Abstract
Ewing's family tumors (EFTs) are characterized by recurrent chromosomal translocations that produce chimeric fusions between the EWS gene and one of five ETS transcription factors. The expression of EWS/FLI1, the predominant fusion product in EFTs, is believed to deregulate downstream target genes in an undefined tissue type and leads to development of EFTs. Attempts to generate model systems that represent EFTs have been hampered by an unexpected toxicity of the fusion gene. In the present study, we used gene expression analysis to identify tissue types based on the similarity of their expression profiles to those of EWS/FLI1-modulated genes. The data obtained from this screen helped to identify IMR-90 cells, a human fetal fibroblast, that upon further manipulation can maintain stable EWS/FLI1 expression without the reported toxicity. In addition, gene expression profiling of these cells revealed a significant overlap of genes that have been previously reported to be targets of EWS/FLI1. Furthermore, we show, for the first time, a partial transformation of these human primary fibroblasts with EWS/FLI1 expression. The experiments presented here provide a solid foundation for generation of a new model system for studying Ewing's sarcoma biology.
Similar content being viewed by others
Main
Somatic mutation is a hallmark of human cancer. There are hundreds of tumor-associated chromosomal translocations that have been described in a wide range of malignancies. The most common outcome of such genomic rearrangements is the formation of chimeric fusion genes in which at least one of the translocation partners encodes a transcription factor. This suggests that many of these fusion genes may produce aberrant transcription factors that are important in the initiation and progression of tumors.
Efforts to create model systems in which to study these fusion genes have been hampered by unexpected cellular toxicity. Stable expression of these chimeric oncoproteins in human or rodent cells frequently induces growth arrest or apoptosis. This suggests that both a permissive cellular context as well as possible additional complementing genomic mutations are necessary for these fusion genes to exert full oncogenic effect.1, 2 From a pragmatic point of view, defining cellular conditions in which fusion genes are stably tolerated is a prerequisite for prospectively studying how these oncoproteins function.
The EWS/FLI1 fusion found specifically in Ewing's family tumors (EFTs), is a well-studied example of a chimeric oncoprotein that has the primary structure and biochemical characteristics of an aberrant transcription factor (for review see Arvand and Denny3). When stably expressed in NIH 3T3 murine fibroblasts, EWS/FLI1 induces cellular transformation and accelerates tumorigenesis.4 However, most cell lines do not tolerate stable expression of EWS/FLI1. For example, when EWS/FLI1 is transduced into primary mouse embryonic fibroblasts (MEFs), these cells undergo cell-cycle arrest and apoptosis.2 A similar effect was seen when EWS/FLI1 was expressed in TERT-immortalized human foreskin fibroblasts.1 In both instances, stable expression of EWS/FLI1 was achieved by modulating the ARF-p53 pathway indicating that additional genetic alteration can overcome EWS/FLI1-mediated cellular toxicity in tissue culture.
EFTs pose an additional degree of difficulty in that their cell of origin is unknown. The fact that these tumors arise from bone and soft tissue would suggest that a mesenchymal progenitor was the culprit. However, the finding that EFT cells express neural markers supported the long-held belief that EFTs were neuroectodermal in origin.5, 6, 7 Though recent data favor the former hypothesis, there are as yet no EWS/FLI1-based transgenic animal models of EFT to provide definitive proof.
Both the inherent cellular toxicity of EWS/FLI1 and the continued ambiguity surrounding the EFT cell of origin has prompted the development of model systems using siRNA technologies. Using this strategy EWS/FLI1 is studied in its native cellular context by transducing anti-EWS/FLI1 shRNA constructs into EFT tumor-derived cell lines. The effects of EWS/FLI1 are inferred by assaying the effects that are lost or gained concurrent to downregulation of the fusion. Although much useful information has been gleaned from these model systems, there are inherent limitations. First, there is the concern to what degree each shRNA is downregulating genes other than EWS/FLI1, so called off-target effects. Second, the amount of EWS/FLI1 reduction is usually limited to ∼80% of baseline. Finally, downregulation of EWS/FLI1 results in cell death in most EFT cell lines.
In this paper we present data resulting from our efforts to construct a heterologous EWS/FLI1 model system. Our previous experience suggested that EWS/FLI1, when expressed in NIH 3T3 fibroblasts, can induce phenotypic features reminiscent of EFT.4 However, NIH 3T3 cells have an unstable genetic background and EWS/FLI1 modulates a repertoire of target genes in NIH 3T3 cells that is significantly different from those found in an EFT background. In this study, we use an shRNA-mediated EWS/FLI1 knockdown in EFT cell lines to identify candidate cell hosts. We then show that further genetic alterations are necessary to achieve stable EWS/FLI1 expression. We then assess the in vitro and in vivo tumorigenic potential of this novel heterologous model system.
MATERIALS AND METHODS
Plasmid Constructs, Viruses, and Tissue Culture
All EWS/FLI1 shRNA constructs were previously described.8 The p53 shRNA construct was cloned into the CSCG lentiviral vector (sequence available upon request). The retroviral triple FLAG epitope-tagged EWS/FLI1 was previously described.4 Lentiviral expression construct design, viral stock preparation, infection procedures, and cell culture techniques are described in Supplementary Data.
Anchorage Independent Growth Assay
Approximately 2 × 105 cells per 60 mm dish in semisolid Iscove's media with high serum (20% FBS). The cultures were refed every 7 days with fresh media. Images were captured using × 200 magnification at day 45 and analyzed with NIH ImageJ software v1.38x.
Western Blot and Quantitative Real-Time PCR
Standard western blot analysis was used to detect protein detection. Monoclonal anti-FLAG M2 antibody (Sigma) was used to detect FLAG epitope-tagged EWS/FLI1. Anti-p53, -p16, and -actin polyclonal antibodies were purchased from Santa Cruz Biotechnology.
Total RNA was isolated using RNeasy mini kit (Qiagen) according to manufacturer's protocol. Approximately 5 μg of the total RNA was used to synthesize the first strand cDNA using the first-strand cDNA synthesis kit (Invitrogen) according to manufacturer's protocol. Quantitative real-time PCR was performed using site-specific oligonucleotide primers as previously described (Potikyan et al8). Primer sequences are available upon request.
Microarray Expression Analysis
A complete description of microarray expression analyses is provided in the Supplementary Data.
RESULTS
Expression profile of EWS/FLI1-Silenced EFT Cells is Similar to that of Cells of Mesenchymal Lineage
To gain insight into the origin of EFTs, we silenced the expression of EWS/FLI1 in EFT cell lines and compared their expression profiles with those of various tissue types available from the Celsius database.9 We reasoned that by inhibiting EWS/FLI1, the cells may regain some of the genetic characteristics of the primary cells that gave rise to EFTs. Cell types with similar expression profiles could then be assessed for their tolerance and capacity to be transformed by EWS/FLI1 expression. This approach could potentially help design a new heterologous model system with a better representation of EFTs than the current NIH3T3-based model system.
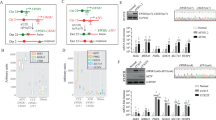
Previously reported FLI1 shRNAs (Figure 1a) were used to knockdown the expression of EWS/FLI1 in EFT cell lines.8 To eliminate idiosyncratic effects from the expression profile due to off-target effects of RNAi, we performed the knockdown in a redundant fashion. Two EFT cell lines (A4573 and TC32) were transduced with two different FLI1 shRNAs (818 and EF4) or their associated empty vector controls. Total RNA and protein from A4573 and TC32 cells were harvested 48 h after lentiviral transduction and subjected to quantitative real-time PCR analysis (Figure 1b). We detected over 80% knockdown of EWS/FLI1 mRNA levels in A4573 and TC32 cells transduced with the 818 shRNA, over 60% knockdown in EF4 transduced A4573 cells, and about 40% knockdown in EF4 transduced TC32 cells. The low knockdown levels by the EF4 shRNA were attributed to low transduction efficiency of this construct determined by GFP expression from the lentiviral vector (data not shown).
Short hairpin RNA-mediated knockdown of EWS/FLI1 in EFT cell lines. (a) Schematic diagram of the EWS/FLI1 transcript with the approximate RNAi target locations; 818 is downstream of the breakage point of the fusion gene and upstream of the DNA binding domain; EF4 targets the 3′-untranslated region of FLI1. (b) Quantitative real-time PCR analysis of EWS/FLI1 transcript expression levels in EFT cell lines 48 h after transduction with the FLI1 shRNA constructs or their associated vector controls. (c) Phase contrast and fluorescence microscopy of A4573 cells transduced with EF1 shRNA or vector control 2 and 6 days after lentiviral transduction. At 6 days after lentiviral transduction A4573 cells exhibit large flat cell morphology and slowing of proliferation rate, a characteristic of senescence. GFP is expressed from the lentiviral vector containing the FLI1 shRNA or control constructs and its expression is used as a measure of transduction efficiency. All images were captured at × 200 magnification. (d) Growth analysis using a 3T5 assay1 demonstrating significant reduction in proliferation of A4573 and TC32 cells transduced with 818 and EF4 FLI1 shRNA.
Previous reports suggest that most EFT cell lines are growth inhibited upon EWS/FLI1 knockdown.10 To assess the effects of our shRNA constructs on the growth potential of A4573 and TC32 cell lines, the shRNA and empty vector control transduced cells were maintained in culture for up to 4 weeks. At 7–10 days after transduction with either shRNA but not empty vector control, cells exhibited growth inhibition and displayed a large flat morphology consistent with cellular senescence (Figure 1c and d). Staining for β-galactosidase activity, a marker of senescence revealed that the EWS/FLI1-silenced cells had undergone senescence (Supplementary Figure S1). Active growth of GFP-negative cells was detected about 3 weeks after transduction, which ultimately overtook the culture, suggesting either overgrowth of untransduced cells in culture, or nonspecific silencing of the shRNA constructs (data not shown).
To generate expression profiles before the onset of these global effects, cells were harvested for total RNA isolation 48 h after transduction with the shRNAs or their corresponding vector controls and subjected to microarray expression profile analysis using the Affymetrix HGU133A arrays. Standard two-group comparison analysis identified 915 probe sets (746 genes) in 818 transduced A4573 cells, 769 probe sets (554 genes) in EF4 transduced A4573 cells, and 467 probe sets (315 genes) in 818 transduced TC32 cells that were differentially expressed in EWS/FLI1 inhibited vs empty vector control cells by at least twofold (P<0.05) (Figure 2a). Permutation analysis of each condition revealed median false discovery rates (FDRs) of below 0.02, indicating that the identified differentially expressed genes are not due to randomness. We found 214 probe sets that correspond to 162 genes commonly shared between all experimental conditions that are modulated by EWS/FLI1 (FDR=0.014) (Figure 2a; Supplementary Table S1). Of the 162 modulated genes, 93 were downregulated and 69 upregulated by EWS/FLI1. Among this list, we found several genes that were previously reported as targets of EWS/FLI1 in EFT cells, including NR0B1, DKK1, DAB1, CNTNAP2, cMYC, SOX2, CXCR4, NGFR, IGFBPs, and IGF1.11, 12, 13, 14
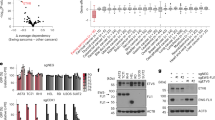
Microarray analysis of EFT cell lines with EWS/FLI1 RNAi compared to IMR-90 fibroblasts. (a) Venn diagram showing the number of differentially expressed genes in EFT cell lines transduced with FLI1 shRNA constructs. Gene expression of A4573 and TC32 cells transduced with 818, EF4 shRNA or their corresponding empty vector controls was analyzed using Affymetrix HGU133A chipset. Genes downregulated in shRNA-transduced cells are upregulated by EWS/FLI1, and genes upregulated in shRNA-transduced cells are downregulated by EWS/FLI1. Of the 746, 554, and 467 (fold change >2, P-value <0.05, median false discovery rate <2%) genes regulated in A4573 (818), A4573 (EF4), and TC32 (818), 162 genes are commonly regulated in the two cell lines, respectively. The complete list of genes regulated by EWS/FLI1 is listed in Supplementary Table S1. (b) Left panel, correlation plot of the Manhattan distance metric of the mean expression values of the 162 commonly regulated genes in all 6900 samples (top) and the first 200 samples (bottom). The plots demonstrate that the top 50 samples have the shortest distance from the mean expression values of the modulated genes in EWS/FLI1-silenced EFT cell lines, thus represent the tissue types with the most similar expression signatures. Right panel, list of tissue types with the most similar expression signatures of the 162 modulated genes to EWS/FLI1-silenced EFT cell lines. The list is ranked by the Manhattan distance metric calculated between the mean expression values of each gene. Included in this list are the tissue types from the top 50 samples (above dotted line) and the distance values of EFT cell lines (below dotted line).
The average expression values of the 162 modulated genes in EWS/FLI1-silenced EFT cells were used to calculate the Manhattan distance to the expression values of those same genes to 6900 individual samples from the Celsius database.9 This distance metric allows the ranking of all 6900 samples in the database based on their similarity to the expression signature of EFT cells with EWS/FLI1 knockdown. Samples with the most similarity will have the closest distance. All 6900 samples used in this analysis were microarray experiments performed on the HGU133A platform. A total of 11 different tissue types were represented in the top 50 samples with the most similar expression signatures. Of the 11 tissue types, IMR-90 cells showed the most similarity to EWS/FLI1-silence EFTs (Figure 2C). IMR-90 cells are normal diploid fibroblasts isolated from human fetal lungs.15 These results are consistent with the current view of the mesenchymal origin of EFTs.12
Expression of EWS/FLI1 in IMR-90 Cells Induces Growth Arrest
Expression of EWS/FLI1 has been shown to induce growth arrest in most normal human primary cells.1 To assess whether the IMR-90 cell background is permissive for EWS/FLI1 expression, we transduced TERT immortalized IMR-90 cells (IMR-90T) with FLAG epitope-tagged EWS/FLI1 or empty vector. Telomerase has been previously shown to be activated in Ewing's tumors.16 After a 7-day antibiotic selection, cells transduced with EWS/FLI1 exhibited a dramatic growth inhibition and displayed a large flat cell morphology consistent with cellular senescence (Figure 3a). This observation is consistent with previous reports of induction of senescence of IMR-90 cells in response to oncogenic stress by Ras.17
Effects of ectopic EWS/FLI1 expression in IMR-90T fibroblasts. (a) Phase contrast (PC) and immunofluorescence (IF) microscopy images of IMR-90T cells 12 days after transduction with 3 × FLAG-tagged EWS/FLI1 (right panels) and empty vector control (left panels). Cells expressing EWS/FLI1 show nuclear localization of the fusion protein with immunofluorescent staining with anti-FLAG antibody. These cells show a marked decrease in cell proliferation rate and exhibit senescent cell morphology, whereas vector control transduced cells retain the elongated spindle shaped morphology. All images were captured at × 200 magnification. (b) Western blot analysis of IMR-90T cells transduced with EWS/FLI1 or vector control. Whole cell lysates were prepared after 4 or 18 days after retroviral transduction and subjected to immunoblot analysis using anti-FLAG, p53, and p16INK4A antibodies. Increased expression of p53 and p16INK4A proteins 18 days post-retroviral transduction suggests induction of senescence by the p16INK4A-p53-RB pathway. Actin is shown as a loading control.
Oncogene-induced senescence has been shown to involve activation of the ARF-p53 pathway, a mechanism by which normal mammalian cells evade transformation (for review see Bardeesy and Sharpless18). To determine whether the observed growth inhibition is due to activation of the ARF-p53 tumor suppressor pathway, protein lysates were harvested 4 and 18 days after transduction with EWS/FLI1 or empty vector. Immunoblot analysis revealed a dramatic increase in both p53 and ARF protein levels 18 days after transduction with EWS/FLI1 but not empty vector, suggesting that oncogenic stress exerted by EWS/FLI1 induces p53-mediated senescence in IMR-90 cells (Figure 3b).
Targeted Silencing of p53 Permits Stable EWS/FLI1 Expression in IMR-90T Cells
Stabilization and activation of p53 tumor suppressor is one of the key events in oncogene-induced senescence. Work from our laboratory had shown that interruption of the ARF-p53 pathways is a critical step in establishing stable EWS/FLI1 expression in mouse embryonic fibroblasts.2 In a parallel experiment, we also showed that EWS/FLI1 expression in spontaneously transformed mesenchymal progenitor cells can be stabilized by inhibition of p53 with pifithrin-α (data not shown). Furthermore, EFT cell lines, including A4573 and TC32, that efficiently grow in tissue culture have deletion or missense mutations in p53 or p16 loci.19, 20 This is in contrast to primary tumors where only about 10 and 30% have such mutations respectively, suggesting that disruption of the ARF-p53 pathway may be a consequence of ex vivo growth of cells expressing the EWS/FLI1 fusion gene.
To test this hypothesis, IMR-90T cells were transduced with an shRNA construct targeting p53 or empty vector control. At 2 days after efficient transduction, total RNA was harvested and subjected to quantitative real-time PCR analysis to determine the level of p53 knockdown. We detected over 90% reduction of endogenous p53 transcript levels as compared to control-infected cells (Figure 4a). Although we did not observe any morphological changes of the p53 shRNA-transduced cells (designated IMR-90Tp), there was a significant increase in proliferation rate compared to the empty vector infected or IMR-90T parental cells (Figure 4e). A similar increase in proliferation rate was also observed in IMR-90T cells transduced with SV40 large T antigen, which also targets the ARF-p53 pathways (Supplementary Figure S2).
Stable knockdown of p53 and expression of EWS/FLI1 in IMR-90T fibroblasts. (a) Quantitative real-time PCR using gene-specific oligonucleotide primers for p53 and GAPDH demonstrates a marked decrease in p53 mRNA transcript levels 48 h after lentiviral transduction (P-value <0.001). GAPDH mRNA transcript levels are included as loading control. (b) Western blot analysis of IMR-90Tp cells 60 days after transduction with RFP IRES 3 × FLAG-tagged EWS/FLI1 (IMR-90TpRIE) or empty vector control using anti-FLAG antibody. Actin is shown as a loading control. (c) Phase contrast and fluorescence microscopy of RFP and RIE transduced IMR-90Tp cells 30 days and 4 months after infection with the lentivirus. Green fluorescence protein expression is shown to indicate p53 shRNA transduction efficiency (middle row). The expression of RFP and RFP-IRES-EWS/FLI1 is assessed by the expression of red fluorescent protein (bottom row) as both genes are expressed from the same mRNA transcript. Images were captured at × 100 and × 200 magnifications, as indicated. (d) Quantitative real-time PCR demonstrating expression of EWS/FLI1 mRNA in IMR-90Tp, IMR-90TpRFP, and IMR-90TpRIE cells 50 days and 4 months after transduction with the lentivirus. Note the insignificant change in mRNA expression levels during prolonged culturing of the cells. GAPDH expression is shown as loading control. The data represent expression values in cells generated in two independent experiments. (e) Population doubling times of parental IMR-90T, IMR-90Tp, and IMR-90TpRIE cells. A significant increase in cell proliferation rate was observed in cells with p53 knockdown compared to the parental IMR-90T cells. Expression of EWS/FLI1 in IMR-90Tp cells does not result in a significant change of proliferation rate. The data represent three independent experiments.
To determine if shRNA-mediated knockdown of p53 is sufficient to stably express EWS/FLI1, IMR-90Tp cells were transduced with lentiviral constructs containing an RFP-IRES-EWS/FLI1 (RIE) or RFP cassettes under the transcriptional control of a CMV promoter. Cells transduced with RIE (IMR-90TpRIE) but not RFP displayed an initial phase of apoptosis in the first 10–14 days (data not shown). All of the cells that were apoptotic were also positive for RFP, suggesting that EWS/FLI1 was inducing apoptosis in IMR-90Tp cells possibly through a p53-independent pathway. However, we consistently detected recovery of RFP-positive cells in the IMR-90TpRIE cultures after 20 days after transduction (Figure 4c, middle panel). These cells were maintained in culture for over 4 months (approximately 60 doublings) with no apparent decrease in the number of RFP-positive cells, suggesting that the cellular environment in IMR-90Tp cells may be suitable for stable EWS/FLI1 expression (Figure 4c, right panel).
Expression of EWS/FLI1 was determined by western blot and quantitative RT-PCR analyses. Cells were harvested for protein and total RNA at various time points. Immunoblot analysis with anti-FLAG antibody revealed expression of EWS/FLI1 in IMR-90TpRIE but not IMR-90TpRFP cells at 60 days after transduction without a significant expression loss (Figure 4b). Similar immunoblots using anti-FLI1 antisera detected EWS/FLI1 protein levels in IMR-90TpRIE cells that were at or slightly below that seen in EFT cell lines (Supplementary Figure S3). Quantitative real-time PCR analysis with oligonucleotide primers specific for FLI1 revealed over 100-fold increase in EWS/FLI1 mRNA levels at 50 days and 4 months postinfection with the RIE but not RFP control lentivirus (Figure 4c). These results suggest that inhibition of p53 permits stable expression of EWS/FLI1 in IMR-90T cells.
Expression of EWS/FLI1 Promotes Limited Transformation in IMR-90 Cells
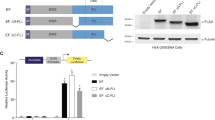
The ability of cells to grow anchorage independently is one of the hallmark characteristics of oncogenic transformation. To determine if EWS/FLI1 expression is sufficient to transform IMR-90 cells, we assessed the colony-forming capacity of IMR-90TpRIE and IMR-90TpRFP cells in three independent soft agar assays using two independently transduced cell populations. Cells (2 × 105) were seeded in soft agar with high serum (20% FBS) conditions and colony formation was monitored microscopically every 5 days. Colonies first appeared 15 days after seeding with IMR-90TpRIE cells and continued to grow, reaching their maximum size 45 days later (Figure 5a). The vector control transduced IMR-90Tp cells did not form colonies after extended incubation times. To quantify the size difference between RIE vs control transduced colonies, a total of 30 images from each plate were captured sequentially at × 100 magnification. Average size of the colonies was measured using NIH ImageJ software, which revealed that the RIE colonies were at least five times larger both in area and size than RFP colonies (Figure 5b). Within the 30 fields captured from each plate we found 45 colonies in the RIE-transduced cells and 5 colonies in the RFP-transduced cells (data not shown).
Anchorage independent growth of IMR-90Tp cells with EWS/FLI1 expression. (a) IMR-90Tp cells transduced with RIE or empty vector were seeded at 2 × 105 cells per 60 mm dish in semisolid media with high serum (20% FBS) condition. Cultures were refed every 7 days with fresh media/serum. The capacity to form colonies was assessed microscopically every 5 days. Four representative fields at × 200 magnification were captured for each condition 45 days after seeding. This experiment was repeated three times with cells generated in two independent experiments. (b) The average area and size of the colonies formed in soft agar by RIE and RFP transduced IMR-90Tp cells. The data represent two plates of each cell line. A total of 30 sequential fields were captured from each plate at × 100 magnification and analyzed with NIH ImageJ software (P<0.05). (c) Immunoblot analysis using monoclonal anti-FLAG M2 antibody shows expression of EWS/FLI1 in RIE-transduced colonies. Each lane (agar colonies) represents five colonies of approximately same size extracted from soft agar. Total protein lysate from IMR-90Tp cells transduced with RIE (RIE) and empty vector control (C) are also shown. Anti-actin immunoblot is shown as loading control.
It is important to note that although the RIE cultures contained both RFP-positive and negative cells, colonies were only formed by cells that were RFP positive. As both RFP and EWS/FLI1 are expressed off of the same mRNA transcript, these results suggest that only EWS/FLI1 expressing cells were able to form colonies. To confirm that the cells maintained EWS/FLI1 expression, a total of 15 colonies were harvested, 5 colonies were pooled per sample, and subjected to SDS–PAGE. Immunoblot analysis confirmed EWS/FLI1 expression in all harvested colonies (Figure 5c). Taken as a whole, these data suggest that limited transformation can be achieved in IMR-90Tp cells expressing EWS/FLI1.
EFT cell lines, when injected into immunodeficient mice, form tumors within 20–25 days and retain their characteristic phenotype. To determine if IMR-90TpRIE cells have the capacity to form tumors in vivo, long-term cultured (3 months) or freshly transduced (48 h after transduction) IMR-90TpRIE and IMR-90TpRFP cells were injected subcutaneously with and without Matrigel into immunodeficient mice. None of the mice developed tumors in the 180 days of monitoring after injections. Although we have shown that EWS/FLI1 induces apoptosis in SV40 LT antigen transduced IMR-90T cells, we sought to determine whether the apoptotic response is a consequence of growth in tissue culture and may be prevented by in vivo growth. Freshly transduced IMR-90TLT-RIE cells injected subcutaneously into immunodeficient mice failed to form tumor, suggesting that despite the strong transforming capacity of the SV40 LT antigen, coexpression of EWS/FLI1 is not sufficient to render IMR-90 cells tumorigenic.
Perturbation of the ARF-p53 Pathways with SV40 T Antigen Leads to EWS/FLI1-Mediated Cell Death in Human Fibroblasts
We had previously shown that expression of EWS/FLI1 in ARF-null MEFs does not support tumorigenesis. However, abrogation of tumor suppressor pathways by SV40 large T antigen (LT) was shown to be sufficient to support tumor formation in mice.2 Therefore, we sought to determine whether inhibition of ARF-p53 pathways in IMR-90T cells using LT is sufficient to promote EWS/FLI1-mediated tumor formation. IMR-90T cells were transduced with a retroviral construct containing LT. After a brief antibiotic selection, IMR-90TLT cells were transduced with RIE or RFP. As before, the RFP-positive cells in the RIE but not RFP-transduced cultures rounded up and lost adherence to the substrate, ultimately floating in growth media (Supplementary Figure S2). Trypan blue exclusion analysis confirmed that all of the nonadherent cells in culture had undergone cell death (data not shown). In contrast to IMR-90Tp cells, no RFP-positive cells were recovered from RIE-transduced IMR-90TLT cultures. In addition to its transforming properties, LT appears to also induce cell death in some cell types. LT-mediated apoptosis in human primary epithelial cells was shown to occur by overexpression of c-jun.21, 22 Although we were unable to determine the expression level of c-jun in IMR90TLT-RIE cells due to rapid cell death, it is possible that LT induces apoptosis in IMR-90TLT cells by a comparable mechanism.
Microarray Expression Profiling of IMR-90 Cells with EWS/FLI1 Expression
To assess the effects of EWS/FLI1 on the expression profile of IMR-90 cells, we performed a microarray analysis of long-term cultured IMR-90TpRIE and IMR-90TpRFP cells. Cells were harvested approximately 3 months after lentiviral transduction for isolation of total RNA and subjected to microarray expression analysis. We found 578 probe sets corresponding to 501 unique genes regulated by EWS/FLI1 in IMR-90 cells (fold change >1.5, P<0.05, median FDR=0). Of the 501 regulated genes, 249 were upregulatedand 252 were downregulated by EWS/FLI1 (see Supplementary Table S2 for a complete gene list).
A gene ontology-based analysis (L2L; University of Washington) revealed that although a significant number of the upregulated genes encode for proteins involved in cell-cycle progression, many of the downregulated genes are involved in cell senescence, apoptosis, and differentiation (Supplementary Table S3). In addition, many of the upregulated genes have been shown to be highly expressed in various cancers that exhibit an undifferentiated morphology. These analyses reveal that the specific expression pattern of the modulated genes is consistent with transformed cells.
In order to assess the similarity between the expression profiles of IMR-90TpRIE and EFT cell lines, we compared the genes that are regulated by EWS/FLI1 in IMR-90Tp cells to those regulated in A4573 and TC32 cells. Of the original 162 commonly regulated genes in EFT cell lines, 25 were also regulated by EWS/FLI1 in IMR-90Tp cells (expected 4.50, P-value=3.47 × 10−12) (Figure 6a). In addition, 74 genes were commonly regulated between IMR-90Tp and A4573 cells (expected 14.60, P-value=1.75 × 10−31), and 48 genes were shared between IMR-90Tp and TC32 cells (expected 13.02, P-value=5.6 × 10−15). Most importantly, all of the genes behave consistently, such that genes that were shown to be up- or downregulated by EWS/FLI1 in EFT cell lines were also respectively up- or downregulated in IMR-90TpRIE cells. Taken together, these data demonstrate similarities in the expression patterns of the modulated genes in IMR-90 and EFT cell lines and also indicate cell-specific differences in the repertoire of genes regulated by EWS/FLI1.
Microarray expression analysis of IMR-90Tp and IMR-90TpRIE cells. (a) Venn diagram showing the number of differentially expressed genes in IMR-90TpRIE cells compared to A4573 and TC32 control vector-transduced cells. Among the 513 genes regulated by EWS/FLI1 (fold change >1.5, P-value <0.05, median false discovery rate=0) in IMR-90Tp cells, 222 genes are upregulated and 291 are downregulated. Of the modulated genes, 74 and 48 genes are also regulated in A4573 and TC32 cells, respectively, and 25 of those are commonly regulated in the three cell lines (complete gene list is provided in Supplementary Table S2). (b) Differentially expressed genes in IMR-90TpRIE and IMR-90Tp cells also distinguish between EFT cell lines and IMR-90 fibroblasts. Of the 578 probe sets regulated by EWS/FLI1 in IMR-90Tp cells, 177 are differentially expressed between EFT cell lines and IMR-90 cells. Of the 177 probe sets, 100 were upregulated and 77 were downregulated by EWS/FLI1 in IMR-90Tp cells. Only one of the downregulated genes is upregulated in EFTs, indicating a strong correlation between gene regulation patterns in IMR-90Tp cells and EFT cell lines (represented in the table). P-values were calculated using hypergeometric cumulative distribution. The expected values are the mean of hypergeometric distribution.
To further investigate the relatedness of IMR-90TpRIE to EFTs, we compared the list of genes modulated in IMR-90TpRIE to genes differentially expressed between IMR-90 cells and EFT cell lines. This analysis identified 2615 probe sets (2063 genes) differentially expressed between IMR-90Tp cells and EFT cell lines with an absolute fold difference greater than 2 (P-value <0.05, median FDR <1%). Among these genes 177 were found to be modulated by EWS/FLI1 in IMR-90 by at least twofold (expected 57.52, P-value=5.6 × 10−15) (Figure 6b). Of these genes, 100 that were upregulated in IMR-90TpRIE were overexpressed in EFT cell lines, and 76 that were downregulated in IMR-90TpRIE were underexpressed in EFT cell lines. Only one gene was inconsistent with the expression pattern in EFT cell lines. Taken as a whole, these data indicate an overall shift of the expression profile of IMR-90TpRIE towards EFT cells.
DISCUSSION
Stable expression of EWS/FLI1 in primary human cells has been difficult to achieve due to toxicity of the chimeric oncoprotein. However, the ability of EFT cells to thrive in presence of EWS/FLI1 implies the existence of certain cell types that can tolerate its expression. In addition, complementing genetic and epigenetic alterations may be necessary to create a suitable host with reduced cytotoxicity to the EWS/FLI1 fusion gene. Identification of such cells and elucidation of these secondary events is required to generate a new model system that provides a better approximation of the biology of EFTs than the current NIH 3T3 model.
By suppressing EWS/FLI1 expression in EFT cell lines, we postulate that the cells will recover some of the genetic characteristics of the primary cells that gave rise to EFTs. There are certain considerations that should be taken into account when comparing the characteristics of EFT cell lines to those of normal tissue. First, these are transformed cells that have secondary mutations acquired during in vivo growth. Trisomies in chromosomes 8 and 12, unbalanced translocations between chromosomes 1 and 16, and many others secondary genetic defects are frequently found in EFTs.23 Second, loss of function mutations of p16 and p53 tumor suppressors are frequently found in EFT cell lines and are likely to have effects on the genetic profile of these cells. For this reason, we compared the expression signature of only EWS/FLI1 modulated genes (162 genes) to the expression signature of the same genes in various tissue types to minimize the effects of secondary mutations. In scanning the Celsius database, we found that the profile of these 162 genes in non-EWS/FLI1-silenced EFT cells (U6 and H1 controls) were similar to SK-N-MC, another EFT cell line (data not shown). This observation suggests that the signature of this subset of genes can be used to accurately identify tissue types similar to EFTs.
Many of the 162 modulated genes in A4573 and TC32 cells are previously reported targets of EWS/FLI1 (see Supplementary Table S1 for complete gene list). Among these were several previously reported genes involved in neural differentiation (eg NGFR, EPHB3, DAB, SOX2, GFRA2), supporting the growing evidence that the neural-specific markers of EFTs are a result of EWS/FLI1 upregulation rather than specific to EFT precursors.24 Of particular interest is SOX2, which is not only highly expressed in neural progenitors, but it is also one of the four genes (SOX2, cMYC, KLF4, and OCT3/4) recently shown to be required for maintenance of pluripotency and self-renewal of stem cells.25 As cMYC is also an EWS/FLI1 target and is highly expressed in EFTs, a more thorough examination is warranted to assess the precise role of these genes in EFT biology. Moreover, genes involved in differentiation of tissue types in the mesenchymal lineage are repressed by EWS/FLI1 (eg IGFBP3, IGFBP7, DKK1, CYR61).26, 27, 28 These findings are consistent with recent reports that demonstrate EWS/FLI1 inhibition of mesenchymal differentiation programs.11, 12
The identification of a primary fibroblast that has a similar expression profile to that of EWS/FLI1-silenced EFTs is consistent with the fact that most Ewing's tumors are localized to bone or soft tissue, both of which are abundant sources of cells of mesenchymal lineage. It is striking that within the top 50 samples representing 11 tissue types, there was a preponderance of mesenchymal and germ cell lines. These include cells derived from osteosarcoma, myoblasts, meningioma, teratoma, and germ cell carcinoma. MCF10A, a breast cancer cell line was also found which may in part be due to gene expression changes associated with epithelial–mesenchymal transition.29 What is striking is the near absence of neural tissues and cell lines in the top 50 other than ependymoma, a nonneuronal glial tumor.30 Taken together, these data point towards a mesenchymal origin of EFTs rather than the long-standing notion that these tumors arise from neuroectoderm.
Induction of senescence by EWS/FLI1 in IMR-90T cells is not an EWS/FLI1-specific toxicity. Normal diploid fibroblasts have previously been reported to undergo cell-cycle arrest and senescence in response to strong oncogenic stress.1, 17 A possible mechanism is through the activation of ARF/p53 tumor suppressor pathways by upregulation of cMYC, a frequent target of oncogenes and one of the genes induced by EWS/FLI1 (for a review see Bringold and Serrano31). Thus, the senescence response is an intrinsic mechanism that inhibits cell proliferation activated through inappropriate signaling pathways. A more striking effect is seen when p53 is inhibited by either shRNA-mediated knockdown or by expression of SV40 large T (LT) antigen followed by expression of EWS/FLI1. Induction of apoptosis in these cells suggests that there is a redundant mechanism that inhibits cell proliferation in response to oncogenic stimuli. Further studies should be conducted to identify such mechanisms, which could provide potential therapeutic targets.
Though both IMR-90Tp and IMR-90TLT cells undergo apoptosis in response to EWS/FLI1 expression, a subpopulation of IMR-90Tp cells that were EWS/FLI1-positive were eventually recovered in prolonged cultures. EWS/FLI1-positive cells were generated this way in two independent experiments, suggesting that a common mechanism may be contributing to this effect. A possible explanation for these observations is that the EWS/FLI1-positive cells acquired secondary mutations due to genetic instability introduced by the lack of p53 expression. In addition, the stress exerted by the low-density culture conditions (after initial cell death) could have selected for a subset of cells that can sustain expression of the fusion gene. Another explanation is that EWS/FLI1 expression can only be tolerated up to a certain level. Evidence supporting this hypothesis can be seen in experiments attempting to overexpress EWS/FLI1 in EFT cell lines. In these experiments, additional expression of ectopic EWS/FLI1 is not tolerated by EFT cell lines (unpublished observation). Therefore, it is possible that cells that underwent apoptosis were expressing EWS/FLI1 above this tolerable level.
The fact that many of the modulated genes in A4573 and TC32 cells were also modulated in IMR-90TpRIE, suggests a high degree of correlation between these fibroblasts and EFT cell lines. Furthermore, we found several genes to be regulated by EWS/FLI1 in IMR-90 cells that were not found to be modulated in A4573 or TC32 cells but were reported by other studies to be targets of EWS/FLI1 (ie ID2, SOX9, UPP1).12, 32, 33 Moreover, several new genes that had not been previously identified as EWS/FLI1 targets were markedly induced or repressed in IMR-90TpRIE, a significant number of which are genes that encode for proteins involved in cell-cycle progression. These include CCND2, CDC2, TOP2A, CDC25B, CKS2, p27, and P16INK4A. In addition, the expression of some p53 target genes was also repressed by EWS/FLI1 in IMR-90Tp cells (BAX, CDKN1A, and TNFRSF10B). Overall, the expression patterns of these genes are indicative of cells with a transformed phenotype.
Although only 25 out of the original 162 genes were modulated by EWS/FLI1 in IMR-90 cells, statistically the overlap is still quite significant (P-value=3.47 × 10−12). Most importantly, the pattern of regulation of those genes was consistent with that seen in the EFT background. In addition, a significant number of differentially expressed genes between IMR-90 cells and EFT cell lines are also modulated in a consistent pattern by EWS/FLI1 in IMR-90TpRIE cells (177, P-value=5.6 × 10−15), suggesting an overall shift in the expression profile of IMR-90 cells towards EFTs.
Though shRNA-mediated inhibition of p53 was sufficient for stable expression of EWS/FLI1 and limited growth capacity in soft agar, IMR-90TpRIE cells did not form tumors when injected into SCID mice. The fact that p53 mutations are found in a minority of EFTs argues that other mechanisms may contribute to the overall oncogenic phenotype of the tumors. It is possible that EFTs that do not harbor inactivating mutations in p16 or p53 loci circumvent the inhibitory effects of these pathways through promoter hypermethylation. Additional undefined genetic and epigenetic alterations are likely to contribute to the EFT phenotype as well. Thus, it is not surprising that inhibition of p53 alone is not sufficient to confer the full oncogenic phenotype of EFTs.
Although IMR-90 cells cannot be considered to be the true origin of EFTs, they are a step closer than previous heterologous model systems and can provide an important alternative for studying Ewing's sarcoma biology. The experiments reported here provide the first steps in generation of a new model system based on a human primary cell line. Such a model system with well-defined alterations and a stable genetic background could provide greater insight into EFT pathogenesis.
References
Lessnick SL, Dacwag CS, Golub TR . The Ewing's sarcoma oncoprotein EWS/FLI induces a p53-dependent growth arrest in primary human fibroblasts. Cancer Cell 2002;1:393–401.
Deneen B, Denny CT . Loss of p16 pathways stabilizes EWS/FLI1 expression and complements EWS/FLI1 mediated transformation. Oncogene 2001;20:6731–6741.
Arvand A, Denny CT . Biology of EWS/ETS fusions in Ewing's family tumors. Oncogene 2001;20:5747–5754.
Thompson AD, Teitell MA, Arvand A, et al. Divergent Ewing's sarcoma EWS/ETS fusions confer a common tumorigenic phenotype on NIH3T3 cells. Oncogene 1999;18:5506–5513.
Lizard-Nacol S, Lizard G, Justrabo E, et al. Immunologic characterization of Ewing's sarcoma using mesenchymal and neural markers. Am J Pathol 1989;135:847–855.
Cavazzana AO, Magnani JL, Ross RA, et al. Ewing's sarcoma is an undifferentiated neuroectodermal tumor. Prog Clin Biol Res 1988;271:487–498.
Cavazzana AO, Miser JS, Jefferson J, et al. Experimental evidence for a neural origin of Ewing's sarcoma of bone. Am J Pathol 1987;127:507–518.
Potikyan G, Savene RO, Gaulden JM, et al. EWS/FLI1 regulates tumor angiogenesis in Ewing's sarcoma via suppression of thrombospondins. Cancer Res 2007;67:6675–6684.
Day A, Carlson MR, Dong J, et al. Celsius: a community resource for Affymetrix microarray data. Genome Biol 2007;8:R112.
Dohjima T, Lee NS, Li H, et al. Small interfering RNAs expressed from a Pol III promoter suppress the EWS/Fli-1 transcript in an Ewing sarcoma cell line. Mol Ther 2003;7:811–816.
Riggi N, Suva ML, Suva D, et al. EWS-FLI-1 expression triggers a Ewing's sarcoma initiation program in primary human mesenchymal stem cells. Cancer Res 2008;68:2176–2185.
Tirode F, Laud-Duval K, Prieur A, et al. Mesenchymal stem cell features of Ewing tumors. Cancer Cell 2007;11:421–429.
Kinsey M, Smith R, Lessnick SL . NR0B1 is required for the oncogenic phenotype mediated by EWS/FLI in Ewing's sarcoma. Mol Cancer Res 2006;4:851–859.
Baird K, Davis S, Antonescu CR, et al. Gene expression profiling of human sarcomas: insights into sarcoma biology. Cancer Res 2005;65:9226–9235.
Nichols WW, Murphy DG, Cristofalo VJ, et al. Characterization of a new human diploid cell strain, IMR-90. Science 1977;196:60–63.
Fuchs B, Inwards C, Scully SP, et al. hTERT is highly expressed in Ewing's sarcoma and activated by EWS-ETS oncoproteins. Clin Orthop Relat Res 2004;426:64–68.
Serrano M, Lin AW, McCurrach ME, et al. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 1997;88:593–602.
Bardeesy N, Sharpless NE . RAS unplugged: negative feedback and oncogene-induced senescence. Cancer Cell 2006;10:451–453.
Lopez-Guerrero JA, Pellin A, Noguera R, et al. Molecular analysis of the 9p21 locus and p53 genes in Ewing family tumors. Lab Invest 2001;81:803–814.
Tsuchiya T, Sekine K, Hinohara S, et al. Analysis of the p16INK4, p14ARF, p15, TP53, and MDM2 genes and their prognostic implications in osteosarcoma and Ewing sarcoma. Cancer Genet Cytogenet 2000;120:91–98.
Chen SL, Tsao YP, Chen YL, et al. The induction of apoptosis by SV40 T antigen correlates with c-jun overexpression. Virology 1998;244:521–529.
Kolzau T, Hansen RS, Zahra D, et al. Inhibition of SV40 large T antigen induced apoptosis by small T antigen. Oncogene 1999;18:5598–5603.
Zielenska M, Zhang ZM, Ng K, et al. Acquisition of secondary structural chromosomal changes in pediatric Ewing sarcoma is a probable prognostic factor for tumor response and clinical outcome. Cancer 2001;91:2156–2164.
Smith R, Owen LA, Trem DJ, et al. Expression profiling of EWS/FLI identifies NKX2.2 as a critical target gene in Ewing's sarcoma. Cancer Cell 2006;9:405–416.
Takahashi K, Yamanaka S . Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006;126:663–676.
Kiepe D, Ciarmatori S, Haarmann A, et al. Differential expression of IGF system components in proliferating vs. differentiating growth plate chondrocytes: the functional role of IGFBP-5. Am J Physiol Endocrinol Metab 2006;290:E363–E371.
van der Horst G, van der Werf SM, Farih-Sips H, et al. Downregulation of Wnt signaling by increased expression of Dickkopf-1 and -2 is a prerequisite for late-stage osteoblast differentiation of KS483 cells. J Bone Miner Res 2005;20:1867–1877.
Noth U, Schupp K, Heymer A, et al. Anterior cruciate ligament constructs fabricated from human mesenchymal stem cells in a collagen type I hydrogel. Cytotherapy 2005;7:447–455.
Vernon AE, LaBonne C . Tumor metastasis: a new twist on epithelial–mesenchymal transitions. Curr Biol 2004;14:R719–R721.
Halata Z, Grim M, Christ B . Origin of spinal cord meninges, sheaths of peripheral nerves, and cutaneous receptors including Merkel cells. An experimental and ultrastructural study with avian chimeras. Anat Embryol (Berl) 1990;182:529–537.
Bringold F, Serrano M . Tumor suppressors and oncogenes in cellular senescence. Exp Gerontol 2000;35:317–329.
Deneen B, Hamidi H, Denny CT . Functional analysis of the EWS/ETS target gene uridine phosphorylase. Cancer Res 2003;63:4268–4274.
Fukuma M, Okita H, Hata J, et al. Upregulation of Id2, an oncogenic helix-loop-helix protein, is mediated by the chimeric EWS/ets protein in Ewing sarcoma. Oncogene 2003;22:1–9.
Acknowledgements
We thank the staff of the UCLA Microarray Core facility for sample preparation, hybridization, and data acquisition, Stephen Lessnick for FLI1 shRNA construct, and Jenny Andersen and Matteo Pellegrini for help with statistical analysis. We also thank Arnold Berk for providing IMR-90 fibroblasts. This work was supported by National Cancer Institute Grant CA087771 to CTD.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on the Laboratory Investigation website (http://www.laboratoryinvestigation.org)
Supplementary information
Rights and permissions
About this article
Cite this article
Potikyan, G., France, K., Carlson, M. et al. Genetically defined EWS/FLI1 model system suggests mesenchymal origin of Ewing's family tumors. Lab Invest 88, 1291–1302 (2008). https://doi.org/10.1038/labinvest.2008.99
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/labinvest.2008.99
Keywords
This article is cited by
-
ETV6 dependency in Ewing sarcoma by antagonism of EWS-FLI1-mediated enhancer activation
Nature Cell Biology (2023)
-
Inhibitor of DNA binding 2 (ID2) regulates the expression of developmental genes and tumorigenesis in ewing sarcoma
Oncogene (2022)
-
Modeling the initiation of Ewing sarcoma tumorigenesis in differentiating human embryonic stem cells
Oncogene (2016)
-
Mesenchymal Stem Cells During Tumor Formation and Dissemination
Current Stem Cell Reports (2016)
-
The Adolescent and Young Adult with Cancer: State of the Art - Bone Tumors
Current Oncology Reports (2013)