (Figure 1e).
In vivo enrichment of MAIT cells p.i. requires Ag derived from a bacterial riboflavin pathway
To confirm the requirement for microbial riboflavin metabolites in driving the in vivo MAIT cell response, we constructed a mutant strain of BRD509 in which ribD and ribH genes, which are co-located in the Salmonella genome, were deleted: BRD509ΔribDH. Although deficiency of the ribH gene does not impact on bacterial activation of MAIT cells,2 the ribD gene is essential for production of the 5-A-RU precursor to MAIT cell Ags 5-OP-RU and 5-OE-RU.2 The mutant BRD509ΔribDH bacteria were first tested in vitro, and bacterial supernatants were found to be deficient in their ability to stimulate reporter cells expressing a MAIT TCR (Jurkat.MAIT)4 in an MR1-dependent activation assay (Figure 2a). By comparison, BRD509 supernatant (WT) caused upregulation of CD69 on Jurkat.MAIT reporter cells (Figure 2a). Supernatant from the BRD509ΔribDH bacteria reconstituted with ribD and ribH genes (BRD509ΔribDH+ribDH) activated Jurkat. MAIT cells to WT levels (Figure 2a), confirming previous in vitro results obtained with mutants of another Salmonella strain.2
Mucosal-associated invariant T (MAIT) cells response depends on both specific riboflavin synthesis–derived antigen and co-stimulation. (a) In vitro activation. Jurkat.MAIT cells were incubated for 6 h with filtered culture supernatant from BRD509 or BRD509ΔribDH mutant (ΔribDH) or reconstituted mutant BRD509ΔribDH+RibDH S. Typhimurium in the presence of C1R.MR1 cells. Activation was detected by staining with anti-CD69. Data show mean fluorescent intensity (MFI) of gated Jurkat.MAIT cells, with s.e.m. as error bars. The experiment was performed more than three times with similar results. (b, c) In vivo accumulation of MAIT cells by BRD509 and BRD509ΔribDH S. Typhimurium. (b) Representative plots and (c) MAIT cells as a percentage of αβ-T cells from the lungs of mice immunized with BRD509 (106) or BRD509ΔribDH (107) Salmonella (intranasally (i.n.) once) in combination with 5-(2-oxopropylideneamino)-6-D-ribitylaminouracil (5-OP-RU) or 6-formyl-pterin (6-FP) (50 μl of 1.52 μM i.n.) three times at d1, d3, and d5 postinfection (p.i.). Day 7 p.i. data are shown. Three mice per group were examined (mean±s.e.m., Student’s t-test, **P<0.05). The experiment was performed twice with similar results. (d) MAIT cells as a percentage of αβ-T cells in the lungs of mice immunized i.n. with S. Typhimurium BRD509 (106) or combinations of S-[2,3-bis(palmitoyloxy)propyl] cysteine (Pam2Cys; 20 nmol, i.n.), CpG (20 μg per mouse, i.n.), or polyinosinic:polycytidylic acid (poly I:C; 50 μg per mouse, i.n.) and/or 5-OP-RU or 6-FP (50 μl containing 76 pmol i.n.). 5-OP-RU or 6-FP was administered twice i.n. 1 and 3 days after Pam2Cys, CpG, or poly I:C administration. Day 7 postinoculation data shown for six individual mice with mean±s.e.m. The experiment was performed twice with similar results. (e) Mice were administered i.n. with combinations of Pam2Cys (20 nmol/5 μl per mouse) and/or MAIT cell ligands (5-OP-RU, 6-FP, 45 μl of 15.2 (hi), 3 (med) or 0.76 (low) μM, or 45 μl of PBS, per mouse). Data represent MFI of CD69 expression on MAIT and non-MAIT T cells for individual mice and mean±s.e.m., 2 h after inoculation. (f) Mice were inoculated i.n. with the indicated MR1 ligands and/or co-stimulator as shown in the schematic. Percentages of MAIT cells of αβ-T cells are shown, from two mice (control groups): uninfected, Pam2cys (day 0) plus 5-OP-RU/6FP (days 0, 1, 2, and 4), or three mice each: six times of 5-OP-RU/6FP alone (days 0, 1, 2, 3, 4, and 5). WT, wild type.
In contrast to S. Typhimurium BRD509, infection with BRD509ΔribDH bacteria did not evoke MAIT cell accumulation beyond the resident MAIT cell numbers in the uninfected lungs (Figure 2b,c). This lack of MAIT cell accumulation was unlikely the result of the reduced growth rate of BRD509ΔribDH as a 10-fold higher dose of BRD509ΔribDH bacteria was used, resulting in equivalent bacterial counts at day 7 (see Supplementary Figure S2), and non-MAIT αβ-T cells still accumulated (Figure 2b). Moreover, the non-responsiveness of MAIT cells to infection with BRD509ΔribDH Salmonella could be rescued if the bacteria were co-administered i.n. with synthetic MAIT cell Ag 5-OP-RU delivered i.n. (Figure 2b,c) or intravenously (see Supplementary Figure S3). In contrast, co-administration of the BRD509 strain and the MR1-ligand 6-FP,4 which does not activate MAIT cells, did not cause MAIT cell accumulation in the lungs (Figure 2c, see Supplementary Figure S3), confirming the specificity of the response. Of note, 5-OP-RU alone caused no MAIT cell accumulation at any dose or time point tested, when delivered either i.n. or intravenously, suggesting that synthetic Ag alone is insufficient to induce the MAIT cell accumulation (Figure 2f, see Supplementary Figure S3). This could reflect a requirement for co-stimulation provided by the BRD509ΔribDH bacteria or the unstable nature of free 5-OP-RU failing to produce sustained MAIT cell stimulation. To examine whether a co-stimulus could be furnished through TLRs, we inoculated mice with 5-OP-RU and TLR agonists S-[2,3-bis(palmitoyloxy)propyl] cysteine (Pam2Cys), CpG oligonucleotides, or polyinosinic:polycytidylic acid (poly I:C). Pam2Cys is a synthetic mimic of the lipid component of TLR2 and TLR6 ligand macrophage-activating lipopeptide-2 (MALP-2) and has been shown to be an agonist for TLR2.27, 28 Poly I:C is a TLR3 agonist similar to double-stranded RNA.29 CpG oligonucleotides stimulate through TLR9.30 Challenge with each TLR agonist by itself, or with the non-activating MR1 ligand 6-FP, led to a lesser (2–3-fold) enrichment for MAIT cells in the lungs, suggesting that co-stimuli alone can exert some influence on MAIT cells (Figure 2d). In addition, although i.n. challenge with Ag alone did not induce accumulation of lung MAIT cells, it did induce rapid upregulation of CD69 on MAIT cells in vivo as early as 2 h following inoculation (Figure 2e). However, the i.n. co-administration of synthetic Ag 5-OP-RU and Pam2Cys, poly I:C, or CpG led to a 15–25-fold enrichment of MAIT cells in the lung in the absence of any bacterial challenge (Figure 2d,f). At least for Pam2Cys plus 5-OP-RU, this response was sustained over time (not shown) and was largely blocked by administration of anti-MR1 mAb (see Supplementary Figure S1b). Thus the failure of i.n. 5-OP-RU alone to drive MAIT cell enrichment was not necessarily the result of the lability of the Ag, suggesting that MAIT cell accumulation and enrichment require stronger signals than those needed for activation. Indeed, repeated (6 ×) i.n. administration of 5-OP-RU alone over a 7-day period was still an insufficient stimulus to induce MAIT cell accumulation, despite sustained Ag exposure (Figure 2f). Taken together, these findings demonstrate that rib-defective (and thus MAIT cell Ag deficient) bacteria and TLR agonists are independently capable of providing a co-stimulus that helps to drive MAIT cell accumulation and enrichment in response to 5-OP-RU Ag in vivo (Figure 2c,d).
Infection induces interleukin (IL)-17 production, an increase in T-bet expression, and the long-lived retention of phenotypically effector/memory MAIT cells in situ
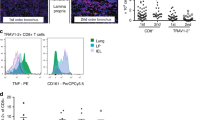
Upon activation in vitro, MAIT cells produce interferon-γ (IFNγ), tumor necrosis factor (TNF), and IL-17.4, 8, 13, 31 This pattern of cytokine expression was mirrored following i.n. inoculation with S. Typhimurium (Figure 3, see Supplementary Figures S5 and S6). Most striking was the production of IL-17, which was detected in approximately 30% of lung MAIT cells directly ex vivo at d7 without further stimulation (Figure 3). Interestingly, MAIT cells still appeared ‘primed’ to rapidly secrete IL-17 long-term p.i. (Figure 3), suggesting that MAIT cells acquire rapid responsiveness as they differentiate into their memory phase. In contrast, a higher proportion of non-MAIT αβ-T cells than MAIT cells produced IFNγ and TNF 7 days p.i. (Figure 3, see Supplementary Figure S4).
Cytokine profiling and phenotyping of mucosal-associated invariant T (MAIT) cells upon infection with S. Typhimurium BRD509. Intracellular cytokine staining of MAIT and non-MAIT αβ-T cells at d7 postinfection detected directly ex vivo or following stimulation with phorbol myristate acetate and ionomycin. (a) Representative plots and (b) scatter plots showing individual mice and mean±s.e.m. from three mice (pooled for uninfected). The experiment was performed twice with similar results. MAIT and non-MAIT T cells are defined as live, CD19−, TCRβ+, Tet+ and live, CD19−, TCRβ+, Tet− cells, respectively. IFN, interferon; IL, interleukin; PMA, phorbol myristate acetate; TCR, T-cell receptor; TNF, tumor necrosis factor.
The transcriptional factor promyelocytic leukemia zinc finger (PLZF) is a master regulator of innate-like T-cell development, including natural killer T cells and some γδT cells.32 Both human and mouse MAIT cells express PLZF.16, 21 Notably, we found that PLZF was expressed by nearly all lung MAIT cells both before and after infection, along with the orphan nuclear receptor, retinoic acid–related orphan receptor γt (RORγt), which has a key role in differentiation of T helper type 17 (Th17) cells33 and is expressed by human and mouse MAIT cells21, 31 (Figure 4a, see Supplementary Figure S5). In addition, we examined the expression of the transcription factor T-bet, a key regulator of Th1 type immunity.34 T-bet is reported to suppress IL-17 expression and promote Th1 development, whereas loss of T-bet expression can induce commitment to Th2 and loss of IFNγ expression.35, 36, 37 The expression of RORγt declined slightly in MAIT cells as T-bet expression levels increased from d7 to d142 p.i. Interestingly, the majority of cells long-term p.i. co-expressed the two transcription factors (Figure 4b, see Supplementary Figure S5). The production of IL-17 by MAIT cells d7 p.i. either directly ex vivo or following phorbol myristate acetate (PMA) stimulation, was observed in the presence of either T-bet or RORγt (or both) (Figure 4c, see Supplementary Figure S6). In contrast, at the same time point, a much smaller proportion of MAIT cells produced IFNγ after PMA stimulation and most of these expressed T-bet. This pattern was also true for IFNγ production by non-MAIT cells (Figure 4c). IL-17 and IFNγ appeared to be independently expressed by MAIT cells, as shown by co-staining (see Supplementary Figure S7).
Mucosal-associated invariant T (MAIT) cell expression of transcription factors promyelocytic leukemia zinc finger (PLZF), retinoic acid–related orphan receptor γt (RORγt), and T-bet. (a) Expression of transcription factors on MAIT cells in uninfected or infected mice at the indicated time points. (b) Co-staining of cells with RORγt and T-bet shown as representative plots and individual mice from one experiment. DP; T-bet, RORγt double positive, DN; T-bet, RORγt double negative. (c) Cytokine profile as in Figure 3 but showing RORγt and T-bet expression. The experiment was performed three times with similar results. IFN, interferon; IL, interleukin; PMA, phorbol myristate acetate.
Even before infection, CD44, a cell adhesion molecule highly expressed on memory T cells, was expressed by most MAIT cells. In contrast, a smaller proportion of non-MAIT αβ-T cells expressed CD44 preinfection and p.i. (Figure 5). CD62L (L-selectin) is highly expressed on naive T cells and is used to further distinguish central memory (CD62L+) from effector memory (CD62L−). The regulation of CD62L has a pivotal role in controlling the traffic of conventional T lymphocytes through peripheral LNs, with CD62L shed from the cell membrane following activation.38 Not surprisingly, CD62L remained low on lung MAIT cells at all three phases: homeostatic MAIT cells in the absence of infection, activated MAIT cells (during infection), and Ag experienced MAIT cells (>d30 p.i.) (Figure 5). These observations are consistent with the effector/memory phenotype previously described for human MAIT cells.31 However, the proportion of MAIT cells expressing the activation marker CD69 increased from ∼20% preinfection to 90% p.i. (Figure 5b), consistent with our other results showing that further activation of MAIT cells occurred following infection. MAIT cells from the lungs of uninfected mice also showed CD25 expression (not shown).
Mucosal-associated invariant T (MAIT) cells maintain an effector memory phenotype following infection. Analysis of phenotypic and activation marker expression on MAIT cells isolated from the lungs of mice infected intranasally with 106 S. Typhimurium BRD509 at the indicated time points. (a) Representative plots. (b) Percentages among MAIT and non-MAIT cells expressing markers for individual mice. The experiment was performed twice with similar results. p.i., postinfection.
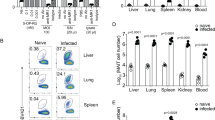
One possibility is that the enrichment of MAIT cells in the lungs is due to recruitment from blood and other sites. To evaluate this possibility, we examined MAIT cell enrichment in lungs and draining LNs vs. possible loss in other organs over 7 weeks following infection with BRD509 (Figure 6a,b).1,12 The enrichment of MAIT cells as a proportion of all αβ-T cells was most evident in the lungs of infected mice (d7, 35–50% of αβ-T cells) but was only evident in the draining LNs and spleen at week 7 (Figure 6a,b), suggesting redistribution to these sites during the resolution phase of the infection (Figure 6c). No MAIT cell loss was observed in the spleen or draining LNs, where the percentage of MAIT cells remained stable throughout the infection course and increased slightly during the late course (Figure 6a,b).
Mucosal-associated invariant T (MAIT) cells accumulate in the lungs, spleen and draining lymph nodes (dLNs) in response to intranasal (i.n.) infection. (a) Kinetics of MAIT cell accumulation in the lungs, mediastinal draining LNs, and spleen at the indicated times postinfection (p.i.) from C57BL/6 inoculated with BRD509 i.n. The experiment was performed three times with similar results. Data represent mean±s.e.m. from five mice per group. Data from C57BL/6.MR1−/− mice are not shown. (b) MAIT cell percentage in various organs 7 weeks after i.n. S. Typhimurium BRD509 inoculation. Same data as in a, shown in bar graphs for ease of comparison. (c) Bacterial load (colony-forming units (CFU)) recovered from the lungs of C57BL/6 or C57BL/6.MR1−/− mice inoculated i.n. with 106 S. Typhimurium BRD509. Individual mice (five per group) and mean are shown. The experiment was performed three times with similar results.
Together, our data show a rapid MAIT cell response to bacterial lung infection, which was dependent on MR1 presentation of riboflavin biosynthesis-derived ligands. MAIT cell accumulation and proliferation, but not CD69 upregulation, was dependent on second signals, which could be furnished through TLR ligation or by bacteria. Cell surface markers and cytokine production were consistent with an effector/memory phenotype.
Discussion
We have developed a model bacterial infection in mice to evaluate the importance of the riboflavin synthesis pathway in driving MAIT cell responses in vivo and to define the phenotypic and functional characteristics of the responding MAIT cells. The generation of highly specific MR1-Ag tetramers2, 8 has allowed the precise Ag-dependent definition of MAIT cells, which are much less abundant in WT mice than humans,9 overcoming potential confusion engendered by using surrogate markers of MAIT cells or transgenic mice. Using MR1-Ag tetramers, we show that MAIT cells are readily detectable, albeit in low numbers, in WT C57BL/6 mice, being most abundant in the reproductive tract, lung, and liver. We exploited the capacity of S. Typhimurium, which we previously showed to potently activate MAIT cells in vitro2, 4 as a model to analyze MAIT cell responses and activation requirements in vivo, with the respiratory tract providing an accessible organ to assess MAIT activation. S. Typhimurium is a gastrointestinal pathogen of humans and a systemic pathogen of mice. Clearance of S. Typhimurium in mice is achieved through a number of redundant cellular mechanisms, including CD4+, and to a lesser extent CD8+, T cells.23 In our respiratory model, lung clearance was independent of the presence of MAIT cells, suggesting their redundancy as protective T cells in this model too. It would be interesting to establish whether the type or kinetics of the non-MAIT immune response differed in the absence of MAIT cells. Nevertheless, i.n. infection of mice led to a substantial increase in MAIT cells in the lung to up to 50% of all pulmonary αβ-T cells (a ∼200-fold increase in absolute numbers vs. ∼5-fold for non-MAIT cells). This enrichment peaked at d7 and was MR1 dependent, as shown genetically (in MR1−/− mice) and by anti-MR1 mAb blocking, consistent with previous in vitro data.1, 2, 3, 4, 5, 6 The accumulation of MAIT cells was directly related to the size of the bacterial inoculum. Interestingly, expanded MAIT cells were long lived in the lung and other tissues, persisting in higher numbers than found in uninfected mice long after S. Typhimurium was cleared to undetectable levels (∼10 weeks). It is interesting to speculate that a history of infection in humans could explain the higher proportion of MAIT cells compared with specific pathogen-free–housed mice, and it will be exciting to discover the role of MAIT cells in immune protection against a range of microbial respiratory pathogens.
Genetic manipulation of S. Typhimurium allowed us to demonstrate that responsiveness of MAIT cells in vivo required an intact riboflavin pathway and hence was abolished upon disruption of the key ribD gene responsible for generating the precursor 5-A-RU, which generates the dominant MAIT cell ligand, 5-OP-RU. Notably, the BRD509ΔribDH mutant could still induce weak, transient upregulation of MAIT cell CD69 in vitro but did not induce accumulation or enrichment of MAIT cells in vivo. It remains unclear whether there are natural MAIT cell Ags derived from pathways other than riboflavin synthesis. The variability of MAIT TCRs from the semi-invariant MAIT TCR combinations7, 8, 18 suggests that these receptors may be selected by alternate ligands and MAIT cells have an apparent capacity o discriminate between pathogen-derived ligands in a clonotype-dependent manner.20 On the other hand, the semi-invariant α and restricted β chain usage, the importance of the conserved Tyr 95α residue across the TRAJ33, TRAJ20 and TRAJ12 gene segments tested to date, and the stereotypic mode of binding of the MAIT TCR to all known ligands and analogues studied1, 2, 4, 17, 18 suggest that a limited ligand family is recognized by MAIT cells. Accordingly, the abrogation of MAIT cell accumulation and activation following mutagenesis of the riboflavin synthesis pathway implies that this pathway furnishes the dominant, and likely exclusive, family of natural Ags recognized by MAIT cells at least for S. Typhimurium.
Previous studies have shown that MAIT cells can be directly activated via non-TCR signals, including IL-12 and IL-18,39, 40 and control of Francisella tularensis intracellular growth required the IL-12 40-kDa subunit, implying a need for additional signals from infected Ag-presenting cells (APCs).13 These observations are consistent with our finding that a co-stimulus was required to induce MAIT cell accumulation in the lungs following i.n. administration of synthetic MAIT cell Ag, 5-OP-RU, even when delivered repeatedly over 7 days, and despite inducing MAIT cell upregulation of CD69. In contrast, i.n. administration of 5-OP-RU plus the TLR agonists Pam2Cys (TLR2/6), poly I:C (TLR3), or CpG (TR9) induced enrichment of lung MAIT cells to the approximate level achieved by S. Typhimurium infection. Importantly, addition of 5-OP-RU during i.n. challenge with bacteria lacking the riboflavin synthesis pathway (BRD509ΔribDH mutant) restored their capacity to induce MAIT cell accumulation in the lungs, indicating that co-stimulatory capacity is inherent in bacterial infection and has a role in the expansion and responsiveness of MAIT cells.
The phenotype of MAIT cells that expand in response to infection with S. Typhimurium is consistent with previous studies of in vitro-stimulated MAIT cells, in that they express activation markers even before infection and produce pro-inflammatory cytokines IL-17, IFNγ, and TNF upon activation by microbes. This is also consistent with a study of pulmonary infection of mice with F. tularensis,13 although, of note, this study defined MAIT cells as DN thus not addressing a potential role for CD8+ or CD4+ MAIT cells in response to infection. The expression of CD69 on 20% of MAIT cells, compared with ∼4% of non-MAIT cells, in the steady state is intriguing and could suggest that there may be stimulation of MAIT cells through the presence of commensal microbes providing Ag at mucosal sites. As well as being described as an early-activation marker, CD69 expression has been previously shown on human tissue resident memory T cells41 and to have a role in tissue retention of T cells.42 However, further stimulation of MAIT cells, and CD69 upregulation, could be induced by infection or provision of Ag plus co-stimulatory signals. The expansion of MAIT cells following infection may be a consequence of the lower number of MAIT cells in mice vs. humans. Interestingly, once expanded, the MAIT cells are long lived in primary tissues, preserving their activation and homing phenotype and maintaining expression of their transcriptional signature PLZF and RORγt but with gradual increase in the level of T-bet, known to co-ordinate developmental and effector programs some of which are conserved across adaptive and innate T cells.
Our study presents a picture of MAIT cell responsiveness to pathogens that bridges attributes of innate and adaptive immunity but that is dependent on specific Ag expressed by the infecting pathogen.43 It will be fascinating to unravel whether these cells have evolved in response to a range of pathogens with shared Ag(s) or whether they have a more focused role, perhaps involving a single pathogen family possibly at a key site.
Methods
In vitro activation assay. Jurkat cells expressing a MAIT TCR comprising the TRAV1-2-TRAJ33 α-chain and TRBV6-1 β-chain (Jurkat.MAIT) were co-incubated with filtered bacterial culture supernatant and C1R APCs expressing MR1 (CIR.MR1) for 16 h and stained with anti-CD3-PE and anti-CD69-APC Abs before flow cytometric analysis. Activation of Jurkat.MAIT cells was measured by an increase in surface CD69 expression.
Compounds and immunogens. 5-OP-RU was prepared as described previously.2 6-FP was purchased from Schircks Laboratories (Jona, Switzerland). TLR2/6 agonist Pam2Cys44 were chemically synthesized and functionally verified. CpG1688 (Sequence: T*C*C*A*T*G*A*C*G*T*T*C*C*T*G*A*T*G*C*T (*phosphorothioate linkage) nonmethylated cytosine-guanosine oligonucleotides was purchased from Geneworks (Thebarton, Australia). Poly (I:C) (HMW VacciGrade) was purchased from InvivoGen (San Diego, CA).
Bacterial strains and mutants. S. Typhimurium BRD509, (previously described45, 46) harbors deletions in aroA and aroD, resulting in limitation of replication of bacteria24, 45 and has an intact riboflavin synthesis pathway. The BRD509ΔribDH mutant, lacking a gene segment containing ribD and ribH, which encode key enzymes in the riboflavin synthesis pathway, was constructed by lamda red-recombinase-mediated allelic replacement followed by transduction using phage P22 as previously described.24 BRD509ΔribDH cultures were supplemented with 20 μg ml−1 riboflavin.
BRD509ΔribDH complemented by transformation of ribDH genes resulted in strain BRD509ΔribDH+ribDH. Mutation and reconstitution were verified by lack of growth or growth on Luria Agar and by PCR. Bacteria were cultured at 37 °C statically, in Luria Bertani broth with appropriate antibiotics for 16–18 h to log-phase (OD600 0.6–0.9). Supernatant was collected by centrifugation, filtered (0.22 μm) and stored at −80 oC until use. For the infecting inoculum, bacteria were re-inoculated in prewarmed medium for a further 2–4 h static culture (OD600 0.4–0.6). With the estimation that 1 OD600=5 × 108 ml−1, sufficient bacteria were washed and diluted in phosphate-buffered saline (PBS) for i.n. delivery to mice. A sample of inoculum was plated onto Luria Agar with appropriate antibiotics for verification of bacterial concentration by counting colony-forming units.
Mice and inoculations. Mice were bred and housed in the Biological Research Facility of the Peter Doherty Institute (Melbourne, Victoria, Australia). MR1−/− mice were generated by breeding Vα19iCα−/−MR1−/− mice15 (from Susan Gilfillan, Washington University, St Louis School of Medicine, St Louis, MO) with C57BL/6 mice and inter-crossing of F1 mice. The genotype was determined by tail DNA PCR at the MR1 locus and by the presence of intact αβ-T cells in blood using Ab against TCRβ. The absence of MAIT cells was verified by flow cytometry in the retired founder breeder mice. Male mice aged 6–12 weeks were used in experiments, after approval by the University of Melbourne Animal Ethics Committee.
I.n. inoculation with S. Typhimurium (BRD509: 106 or BRD509ΔribDH: 107 unless otherwise stated) or Ags (76 pmol 5-OP-RU or 6-FP alone or plus 20 nmol Pam2Cys, 20 μg CpG, or 50 μg poly I:C) in 50 μl per nares was performed on anesthetized mice. For blocking experiments, mice were given 250 μg anti-MR1 (26.5 or 8F2.F9)25, 26 or isotype control mAbs in 200 μl PBS, once (i.v or intraperitoneally) 1 day prior to inoculation and three times (d1, d3, d5) postinoculation. Mice were killed by administration of CO2 and organs: lung (following heart perfusion with 10 ml cold RPMI), liver (following portal vein perfusion with 10 ml RPMI), mediastinal LNs, blood, spleen, thymus, vagina, and uterus were taken.
Preparation of organs and isolation of cells. To prepare single-cell suspensions, lungs, uterus, and vagina (pooled from three mice) were finely chopped with a scalpel blade and treated with 3 mg ml−1 collagenase III (Worthington, Lakewood, NJ), 5 μg ml−1 DNAse, and 2% fetal calf serum in RPMI for 90 min at 37 °C with gentle shaking. Cells were then filtered (70 μm) and washed with PBS/2% fetal calf serum. Liver lymphocytes were prepared by pushing through metal mesh, followed by Percoll (33.75%) density gradient centrifugation (693 g, 12 min, room temperature). LNs, spleen, and thymus were prepared by pushing the tissues through 70-μm cell strainers. Red blood cells were lysed from lung, liver, and spleen preparations with hypotonic buffer TAC (Tris-based amino chloride) for 5 min at 37 °C. Approximately 1.5 × 106 cells were filtered (40 μm) and used for flow cytometric analysis. Blood cells were prepared by centrifugation and serum removal. Red blood cell lysis was by Lysing buffer (BD Biosciences, North Ryde, NSW, Australia) after flow cytometric staining.
Determination of bacterial counts in infected lungs. Bacterial colonization was determined by counting colony-forming units obtained from plating homogenized organs from infected mice (⩾5 per group) on Luria agar containing appropriate antibiotics and supplements.
Generation of soluble MR1 tetramers. Murine MR1 and β2-Microglobulin genes were expressed in Escherichia coli inclusion bodies, refolded, and purified as described previously.17 MR1-5-OP-RU and MR1-6-FP tetramers were generated as described previously.2
Abs and flow cytometry. Ab against CD19 (1D3, PerCP-Cy 5.5), CD3 (UCHT1, PE or 145-2C11, PE-Cy7), CD4 (GK1.5, APC-Cy7), CD45.2 (104, FITC), CD69 (FN50, APC), IFNγ (XMG1.2, PE-Cy7), TCRβ (H57-597, APC or FITC), CD44 (IM7, PE), TNF (MP6-XT22, PE), and IL-17A (TC11-18H10, AlexaFluor700) were purchased from BD (Franklin Lakes, NJ). Ab against MHC II (M5, AlexaFluor700), CD69 (H1.2F3, PE), CD25 (PC61.5, APC), PLZF (Mags.21F7, PE), RORγt (B2D, APC), and T-bet (4B10, PE-Cy7) were purchased from eBioscience (San Diego, CA). Ab against CD62L (Mel-14, FITC) was purchased from Biolegend (San Diego, CA). Blocking Ab (26.5, 8F2.F9) and isotype controls (3E12, 8A5) were prepared in house. To block non-specific staining, cells were incubated with MR1-6FP tetramer and anti-Fc receptor (2.4G2) for 15 min at room temperature and then incubated at room temperature with Ab/tetramer cocktails in PBS/2% fetal calf serum. 7-aminoactinomycin D (5 μl per sample) was added for the last 10 min. Cells were fixed with 1% paraformaldehyde prior to analysis on LSRII or LSR Fortessa (BD Biosciences) flow cytometers. Data analysis was performed with the FlowJo software (Ashland, OR). For intracellular cytokine staining, Golgi plug (BD Biosciences) was used during all processing steps. Cells stimulated with PMA/ionomycin (20 ng ml−1, 1 μg ml−1, respectively) for 3 h at 37 °C were included as positive controls. Surface staining was performed at 37 °C, and cells were stained for intracellular cytokines using the BD Fixation/Permeabilization Kit (BD, Franklin Lakes, NJ) or transcription factors using the transcription buffer staining set (eBioscience) according to the manufacturers’ instructions.
Statistical analysis. Statistical tests were performed using the Prism GraphPad software (version 6, La Jolla, CA). Comparisons between groups were performed using Student’s t-test, unless otherwise stated.
References
Birkinshaw, R.W., Kjer-Nielsen, L., Eckle, S.B., McCluskey, J. & Rossjohn, J. MAITs, MR1 and vitamin B metabolites. Curr. Opin. Immunol. 26, 7–13 (2014).
Corbett, A.J. et al. T-cell activation by transitory neo-antigens derived from distinct microbial pathways. Nature 509, 361–365 (2014).
Gapin, L. Where do MAIT cells fit in the family of unconventional T cells? PLoS Biol. 7, e70 (2009).
Kjer-Nielsen, L. et al. MR1 presents microbial vitamin B metabolites to MAIT cells. Nature 491, 717–723 (2012).
Le Bourhis, L. et al. Mucosal-associated invariant T cells: unconventional development and function. Trends Immunol. 32, 212–218 (2011).
Treiner, E. et al. Selection of evolutionarily conserved mucosal-associated invariant T cells by MR1. Nature 422, 164–169 (2003).
Lepore, M. et al. Parallel T-cell cloning and deep sequencing of human MAIT cells reveal stable oligoclonal TCRbeta repertoire. Nat. Commun. 5, 3866 (2014).
Reantragoon, R. et al. Antigen-loaded MR1 tetramers define T cell receptor heterogeneity in mucosal-associated invariant T cells. J. Exp. Med. 210, 2305–2320 (2013).
Tilloy, F. et al. An invariant T cell receptor alpha chain defines a novel TAP-independent major histocompatibility complex class Ib-restricted alpha/beta T cell subpopulation in mammals. J. Exp. Med. 189, 1907–1921 (1999).
Chiba, A. et al. Mucosal-associated invariant T cells promote inflammation and exacerbate disease in murine models of arthritis. Arthritis Rheum. 64, 153–161 (2012).
Gold, M.C. et al. Human mucosal associated invariant T cells detect bacterially infected cells. PLoS Biol. 8, e1000407 (2010).
Le Bourhis, L. et al. Antimicrobial activity of mucosal-associated invariant T cells. Nat. Immunol. 11, 701–708 (2010).
Meierovics, A., Yankelevich, W.J. & Cowley, S.C. MAIT cells are critical for optimal mucosal immune responses during in vivo pulmonary bacterial infection. Proc. Natl. Acad. Sci. USA 110, E3119–E3128 (2013).
Cui, Y. et al. Mucosal-associated invariant T cell-rich congenic mouse strain allows functional evaluation. J. Clin. Invest. 125, 4171–4185 (2015).
Kawachi, I., Maldonado, J., Strader, C. & Gilfillan, S. MR1-restricted V alpha 19i mucosal-associated invariant T cells are innate T cells in the gut lamina propria that provide a rapid and diverse cytokine response. J. Immunol. 176, 1618–1627 (2006).
Martin, E. et al. Stepwise development of MAIT cells in mouse and human. PLoS Biol. 7, e54 (2009).
Patel, O. et al. Recognition of vitamin B metabolites by mucosal-associated invariant T cells. Nat. Commun. 4, 2142 (2013).
Eckle, S.B. et al. A molecular basis underpinning the T cell receptor heterogeneity of mucosal-associated invariant T cells. J. Exp. Med. 211, 1585–1600 (2014).
Gherardin, N.A. et al. Diversity of T cells restricted by the MHC class I-related molecule MR1 facilitates differential antigen recognition. Immunity 44, 32–45 (2016).
Gold, M.C. et al. MR1-restricted MAIT cells display ligand discrimination and pathogen selectivity through distinct T cell receptor usage. J. Exp. Med. 211, 1601–1610 (2014).
Rahimpour, A. et al. Identification of phenotypically and functionally heterogeneous mouse mucosal associated invariant T cells using MR1 tetramers. J. Exp. Med. 20, 1095–1108 (2015).
Sakala, I.G. et al. Functional heterogeneity and antimycobacterial effects of mouse mucosal-associated invariant T cells specific for riboflavin metabolites. J. Immunol. 195, 587–601 (2015).
Kupz, A., Bedoui, S. & Strugnell, R.A. Cellular requirements for systemic control of Salmonella enterica serovar Typhimurium infections in mice. Infect. Immun. 82, 4997–5004 (2014).
Strugnell, R. et al. Characterization of a Salmonella typhimurium aro vaccine strain expressing the P.69 antigen of Bordetella pertussis. Infect. Immun. 60, 3994–4002 (1992).
Chua, W.J. et al. Endogenous MHC-related protein 1 is transiently expressed on the plasma membrane in a conformation that activates mucosal-associated invariant T cells. J. Immunol. 186, 4744–4750 (2011).
Huang, S. et al. Evidence for MR1 antigen presentation to mucosal-associated invariant T cells. J. Biol. Chem. 280, 21183–21193 (2005).
Jackson, D.C. et al. A totally synthetic vaccine of generic structure that targets Toll-like receptor 2 on dendritic cells and promotes antibody or cytotoxic T cell responses. Proc. Natl. Acad. Sci. USA 101, 15440–15445 (2004).
Lau, Y.F. et al. Lipid-containing mimetics of natural triggers of innate immunity as CTL-inducing influenza vaccines. Int. Immunol. 18, 1801–1813 (2006).
Alexopoulou, L., Holt, A.C., Medzhitov, R. & Flavell, R.A. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature 413, 732–738 (2001).
Takeshita, F. et al. Cutting edge: role of Toll-like receptor 9 in CpG DNA-induced activation of human cells. J. Immunol. 167, 3555–3558 (2001).
Dusseaux, M. et al. Human MAIT cells are xenobiotic-resistant, tissue-targeted, CD161hi IL-17-secreting T cells. Blood 117, 1250–1259 (2011).
Doulatov, S. et al. PLZF is a regulator of homeostatic and cytokine-induced myeloid development. Genes Dev. 23, 2076–2087 (2009).
Ivanov, I.I. et al. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell 126, 1121–1133 (2006).
Szabo, G., Katz, E. & Bonkovsky, H.L. Management of recurrent hepatitis C after liver transplantation: a concise review. Am. J. Gastroenterol. 95, 2164–2170 (2000).
Lazarevic, V. et al. T-bet represses T(H)17 differentiation by preventing Runx1-mediated activation of the gene encoding RORgammat. Nat. Immunol. 12, 96–104 (2011).
Mathur, A.N. et al. T-bet is a critical determinant in the instability of the IL-17-secreting T-helper phenotype. Blood 108, 1595–1601 (2006).
Villarino, A.V., Gallo, E. & Abbas, A.K. STAT1-activating cytokines limit Th17 responses through both T-bet-dependent and -independent mechanisms. J. Immunol. 185, 6461–6471 (2010).
Yang, S., Liu, F., Wang, Q.J., Rosenberg, S.A. & Morgan, R.A. The shedding of CD62L (L-selectin) regulates the acquisition of lytic activity in human tumor reactive T lymphocytes. PLoS One 6, e22560 (2011).
Ussher, J.E. et al. CD161(++) CD8(+) T cells, including the MAIT cell subset, are specifically activated by IL-12+IL-18 in a TCR-independent manner. Eur. J. Immunol. 44, 195–203 (2014).
Kurioka, A. et al. MAIT cells are licensed through granzyme exchange to kill bacterially sensitized targets. Mucosal Immunol. 8, 429–440 (2015).
Sathaliyawala, T. et al. Distribution and compartmentalization of human circulating and tissue-resident memory T cell subsets. Immunity 38, 187–197 (2013).
Mackay, L.K. et al. Cutting edge: CD69 interference with sphingosine-1-phosphate receptor function regulates peripheral T cell retention. J. Immunol. 194, 2059–2063 (2015).
Gold, M.C. et al. Human thymic MR1-restricted MAIT cells are innate pathogen-reactive effectors that adapt following thymic egress. Mucosal Immunol. 6, 35–44 (2013).
Tan, A.C. et al. Intranasal administration of the TLR2 agonist Pam2Cys provides rapid protection against influenza in mice. Mol. Pharm. 9, 2710–2718 (2012).
Hoiseth, S.K. & Stocker, B.A.D. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature 291, 238–239 (1981).
Newland, J.W., Hale, T.L. & Formal, S.B. Genotypic and phenotypic characterization of an aroD deletion-attenuated Escherichia coli K12-Shigella flexneri hybrid vaccine expressing S. flexneri 2a somatic antigen. Vaccine 10, 766–776 (1992).
Acknowledgements
This research was supported by Program Grants 1016629 and 606788 and Project Grant 1083942 from the National Health and Medical Research Council of Australia (NHMRC). J.R. was supported by an NHMRC Australia Fellowship, D.P.F. was supported by an NHMRC Senior Principal Research Fellowship, and D.C.J. and D.I.G. are supported by NHMRC Research Fellowships. H.W. is supported by a Melbourne International Engagement Award (University of Melbourne). C.D’S. is supported by a Melbourne International Research Scholarship and a Melbourne International Fee Remission Scholarship (University of Melbourne). We thank Dr Wei-Jen Chua and Dr Ted Hansen for their kind provision of 8F2.F9 and 26.5 mAbs.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
Z.C., S.E., D.F., L.L., J.R., J.McC., and A.C. are inventors on patents describing MR1 tetramers. The other authors declared no conflict of interest.
Additional information
SUPPLEMENTARY MATERIAL is linked to the online version of the paper
Supplementary information
Rights and permissions
About this article
Cite this article
Chen, Z., Wang, H., D'Souza, C. et al. Mucosal-associated invariant T-cell activation and accumulation after in vivo infection depends on microbial riboflavin synthesis and co-stimulatory signals. Mucosal Immunol 10, 58–68 (2017). https://doi.org/10.1038/mi.2016.39
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/mi.2016.39
This article is cited by
-
MR1 antigen presentation to MAIT cells and other MR1-restricted T cells
Nature Reviews Immunology (2024)
-
Innate immune responses in pneumonia
Pneumonia (2023)
-
MAIT cell inhibition promotes liver fibrosis regression via macrophage phenotype reprogramming
Nature Communications (2023)
-
Divergent metabolic programmes control two populations of MAIT cells that protect the lung
Nature Cell Biology (2023)
-
Mucosal-associated invariant T cells display both pathogenic and protective roles in patients with inflammatory bowel diseases
Amino Acids (2023)