Abstract
We performed a genome-wide association study on 377 cases of neovascular age-related macular degeneration (AMD) and 1074 controls to determine the association of previously reported genetic variants associated with neovascular AMD in the Thai population. All patients were of Thai ancestry. We confirmed the association of age-related maculopathy susceptibility 2 (ARMS2) rs10490924 (P=7.38 × 10−17), HTRA1 rs11200638 (P=5.47 × 10−17) and complement factor H gene (CFH) rs800292 (P=2.53 × 10−8) with neovascular AMD, all loci passing the genome-wide significance level (P<5.22 × 10−8). We also found association of the previously reported CFH rs10737680 (P=1.76 × 10−6) locus in the discovery sample. Two loci not previously reported to be associated with neovascular AMD were selected for replication in 222 cases and 623 controls. The loci included LINCO1317 rs6733379 and rs2384550 on chromosome 12. LINCO1317 rs6733379 (P=3.85 × 10−2) remained significantly associated with neovascular AMD after replication. In conclusion, we confirm that ARMS2, HTRA1 and CFH variants are associated with neovascular AMD in the Thai population. Findings from this study also suggest that variants contributing to the susceptibility of neovascular AMD in the Thai population are mostly similar to other Asians with additional local genetic risks that may specifically be identified in Thai population.
Similar content being viewed by others
Introduction
Age-related macular degeneration (AMD) is one of the worldwide leading causes of blindness from retinal diseases. It has two distinct clinical manifestations, the dry form and the wet or neovascular form; it is the neovascular form that is responsible for the majority of the blindness from AMD and is the third leading cause of blindness worldwide. Neovascular AMD is characterized by progressive degenerative changes at the macula causing deterioration of central vision by having choroidal neovascularization (CNV). There are two phenotypes of the neovascularization in neovascular AMD, one is typical CNV commonly seen in Caucasians and the other is polypoidal choroidal vasculopathy (PCV) commonly seen in Asians. PCV is said to be distinct from the typical CNV with specific features that are not found in typical CNV, such as the presence of polyp-like subretinal orange nodules at the terminal bulbs of inner choroidal vascular abnormalities seen from clinical examination and focal hyperfluorescence of the polypoidal nodules seen from fundus indocyanine green angiography.1 PCV also had better visual and anatomical outcome when treated with verteporfin photodynamic therapy, compared with typical CNV.
The prevalence of neovascular AMD increases with age. The Thailand National Survey of Visual Impairment found neovascular AMD in 0.2% of people who were >50 years old.2 Neovascular AMD is a complex genetic disorder in which DNA changes make an individual at risk for susceptibility to the disease.3 In 2005, a genome-wide association study (GWAS) provided a clearer view in finding links between AMD and genetic variations, suggesting AMD as a model of common polygenic disease.4 This finding triggered numerous studies of the genetic basis of AMD.5, 6, 7, 8, 9, 10 Though the two phenotypes of neovascular AMD, typical CNV and PCV, have different clinical characteristics, their genotypes are found to be similar. The major genetic risk factors for both dry form and neovascular form of AMD, complement factor H gene (CFH) and age-related maculopathy susceptibility 2 (ARMS2) polymorphisms,11 were also found to be associated with a higher risk for PCV.12, 13 In 2010, the AMD Gene Consortium conducted a meta-analysis, which included 17 100 advanced AMD cases and 60 000 controls throughout Europe and Asia. The study found seven loci associated with AMD surviving the genome-wide significance level. All seven loci resided in the regulatory genes of the complement pathway including COL8A1-FILIP1L, IER3-DDR1, SLC16A8, TGFBR1, RAD51B, ADAMTS9 and B3GALTL.14
Despite the generally low power of discrimination for genetic risk assessment, AMD has distinct phenotypes with high heritability. This suggests promising potential for risk modification. We aimed to analyze the role of human genetics in neovascular AMD conducted through the GWAS in the Thai population in hopes of confirming previous loci and discovering potentially associated variants specific to Thai ethnicity.
Materials and methods
A GWAS was conducted in case–control groups to find genetic associations in neovascular AMD including both typical CNV and PCV. The study protocol was approved by the Institutional Review Board at Rajavithi Hospital in accordance with the Declaration of Helsinki. Written informed consent was obtained from all subjects before participation.
Genome-wide association samples
All cases and controls included in this sample set are of Thai ancestry. Cases were recruited from the database of patients diagnosed with AMD in the Eye Clinic of Rajavithi Hospital. Disease controls were patients from the Eye Clinic diagnosed with other eye diseases without any signs of dry or wet AMD on dilated fundus examination. Population controls were participants who have no visual symptoms or history of eye diseases from the database of the pooled controls at the Department of Medical Sciences of Ministry of Public Health of Thailand. Replication samples were independent from the discovery phase.
Diagnostic criteria for neovascular AMD
All patients diagnosed with neovascular AMD were required to have retinal color photography, fluorescein angiography and indocyanine green angiography of the macular area. The images were recorded and reviewed by retinal specialists who were attending staffs in the Retina Service of the Eye Clinic at Rajavithi Hospital for definite diagnosis. The inclusion criteria were patients of Thai ancestry, diagnosed with neovascular AMD in at least one eye. Typical CNV was confirmed by presence of active leakage from CNV on FA images without evidence of any signs suggesting PCV on either FA or indocyanine green angiography. PCV, on the other hand, was confirmed by presence of typical features, such as leakage of branching vascular networks on FA images and subretinal focal hyperfluorescenct nodules in the early phase of indocyanine green angiography images, and other criteria proposed in the major randomized controlled trial of PCV.1
Patients who did not fulfill all the steps in the study, had a history of allergy to fluorescein or indocyanine green, and patients with macular diseases other than AMD in either eye such as myopic CNV and other causes, were excluded.
DNA extraction
Blood samples of 5 ml were collected into EDTA blood tubes and stored at −20° Celsius for no longer than 2 months before they were sent to the Department of Medical Sciences of Ministry of Public Health for DNA extractions performed by QIAamp DNA, Mini Kit (QIAGEN, Valencia, CA, USA). The quantitative DNA analysis was determined by the Nanodrop ND-1000 UV spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). The purity was determined by calculating the ratio of absorbance at 260–280 nm. These DNA samples were genotyped for their sex status and matched back to the reported gender for screening of the mishandling of samples prior to genotyping. The genotyping was done by the Illumina OmniExpressExome-8 v1.3 platform (Illumina, Inc., San Diego, CA, USA) at Center for Integrative Medical Sciences, Riken Institute.
Genotyping and statistical analysis
We included 377 neovascular AMD patients and 1074 controls who were genotyped by the InfiniumOmniExpressExome-8 v1.3 platform. Principal component analysis of the genotyped data was performed to assess the level of population stratification in the samples. The association analysis were analyzed with the ‘qtscore’ function in the GenABEL package.15 The minimum allele frequency of polymorphism in the analysis was 0.05. The genome-wide significance threshold was calculated for multiple testing by Bonferroni correction taking into account for the 958 497 SNPs that passed quality control (P<5.22 × 10−8). The quality of significant SNPs was evaluated after association analysis by investigating the intensity plots. The quantile–quantile plot was used to assess the overall quality of the GWAS. The required analysis was performed in R statistical environment.
Replication of GWAS findings
Non-synonymous coding SNPs, not previously reported to be associated with AMD but associated with function based on a customer-developed Exome Consortium database, were selected for replication. We included 222 independent cases of neovascular AMD and 623 independent controls. Selected patients were genotyped using the multiplex PCR-based Invader assay (Third Wave Technologies, Inc., Madison, WI, USA), a high-throughput SNP genotyping method developed under RIKEN Center for Genomic Medicine.16 All primer sequences can be provided upon request to the authors. Genotyping signals were detected by Sequence Detection System version 2.1, ABI7900 (Thomas Fisher Scientific, Waltham, MA, USA).
Results
Principal component analysis was performed to qualify the genetics matched between cases and controls. Principal component analysis plots and the lambda statistics suggested that genomic profiles are similar between cases and controls (Figure 1).
Plots of the first two principal components of neovascular AMD, disease controls and population controls. Principal component analysis (PCA) plot illustrating genetics matched between neovascular AMD cases (in dark blue), disease controls (pink) and population controls (in black). All individuals plotted are patients included in the discovery phase of the genome-wide study, all of Thai ancestry. This PCA plot suggests that genomic profiles are similar between cases and controls. A full color version of this figure is available at the Journal of Human Genetics journal online.
Genome-wide association discovery phase
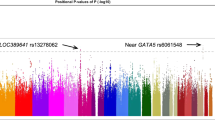
Findings from the genome-wide association data set of significant SNPs after association analysis were plotted on the quantile–quantile plot (Figure 2). The expected P-values of SNPs obtained from the GWAS for neovascular AMD showed no widespread deviation from the null hypothesis, suggesting minimal confounding disease association by population stratification biases. Therefore, no further correction for inflation was applied to the test statistics. Results of all SNPs from the discovery sample were summarized in Figure 3. The top 50 SNPs from the discovery phase were analyzed for allelic and genotypic associations (Supplementary Table 1).
Quantile–quantile (Q–Q) plot of P-values obtained from the genome-wide association study for neovascular AMD. Results of all SNPs are plotted in black. The solid line represents the null model where the observed Fisher’s exact test values matched those of the cases and controls (λ=0.978). A full color version of this figure is available at the Journal of Human Genetics journal online.
A Manhattan Plot of the genome-wide association study results of neovascular AMD patients. Summary of the genome-wide association study results of the discovery sample. P-values for genes that affect the risk of developing age-related macular degeneration (AMD). The blue horizontal line shows the cutoff for P=0.05, suggestive of association. The red horizontal line shows the genome-wide significance level of P<5.22 × 10−8. Statistical strength of association (-log10P) is plotted with each SNP in chromosomal order. A full color version of this figure is available at the Journal of Human Genetics journal online.
Association with previously identified neovascular AMD variants
Data from the discovery phase confirmed previously identified neovascular AMD variants in the ARMS2, HTRA1 and CFH loci reported by previous genetic association analysis in Asians6, 8, 13, 17 and our candidate genes including HTRA1 rs11200638 (P=5.47x10−17), ARMS2 rs10490924 (P=7.38 × 10−17), CFH rs10737680 (P=1.76 × 10−6) and CFH rs800292 (P=2.53 × 10−8). Though the Y402H (rs1061170) polymorphism was not included in the genotyping panel in this study, no significant association was found when analyzing the SNPs found to be in linkage disequilibrium (r2⩾0.9) with rs1061170.
We also analyzed 19 additional SNPs identified as significance in previous GWAS of AMD (typical CNV, PCV or dry AMD) including TNFRSF10A, C2-CFB-SKIV2L, C3, C6orf223, FGD6, SLC44A4, RAD51B, ADAMTS9, TIMP3, VEGFA, LIPC, CFI, COL10A1, IER3, SLC16A8, TGFBR1, B3GALTL, APOE-APOC1 and CETP.11, 17 According to our findings, none of these previously identified SNPs was associated with neovascular AMD based on the genome-wide significance level at 5.22 × 10−8 criteria (Supplementary Table 2). C6orf223 rs2295334, SLC44A4 rs12661281 and FGD6 rs10507047 previously reported to be associated with neovascular AMD in East Asia and Singapore was found to have a P-value of 0.3918, 0.6583 and 0.9336, respectively (Supplementary Table 2).
Association of neovascular AMD with variants not previously identified
In addition to confirming previously identified variants of neovascular AMD and PCV, we identified several SNPs of interest in new regions with the significance level at 10−4 in our discovery data set. We also found significant association at LINCO1317 rs6733379 (P=3.72 × 10−4) and rs2384550 (P=2.77 × 10−4).
Replication phase
The top 20 SNPs reported to have associated function were drawn from a customer-developed Exome Consortium database. (Supplementary Table 3) We performed replication of the two independent SNPs not previously implicated to contribute to the susceptibility of neovascular AMD but reported to have associated function and showed evidence of association surpassing the significance level at 10−4. Replication was done for LINCO1317 rs6733379 and rs2384550 on chromosome 12. Replication of the HTRA1 rs11200638, ARMS2 rs10490924 and CFH rs800292 loci were not performed owing to the significance of the initial association analysis reaching genome-wide significance level of P<5.22 × 10−8.
Though CFH rs10737680 did not pass the significance level, there has been many supporting evidences6, 7, 17 of its association with neovascular AMD, therefore, rs10737680 did not undergo replication as well. Results remained statistically significant after the replication having the significance level at 10−4 for LINCO1317 rs6733379. The second SNP, rs2384550, showed no significant association after replication (Table 1).
Discussion
Extensive genome-wide studies of AMD have been performed throughout many regions of the world.7, 8, 9 However, this genome-wide association analysis is the first study done in patients with neovascular AMD in Thais, a population in Southeast Asia. Two previously identified SNPs of neovascular AMD passed the GWAS significant threshold in this data set including ARMS2 rs10490924 and CFH rs800292 (Figure 4). These two SNPs are among the commonly reported variants of typical CNV and PCV in Asians6, 8 and Europeans.10, 14 Previous findings have suggested that some genetic loci conferring AMD susceptibility in East Asians and Singaporeans are shared with Europeans, yet AMD in East Asians and Singaporeans may also have a distinct genetic signature.17 No association was found in this study for the three reported loci (C6orf223 rs2295334, SLC44A4 rs12661281 and FGD6 rs10507047) of neovascular AMD in East Asia and Singapore where there is more Chinese compared with other countries in Southeast Asia. This may infer that the three new loci identified are specific to the East Asian population,17 not Southeast Asian populations. On the other hand, no studies have found significant association of the two loci (LINCO1317 rs6733379 and rs2384550 on chromosome 12) undergoing replication in this study. Though the function of LINCO1317 is unknown, the significance may be proof of variants associated with neovascular AMD specific to Thais. There may have been false-positive and false-negative results because only a small number of SNPs were carried over from the initial scan into the replication studies. SNPs that survive multiple rounds of replication are often not the most statistically significant associations in the initial scan.18 Therefore, more SNPs from the genome-wide discovery phase should be replicated.
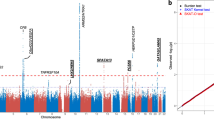
Regional association plots of significant single nucleotide polymorphisms (SNPs) for neovascular age-related macular degeneration (AMD) in the discovery samples. The most significant SNP located in ARMS2 region (a) corresponding with previous genome-wide findings. (b) Plot of significant SNPs for neovascular AMD located in The CFH region. SNPs are plotted as the –log10 of the P-value. A full color version of this figure is available at the Journal of Human Genetics journal online.
Genes (TNFRSF10A, C2-CFB-SKIV2L, C3, C6orf223, FGD6, SLC44A4, RAD51B, ADAMTS9, TIMP3, VEGFA, LIPC, CFI, COL10A1, IER3, SLC16A8, TGFBR1, B3GALTL, APOE-APOC1 and CETP) previously reported as being susceptible to neovascular AMD11, 13, 17 showed no association in the genotype of this population. The role of these genes in AMD may be less significant than those found from GWAS, which are insensitive to rare variants (minor allele frequency <0.05).11 A study including a larger sample size may be able to uncover these rare variants.
The potentially non-significant association of rs1061170, suggested by non-significant association of other SNPs found to be in LD with this SNP, in this study may be due to the low minor allele frequency of rs1061170 in the Thai and Asian populations. A previous comparative analysis of the rs1061170 between Thai and other Asian populations reported that rs1061170 was strongly associated with PCV in the Thai population but the frequency of the risk allele C in PCV was only 15.0% and 10.3% from the Thai and compiled Asian data, respectively, whereas European descents in the United States had a frequency of 49.1%.19 A GWAS of the Japanese population reported that typical neovascular AMD and PCV patients showed stronger association with ARMS2- HTRA1gene region than the CFH gene. The HTRA1 gene was shown to be in linkage disequilibrium with ARMS2.6 SNPs receiving the most significant association in this study was located predominantly on chromosome 10 (Supplementary Table 1), which contained both the ARMS2 and HTRA1 genes. Furthermore, the most significant SNP from the discovery phase was HTRA1 rs11200638, a SNP found to be associated with neovascular AMD in Asians.20 A meta-analysis based on Asian data could possibly yield more genetic risks as the estimated heritability of this disease is high. This study only examined a subtype of AMD, therefore, the results could not be completely compared with previous studies which included both neovascular and dry forms of AMD.7, 9 Studies separately analyzing variants associated with typical CNV and PCV would be useful in identifying distinct genotypes of these two phenotypes of neovascular AMD, which might further explain the difference in treatment responses of the two phenotypes. Previous problems addressed pertaining to the use of GWAS included the lack of interventions available to deal with the disease onset prediction. Although recent studies of gene therapy for neovascular AMD showed promising results,21 genetic variants discovered by genome-wide studies are many steps away from actual clinical application. The primary use for these studies are likely for investigations of biological pathways of disease causation,18 not for treatments.
Another limitation in this study is the lack of ocular examination of the population controls who had neither history of eye disease nor visual symptom and were presumed to have no neovascular AMD. However, some cases of neovascular AMD can have no history of eye disease and still maintain good vision despite having active lesions. Therefore, the possibility of having patients with neovascular AMD in the population controls cannot be ruled out completely. This may somehow cause a reduction of power for detecting the associations in this study. As AMD is relatively rare in Asian populations, however, only a slight reduction of the power may occur.
In conclusion, previously reported loci in the ARMS2, HTRA1 and CFH region were confirmed to be associated with neovascular AMD in this GWAS in the Thai population. Apart from proving that genetic loci conferring to the susceptibility of neovascular AMD in Thais were shared with other Asians and Europeans, we also identified a new locus of LINCO1317 rs6733379 not previously reported. These genotypic variations may be helpful in explaining the prevalence of different phenotypes of neovascular AMD presented in Europeans and Asians.
Although we attempted to classify cases in this study into typical CNV and PCV, the subgroups contained too few samples to draw any conclusions for differentiating genetic risks between CNV and PCV.
A larger sample size through expansion of samples enrollment and meta-analysis could possibly yield more genetic information especially for rare variants pertaining to the Thai population. Further separate studies for association of genetic variants of typical CNV and PCV could assist in the understanding of genetic risks of the two phenotypes leading to discovery of more appropriate treatments.
References
Koh, A., Lee, W. K., Chen, L. J., Hashad, Y., Kim, H., Lai, T. Y. et al. EVEREST study: efficacy and safety of verteporfin photodynamic therapy in combination with ranibizumab or alone versus ranibizumab monotherapy in patients with symptomatic macular polypoidal choroidal vasculopathy. Retina 32, 1453–1464 (2012).
Jenchitr, W., Ruamviboonsuk, P., Sanmee, A. & Pokawattana, N. Prevalence of age-related macular degeneration in Thailand. Ophthalmic Epidemiol. 18, 48–52 (2011).
Kumaramanickavel, G. Age-related macular degeneration: genetics and biology. Asia Pacific J. Ophthalmol. 5, 229–235 (2016).
Klein, R., Zeiss, C., Chew, E., Tsai, J., Sackler, R., Haynes, C. et al. Complement factor H polymorphism in age-related macular degeneration. Science 308, 385–389 (2005).
Seddon, J., Francis, P., George, S., Schultz, D., Rosner, B. & Klein, M. Association of CFH Y402H and LOC387715 A69S with progression of age-related macular degeneration. JAMA 297, 1793–1800 (2007).
Goto, A., Akahori, M., Okamoto, H., Minami, M., Terauchi, N., Haruhata, Y. et al. Genetic analysis of typical wet-type age-related macular degeneration and polypoidal choroidal vasculopathy in Japanese population. J. Ocul. Biol. Dis. Infor. 2, 164–175 (2009).
Neale, B. M., Fagerness, J., Reynolds, R., Sobrin, L., Parker, M., Raychaudhuri, S. et al. Genome-wide association study of advanced age-related macular degeneration identifies a role of the hepatic lipase gene (LIPC). Proc. Natl Acad. Sci. USA 107, 7395–7400 (2010).
Arakawa, S., Takahashi, A., Ashikawa, K., Hosono, N., Aoi, T., Yasuda, M. et al. Genome-wide association study identifies two susceptibility loci for exudative age-related macular degeneration in the Japanese population. Nat. Genet. 43, 1001–1004 (2011).
Cipriani, V., Leung, H. T., Plagnol, V., Bunce, C., Khan, J. C., Shahid, H. et al. Genome-wide association study of age-related macular degeneration identifies associated variants in the TNXB-FKBPL-NOTCH4 region of chromosome 6p21.3. Hum. Mol. Genet. 21, 4138–4150 (2012).
Fritsche, L., Fariss, R., Stambolian, D., Abecasis, G., Curcio, A. & Swaroop, A. Age-related macular degeneration: genetics and biolgy coming together. Annu. Rev. Genomic Hum. Genet. 15, 151–171 (2014).
Black, J. & Clark, S. Age-related macular degeneration: genome-wide association studies to translation. Genet. Med. 18, 283–289 (2015).
Chen, H., Liu, K., Chen, L. J., Hou, P., Chen, W. & Pang, C. P. Genetic associations in polypoidal choroidal vasculopathy: a systematic review and meta-analysis. Mol. Vis. 18, 816–829 (2012).
Ma, L., Li, Z., Liu, K., Rong, S. S., Brelen, M. E., Young, A. L. et al. Association of genetic variants with polypoidal choroidal vasculopathy: a systematic review and updated meta-analysis. Ophthalmology 122, 1854–1865 (2015).
AMD Gene Consortium Seven new loci associated with age-related macular degeneration. Nat. Genet. 45, 433–439 (2013).
Aulchenko, Y. S., Ripke, S., Isaacs, A. & van Duijn, C. M. GenABEL: an R library for genome-wide association analysis. Bioinformatics 23, 1294–1296 (2007).
Ohnishi, Y., Tanaka, T., Ozaki, K., Yamada, R., Suzuki, H. & Nakamura, Y. A high-throughput SNP typing system for genome-wide association studies. J. Hum. Genet. 46, 471–477 (2001).
Cheng, C.-Y., Yamashiro, K., Chen, L. J., Ahn, J., Huang, L., Huang, L. et al. New loci and coding variants confer risk for age-related macular degeneration in East Asians. Nat. Commun. 6, 6063 (2015).
Pearson, T. & Manolio, T. A. How to interpret a genome-wide association study. JAMA 299, 1335–1344 (2008).
Chantaren, P., Ruamviboonsuk, P., Ponglikitmongkol, M., Tiensuwan, M. & Promso, S. Major single nucleotide polymorphisms in polypoidal choroidal vasculopathy: a comparative analysis between Thai and other Asian populations. Clin. Ophthalmol. 6, 465–471 (2012).
DeWan, A., Liu, M., Hartman, S., Zhang, S., Liu, D. T. L., Zhao, C. et al. HTRA1 promoter polymorphism in wet age-related macular degeneration. Science 314, 989–992 (2006).
Constable, I. J., Blumenkranz, M. S., Schwartz, S. D., Barone, S., Lai, C.-M. & Rakoczy, E. P. Gene therapy for age-related macular degeneration. Asia Pacific J. Ophthalmol. 5, 300–303 (2016).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Journal of Human Genetics website
Rights and permissions
About this article
Cite this article
Ruamviboonsuk, P., Tadarati, M., Singhanetr, P. et al. Genome-wide association study of neovascular age-related macular degeneration in the Thai population. J Hum Genet 62, 957–962 (2017). https://doi.org/10.1038/jhg.2017.72
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2017.72
This article is cited by
-
The hypothetical molecular mechanism of the ethnic variations in the manifestation of age-related macular degeneration; focuses on the functions of the most significant susceptibility genes
Graefe's Archive for Clinical and Experimental Ophthalmology (2024)
-
HTRA1 rs11200638 variant and AMD risk from a comprehensive analysis about 15,316 subjects
BMC Medical Genetics (2020)
-
Analysis of genetic polymorphisms for age-related macular degeneration (AMD) in Chinese Tujia ethnic minority group
BMC Medical Genetics (2019)