Abstract
The bacterial communities in the Laccaria proxima mycosphere (soil from beneath fruiting bodies) and the corresponding bulk soil were compared by cultivation-dependent and cultivation-independent methods. To assess the distribution of type III secretion systems (TTSS), a PCR-based system for the broad detection of a highly conserved gene involved in TTSS, that is hrcR, was developed and used to assay the cultured bacteria from the L. proxima mycosphere and surrounding bulk soil. PCR-DGGE based on the 16S ribosomal RNA gene showed the selection of presumably mycosphere-specific bacterial groups in the mycosphere of L. proxima compared to the bulk soil in 3 sampling years. Moreover, plate counts revealed that the numbers of culturable heterotrophic bacteria were increased in the mycosphere as compared to the bulk soil. Strikingly, the percentage of randomly picked isolates that carried the hrcR gene showed a significant increase, from 2.8 in the bulk to 13.4 in the mycosphere soil. The increase could be mainly attributed to the emergence of a hrcR positive Pseudomonas fluorescens, denoted BS053, which constituted the most dominant species in the culturable mycosphere communities. This organism was, together with a hrcR-positive Burkholderia terrae BS110, exclusively found in mycosphere soil. Direct detection of hrcR genes using a cultivation-independent approach showed the selection of several hrcR gene types uniquely in the mycosphere, indicating the selection of several TTSS-harboring bacterial species. Thus, different bacteria were found to be enriched in the L. proxima mycosphere and TTSS can be involved in some of the interactions with the fungal host.
Similar content being viewed by others
Introduction
Bacteria and fungi play important roles in the functioning of soil ecosystems. For instance, either one or both of the two groups can be involved in the degradation of lignin and lignocellulose, the introduction and transport of carbon and other nutrients, the secretion of antibiotics, mineral weathering, organic matter decomposition and in changing pH and soil structure (van Breemen et al., 2000; van Elsas et al., 2006a). A fungal group important for agriculture, forestry and environmental quality is formed by the mycorrhizal fungi, as these fungi can establish symbioses with plants and thus form ‘bridges’ between the bulk soil and the (energy-rich) roots of plants.
In spite of the fact that soil bacteria and fungi have coexisted for a long time in the soil ecosystem, we hitherto understand very little about their putative interactions. These interactions may be most apparent in the mycosphere, that is the narrow zone of soil which is strongly affected by the fungal hyphae (Frey et al., 1997; Johansson et al., 2004; De Boer et al., 2005; Frey-Klett et al., 2005). Bacteria may have evolved in such a way that they can benefit from soil fungi, for example by using the chitin (or other components) of fungal cell walls as an energy and/or carbon source, like recently shown for Collimonas fungivorans in its interaction with Fusarium spp. (De Boer et al., 2004). In other interactions between bacteria and fungi, the two microbial groups appear to benefit from each other (Garbaye, 1994; Johansson et al., 2004; van Elsas et al., 2006b). In such bacterial–fungal interactions, physical contacts between the bacterial cells and the fungal hyphae are often detected (Garbaye, 1994; Toljander et al., 2006), and it is, therefore, likely that these are actually required in the interaction. However, the mechanisms and consequences of the binding of bacterial cells to fungal hyphae are mostly unknown.
A dense network of fungal hyphae is known to be present directly below the fruiting bodies (mushrooms) of mushroom-forming fungi, where a clear soil/fungal tissue interface can be found. The putative influence of the fungal fruiting body on the soil bacterial community is supposedly strongest in this microhabitat (Rangel-Castro et al., 2002) and novel bacterial–fungal interactions can be suspected to exist. Given the paucity of information on the selective processes exerted by fungal fruiting bodies on soil bacteria, we selected these microhabitats as the prime habitats to interrogate in respect of the putative fungal-selective effects on soil bacterial populations.
The highly-evolved type-III secretion system (TTSS), which is often found in bacterial pathogens of both animals and plants, functions as a molecular syringe to introduce effector proteins into the cytoplasm of these eukarya (He et al., 2004). To function, the system is dependent on the physical contact between its host cell and a prey cell. In successful interactions, the effector proteins often interfere with prey cell functions in a way that is beneficial for the prokaryotic partner. TTSSs are widespread among pathogenic and symbiotic proteobacteria like Yersinia spp., Shigella flexneri, Salmonella spp, enteropathogenic Escherichia coli (EPEC), Pseudomonas spp, Bordetella spp., Rhizobium spp., Erwinia spp., Xanthomonas spp., Ralstonia solanacereum, Burkholderia spp., Soldalis spp. and Chlamydia spp. (Hueck, 1998; He et al., 2004; Mazurier et al., 2004, 2006; Rezzonico et al., 2005). Although most of these bacteria exhibit a pathogenic interaction with their host, not all TTSS-positive bacteria are involved in pathogenic relationships. For example, the exact role of TTSS in Rhizobium species is unknown, but the system is certainly necessary in the symbiotic interaction of this organism with its host plant (Marie et al., 2001). An involvement of TTSS in bacterial interactions with fungi has, so far, not been described. We surmised that soil bacteria might make use of this mechanism to interact, in a parasitic/predatory or even mildly commensalistic manner, with fungal hosts that invade their local microhabitats. In this study, the presumptive presence of a TTSS was detected on the basis of the PCR-based detection of a (marker) gene, hrcR. This gene was found to be ‘omnipresent’ in TTSS-containing bacteria based on sequence retrieval and analyses from the NCBI database, and it is in fact the most conserved gene in TTSS (Kaneko et al., 2000; Frederick et al., 2001; Buell et al., 2003; Holden et al., 2004; Feil et al., 2005; Lee et al., 2005).
The aims of this study were as follows: (1) to assess whether the bacterial community in soil is affected by the presence of the ectomycorrhizal fungus Laccaria proxima, and (2) to interrogate the distribution of TTSS in this bacterial community in comparison to the bulk soil. The soil below the feet of naturally growing L. proxima fruiting bodies (mushrooms) was used to access the mycosphere of L. proxima. We assessed the structures of the bacterial communities in these natural mycospheres in comparison with those in the corresponding bulk soils. We also determined the prevalence and presumed nature of the TTSS systems that were locally present on the basis of the hrcR-based molecular marker.
Materials and methods
Sampling locations and soil properties
Different L. proxima fruiting bodies identified by morphology and color were sampled in October of the years 2004, 2005 and 2006 in a deciduous forest in Gieterveen, The Netherlands (52°01′07.39″N, 6°49′54.32″E). Per year, three independent fruiting bodies were sampled, thus representing three replicates per sampling. These replicates occurred in close proximity (<2 m) to hazel trees (Corylus avellana). The samples were obtained by carefully collecting each complete fruiting body with the soil adhering to the hyphal network under the feet (depth 0 to 5 cm). The distance between independent fruiting bodies was at least roughly 1 m. As controls, samples were taken from surrounding topsoil (40 cm lateral distance, 0 to 5 cm depth and one sample per fruiting body) in areas where no fruiting bodies had developed. The sandy soil had a pH of 4.5 and total carbon (C) and total nitrogen (N) contents of 2.8% and 0.8%, respectively.
Identification of ectomycorrhizal fruiting bodies via ITS region sequencing
DNA isolation from selected fruiting bodies was performed using a purification kit for the isolation of DNA from plant material (Promega, Madison, WI, USA) according to the manufacturer's protocol. The internal transcribed spacer (ITS) region between the 18S and 25S ribosomal RNA genes was amplified by PCR, using primer pair ITS1F and ITS4B (Table 1) (Gardes and Bruns, 1993). PCR was performed using the GeneAmp PCR System 9700 (Applied Biosystems, Foster City, CA, USA). The 25 μl PCR mix consisted of 2.5 μl of PCR buffer (Roche, Basel, Switzerland), 200 μM of each deoxynucleoside triphosphate, 200 nM of each primer, 2% DMSO (dimethyl sulfoxide), 0.06 U Taq DNA polymerase (Roche), 19.7 μl H2O and 10 to 50 ng of DNA (1 μl) from the isolation. The following PCR program was used: 94 °C for 3 min; 94 °C for 1 min, 50 °C for 1 min, 72 °C for 3 min (30 cycles); 72 °C for 10 min (1 cycle). The PCR product (0.8 kb) was visualized on a 1% agarose gel to asses size and quality. It was then ligated in the pGEM-T easy vector (Promega) and introduced into E. coli MM294 competent cells (Sylphium Life Sciences, Groningen, The Netherlands). Selected clones were sequenced to identify the ectomycorrhizal fungus. Sequence analyses were performed on clones obtained from three fruiting bodies and identification supported morphological classification.
Sampling of bulk and mycosphere soil
Following removal of loosely attached soil by tapping and careful rinsing with sterile water, tightly adhering soil was obtained from the foot of each of the collected L. proxima fruiting bodies. The fruiting body part was then removed by cutting it off from the remaining (aggregated) soil sample. Using a binocular, we found that the resulting soil sample was heavily under the influence of fungal hyphae (due to the hyphal density) and, thus, could be flagged as the mycosphere. This sample (approximately 0.5 g soil per fruiting body) was used for further processing. The corresponding bulk soil samples obtained were used directly for further processing. The mycosphere and bulk soil samples were tenfold diluted (0.5 g per 5 ml) in sterile 0.85% NaCl. The resulting suspensions were then homogenized by mixing on a Vortex shaker (full speed, three times for 1 min each) followed by a 30 s treatment in an ultrasonic bath (±41.7 kHz). Following this treatment, soil particles were allowed to settle for 1 min on the bench, after which the supernatant was used for plating as well as direct DNA isolation.
Isolation of DNA from bulk and mycosphere soil
Total soil DNA was isolated from all samples in all sampling years with the UltraClean Soil DNA isolation kit (MoBio Laboratories, Carlsbad, CA, USA, catalog number 12800-100) according to the manufacturer's protocol. DNA purification was based on the Wizard DNA Clean-Up System kit (Promega, catalog number 225976), which was used according to the manufacturer's protocol to remove the remaining humic acids. The quality and quantity of the DNA were checked on 1% agarose gels. In all cases, about 5 μg DNA (100 ng μl−1) of 10 kb average fragment size was obtained. The isolated DNA was directly used for PCR-based assessments.
Analysis of bacterial community structure with PCR-DGGE
All bulk and mycosphere soil DNA samples (years 2004 to 2006) were amplified with bacterial primers F968-GC and 1378R (Table 1) (Nübel et al., 1996; Heuer and Smalla, 1997). PCR product (about 450 bp in size) was formed on the GeneAmp PCR System 9700 (Applied Biosystems). The 50 μl PCR mix consisted of 5 μl 10 × Stoffel buffer (Roche), 200 μM of each deoxynucleoside triphosphate, 400 nM of each primer, 1% formamide, 0.05 U Stoffel Taq DNA polymerase (Roche), 31 μl H2O and 1 μl (20 ng) bulk or mycosphere soil DNA. The following touchdown PCR program was used: 94 °C for 3 min; 94 °C for 1 min, 65 °C (decrease per cycle 1 °C) for 1 min, 72 °C for 3 min (10 cycles); 94 °C for 1 min, 55 °C for 1 min, 72 °C for 3 min (35 cycles); 72 °C for 30 min. In addition, a nested Pseudomonas-specific PCR system (Costa et al., 2006) was used to assess the distribution of pseudomonads in the bulk and mycosphere soil samples, in accordance with the authors. In short, primers F311Ps and R1459Ps (Table 1) were used to amplify Pseudomonas sp. 16S rRNA genes, which was followed by a PCR with primers F968-GC and 1378R (Table 1) as described above. PCR products were checked for quantity and quality by electrophoresis in 1% agarose gels. PCR products were used in denaturating gradient gel electrophoresis (DGGE) analyses.
Denaturating gradient gel electrophoresis analyses were performed on a PhorU2 system (Ingeny International, Goes, The Netherlands) according to the manufacturer's protocol, using a gradient (Muyzer et al., 1995) of 40% to 70% denaturants (ureum/formamide) (100% denaturants is 8 M urea plus 40% formamide), a buffer temperature of 60 °C and 110 V for 18 h. Following electrophoresis, the polyacrylamide gels were stained using silver staining (Heuer et al., 2001).
High-throughput growth of bacteria and isolation of DNA
For the 2004, 2005 and 2006 samples, bacterial suspensions from mycosphere and bulk soil samples were dilution-plated, in triplicate, on R2A (Becton Dickinson Company, Sparks, MD, USA) plates (1.5% agar) and incubated at 22 °C until no new colonies were formed (approximately 2 weeks). Following incubation, the colonies were enumerated and counts of colony forming units (CFU) per gram of dry bulk or mycosphere soil were, thus, obtained.
Randomly picked colonies from the 2004 samples (the 105 dilution plate was used for all samples) were transferred to 200 μl R2A liquid medium contained in 96-well microtitre plates. Plates were incubated for 7 days at 22 °C. DNA was then isolated from each well based on the protocol of Klimyuk et al. (1993). Briefly, the 96-well plates were spun for 10 min at 4500 g, the supernatant was discarded from each well and 200 μl 0.25 M NaOH was added per well. After this, the mixtures were boiled for 1 min and neutralized with 200 μl of 0.25 M HCl. Then, 100 μl of 0.5 M Tris-HCl (pH 8.0) with 0.25% Nonidet P-40 was added, after which the mix was boiled for 3 min and spun for 5 min at 4500 g. Aliquots from the resulting supernatant were used for further PCR analyses.
HrcR gene detection
A detection system for the presumed presence of TTSS was developed on the basis of the conserved hrcR gene of the TTSS machinery using known hrcR-like sequences extracted in 2004 from the NCBI database. We found that genes with similar function in the TTSS machinery and sequence similar to that of the hrcR gene of R. solanacearum biovar 1 (GMI1000) have different nomenclature in different bacterial strains. For instance, pscR, spaP, spa24 and escR were hrcR homologs in P. aeruginosa, Salmonella sp., Shigella sp. and enteropathogenic E. coli (EPEC), respectively (He et al., 2004). These hrcR-like genes are commonly found in combination with other genes of the TTSS machinery, as evidenced on the basis of TTSS sequence retrieval from the NCBI database (Kaneko et al., 2000; Frederick et al., 2001; Buell et al., 2003; Holden et al., 2004; Feil et al., 2005; Lee et al., 2005). Thus, detection of a hrcR-like gene seems to be a good indication for the presence of the TTSS machinery in any bacterial strain. The sequences of 30 hrcR-like genes (Figure 1) extracted from the NCBI database and representing a wide variety of soil bacteria, plant and animal pathogens were aligned using MEGA 4 (Kumar et al., 2004). Two primers, denoted hrcRF (basepair position 125 of the R. solanacearum GMI1000 hrcR gene) and hrcRR (basepair position 597 of the GMI1000 hrcR gene) (Figure 1), were then designed on the basis of conserved sequences in this gene. Following extensive testing, an optimized PCR protocol was developed, as follows. A 15 μl PCR mix was established consisting of 1.5 μl 10 × PCR buffer (Roche), 200 μM of each deoxynucleoside triphosphate, 20 nM of each primer, 2% of DMSO, 0.06 U Taq DNA polymerase (Roche), 11.4 μl H2O and 1 μl template DNA (10 ng). The following PCR program was used: 94 °C for 3 min; 94 °C for 45 s, 56 °C for 45 s, 72 °C for 25 s (30 cycles); 72 °C for 10 min. The roughly 460-bp PCR products were visualized following electrophoresis on 1% agarose gels. Further, the specificity of the PCR products obtained was confirmed by cloning/sequencing and BLAST analyses.
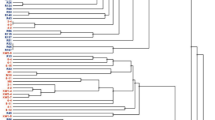
Clustering of hrcR-like genes from selected bacterial strains using the program MEGA, version 4 (Kumar et al., 2004), option Neighbor Joining (bootstrap 400 replicates). Sequences were extracted from NCBI in October 2004 and May 2007. Accession numbers are given behind strain names (brackets). Annealing sites of primers hrcRF (base position 125) and hrcRR (base position 597) are shown. Between the primers, a 410-bp region of low conservation is present. On the basis of database analyses in 2004, primers were designed to include degeneracies (F: one position and R: three positions). In the 2007 analysis, new hrcR type sequences were found, which revealed some mismatches in the primer annealing regions, indicating that possibly not all TTSS-harbouring bacteria are detected by the 2004 primers. Grey boxes: presumably soil/plant associated bacteria, not boxed: other bacteria, mainly consisting of animal-associated ones.
Screening for hrcR-positive bacteria
Randomly picked bacterial isolates from the 2004 L. proxima mycosphere (151 isolates) and corresponding bulk soil samples (288 isolates) were screened for the presence of hrcR using the specific PCR-based detection system.
Detection of hrcR genes in mycosphere and bulk soil
Using mycosphere and bulk soil DNA, hrcR specific PCR was applied followed by DGGE and clone library analyses. Thus, primer pair hrcRF/hrcRR was used for the hrcR clone library, whereas for DGGE, the primer hrcRR was coupled to a GC clamp at the 5′ end (Table 1). The 50-μl PCR mixtures consisted of 10 μl 5 × PCR buffer H (PCR optimizer kit, Invitrogen, Carlsbad, CA, USA), 200 μM of each deoxynucleoside triphosphate, 20 nM of each primer, 2% DMSO, 0.06 U Taq DNA polymerase (Roche), 30.9 μl H2O and 1 μl template DNA (10 ng). The following PCR program was used: 94 °C for 3 min; 94 °C for 45 s, 60 °C for 45 s, 72 °C for 45 s (35 cycles); 72 °C for 10 min. PCR products (470 bp) were visualized on 1% agarose gels, to asses size and quality. They were then used in DGGE analyses (see above) or in clone library construction with the pGEM-T easy vector system (Promega; see below)
Genomic fingerprinting of isolates
All bacterial isolates from the mycosphere and bulk soil samples obtained in 2004, as described in the foregoing, were analyzed by genomic fingerprinting using primer GTG-5 (Table 1) (Versalovic et al., 1994). The PCR products were visualized following electrophoresis on 1% agarose gels and digitized for further statistical analysis by Gelcompar II (see further).
Clone library construction and sequencing
PCR products (16S ribosomal RNA gene, hrcR gene and fungal ITS region) were ligated into the pGEM-T easy vector (Promega) according to the manufacturer's protocol. The ligation products were introduced into E. coli MM294 competent cells (Sylphium Life Sciences) by transformation according to the supplier's protocol, after which cells were plated onto selective media. Following growth at 37 °C, white colonies were randomly picked, and plasmids with inserts of the correct size were isolated using the cetyl trimethyl ammonium bromide isolation method (Delsal et al., 1989). Sequencing reactions were performed on the plasmid material according to the PE ABI Prism protocol (Applied Biosystems) in one direction with the pGEM-T-based primer SP6. Sequence runs were done on an ABI377 DNA sequencer (Applied Biosystems). The sequences obtained were checked by a BLAST search in the NCBI database to identify the gene or isolate sampled. All new sequences generated in this study were deposited in GENBANK under accession numbers EU567676-EU567703 and EU579294-EU579349.
HrcR, hrcS and hrcT detection in P. fluorescens
The validity of using the hrcR gene as a molecular marker for the TTSS machinery was confirmed using a hrcR-positive P. fluorescens isolate from the mycosphere with the Pseudomonas-specific TTSS detection method described by Mazurier et al. (2004). Briefly, the detection system identifies the TTSS genes hrcR, hrcS and hrcT of a wide range of pseudomonads. Gene hrcR detection indeed coincided with positive detection of the three genes, as evidenced by sequencing of the respective PCR products.
Flexirubin pigment identification test
The nature of the pigment of conspicuous yellow colonies obtained from the mycosphere was identified using the test described by Gude (1980). The response was scored as positive if the color changed from yellow to red after dropping 20% KOH on a colony formed on R2A medium. A representative of each yellow-pigmented group (by GTG-5 fingerprinting) was tested and the results were then extrapolated to all members of the same genomic fingerprinting group.
Statistics and gel pattern analyses
Bacterial counts (CFU) were log transformed and used for further comparative analyses. Log CFU and percentages of hrcR-harboring bacteria were analyzed for significance with the Mann–Whitney U-test by using the Statistical Product and Service Solutions for Windows, Rel. 14.0. (SPSS Inc., Chicago Ill, USA). All statistical analyses were performed using as a criterion P⩽0.05.
The DGGE and GTG-5 genomic fingerprinting patterns were evaluated with the program Gelcompar II (Applied Maths, Sint-Martens-Latem, Belgium). Clustering was done using the unweighted pair group method with mathematical averages (UPGMA; Sneath and Sokal, 1973). The Jaccard index was used for clustering hrcR PCR-DGGE profiles with a position tolerance of ±0.5%. The Shannon–Weaver diversity indices, based on the surface area of each peak extracted from Gelcompar (DGGE), or based on the number of members of each clustered bacterial group (GTG-5 genomic fingerprinting), were used to determine the influence of L. proxima on the diversity of the dominant members of the soil bacterial community within each year. In addition, the hrcR sequences generated from the hrcR clone library were analyzed and clustered with the program MEGA 4 (Kumar et al., 2004).
Results
Development of a PCR-based broad hrcR detection system
All ‘hypersensitivity response and conserved’ (hrc) genes (hrcS, hrcR, hrcQ, hrcV, hrcU, hrcJ, hrcN, hrcT and hrcC) of the TTSS machinery from R. solanacearum GMI1000 (Salanoubat et al., 2002) were tested for conservation among different bacterial strains. We found that only the hrcR gene was truly conserved among the bacterial species used in the analysis, that is, members of the genera Burkholderia, Xanthomonas, Erwinia, Pseudomonas, Yersinia, Salmonella and Aeromonas. Clustering of the hrcR-like sequences showed a clear separation between presumed soil/plant- and animal-associated bacteria (Figure 1).
The alignment of the hrcR-like sequences of 39 selected strains further showed the existence of two small regions with conserved sequence across all hrcR-like genes. Moreover, these regions showed a similar separation in sequence between bacteria that are presumably associated with soil and/or plant environments (Figure 1; upper broad cluster) and those mainly associated with animals (lower two broad clusters). This study focused on hrcR-like genes typical for organisms loosely grouped as ‘soil/plant-associated’ bacteria, as these were primarily expected to appear in the mycosphere. Thus, forward primer HrcRF and reverse primer HrcRR (Table 1) were based on these conserved regions (Figure 1). To allow recognition of all soil/plant-associated hrcR-containing strains, both primers were equipped with ambiguous bases.
The specificity of the detection system based on these primers was assessed by (1) BLAST analyses of the primer sequences, and (2) testing of a suite of different bacterial strains. The first analysis revealed that, on a theoretical basis, the TTSS of most soil/plant-associated bacterial strains (17 of 22 species) was detectable with our detection system (Figure 1), whereas detection of the other five strains was doubtful due a mismatch in the second base in one primer. Moreover, 5/15 of the animal-associated strains were, on a theoretical basis, detectable, whereas detection of the other animal-associated strains was questionable due to a mismatch similar to the above. The hrcR detection system was further experimentally tested against a range of strains from our culture collection (Table 2). It produced the expected fragment of about 460 bp for all soil/plant-associated strains that were expected to be TTSS-positive, whereas strains that were expected to lack TTSS did not yield any amplicons. In addition, the TTSS-positive (animal) strain P aeruginosa tested negative with our system, corroborating our in silico findings based on the sequences retrieved from the NCBI database.
Sampling and identification of the ectomycorrhizal fungus
Sequence analyses of the ITS region of the fruiting bodies sampled in 2004 and 2005 showed closest similarity (99%) with the ITS region of L. proxima in the NCBI database. This identification was confirmed by the color (reddish brown) and morphological properties (size, shape, surface texture and spore gills) of the fruiting bodies sampled. The fruiting bodies, of completely similar appearance, and collected at the same location in 2006 were identified by color and morphological characteristics only.
Total bacterial, as well as Pseudomonas, community analysis via PCR-DGGE
Molecular (cultivation-independent) analysis of the total bacterial community was performed in three consecutive years. In all years, there were great similarities between the profiles generated for the three replicate mycosphere soils on the one hand and the three corresponding bulk soils on the other hand. Moreover, these two sample groups showed clear differences between them, yielding separate clusters (for example, Figures 2a and b). For reasons of consistency with the cultivation-dependent data, we further show an in-depth analysis of the 2004 samples. Visual inspection of the bulk and mycosphere-soil-derived DGGE profiles revealed the presence of 39±1.5 and 41±2.6 bands, respectively. Hence, there was no significant difference (P>0.05, t-test) in the numbers of bands between mycosphere or bulk soil samples (Figure 2a). The Shannon–Weaver diversity indices for these bulk and mycosphere soil samples were also not significantly different (P>0.05), with averages of 2.6±0.38 and 2.7±0.17, respectively. However, the patterns differed quite strongly, as several of the intense bands in the mycosphere soil derived DGGE patterns were less intense in, or completely absent from, the bulk soil-derived patterns (Figure 2a). Moreover, some major bands prevalent in the bulk soil samples were absent from the profiles generated from the mycosphere soil. Clustering of the profiles, thus, revealed two broad clusters, separated by habitat, that is, mycosphere versus bulk soil (Figure 2b). The similarity levels between all bulk and mycosphere soil-derived patterns were 82% and 81%, respectively. In contrast, the similarity between, on the one hand, the bulk soil and, on the other hand, the mycosphere soil derived patterns was only 38% (Figure 2b). Hence, the mycosphere—defined by the feet of the L. proxima fruiting bodies—consistently selected bacterial communities in soil that were different from those in the corresponding bulk soil. The selective effect leading to such differences occurred in all sampling years, although we did not obtain precisely the same patterns (data not shown).
Denaturating gradient gel electrophoresis (DGGE) of the bacterial diversity in mycosphere and bulk soil. (a) Analysis of three replicates of bulk (S1–S3) and Laccaria proxima mycosphere soil (M1–M3) collected in October 2004. (b) UPGMA-based clustering using Gelcompar version 4.1 (Applied Maths). Marker: 16S ribosomal RNA gene fragments of (from top to bottom): Listeria innocua, Enterobacter cloaceae, Mesorhizobium sp., Burkholderia cepacia and Arthrobacter sp.
In the light of the putative selection of pseudomonads (see further), we ran the Pseudomonas-specific PCR-DGGE system on the 2004 bulk and mycosphere soil samples. Overall, all Pseudomonas communities were rather simple (Figure 3). Strikingly, several specific Pseudomonas types were found in the L. proxima mycosphere, which were not detected in any of the corresponding bulk soil samples (Figure 3). This indicated the consistent selection of specific pseudomonads in the mycosphere.
Denaturating gradient gel electrophoresis (DGGE) of Pseudomonas diversity in mycosphere and bulk soil. Three replicates of bulk (S1–S3) and the Laccaria proxima mycosphere soil (M1–M3) collected in 2004. Pseudomonas fluorescens BS053 isolate from the culturable fraction from the mycosphere (arrows). Marker: 16S ribosomal RNA gene fragments of (from upper top to bottom): Mesorhizobium sp., Burkholderia cepacia and Arthrobacter sp.
Analysis of culturable bacteria in the mycosphere and bulk soil
On average, the bacterial colony counts on R2A agar showed a trend toward increased numbers of culturable bacteria in the mycospheres as compared to the bulk soils in all sampling years (Figure 4). However, this increase was only significant (T-test, P<0.05) in sampling year 2006, that is, log 7.1 (±0.23) CFU g−1 were measured in the mycosphere versus log 6.4 (±0.66) CFU g−1 in the corresponding bulk soil.
We further observed a dominance among the mycosphere soil isolates of colored colonies (mainly yellow), which consistently ranged from 12% to 16% of the total. This apparent selection of colored colonies was seen in all sampling years and in all sampled fruiting bodies, and they were completely absent from the bacterial communities obtained from the bulk soil samples.
All 439 bacterial isolates from the 2004 mycosphere (151) and bulk soil (288) samples were grouped using GTG-5 fingerprinting analyses. The mycosphere and bulk soil isolates clustered into 85 and 128 GTG-5 groups (or singletons), respectively (data not shown). The Shannon–Weaver indices (1.75 for mycosphere and 2.00 for bulk soil) indicated the existence of a lower diversity in the culturable fraction of the mycosphere soil samples as compared to that of the bulk soil. Fourteen mycosphere-specific groups (encompassing 73% of all isolates), most of which contained at least four members (and which were not found in the bulk soil) were identified via partial 16S ribosomal RNA gene sequencing of one to three representatives. Table 3 shows an overview of these data. The analysis showed that the three GTG-5 groups that contained all pigmented colonies encompassed three species of the genus Chryseobacterium (14% of the total; Table 3). Using three representative isolates (one per group), the yellow pigment in these groups was identified as flexirubin (Gude, 1980). The most abundant species found in the culturable bacterial fractions of the mycosphere, which was not detected in corresponding bulk soil, was affiliated with P. fluorescens (11.0%). However, neither this organism, nor any other one identified (Table 3), could be verified to be a dominant member of the total bacterial communities, as evidenced by comparison of their DGGE products with those generated directly from the mycosphere (data not shown). On the other hand, using this analysis, the P. fluorescens isolate, denoted BS053, was presumptively identified as a dominant member of the Pseudomonas communities in two of the three mycosphere samples via its dominance in the respective Pseudomonas-specific DGGE patterns (Figure 3).
Incidence of hrcR in the culturable bacterial fractions of mycosphere and bulk soils
All isolates from the 2004 mycosphere (151) and bulk soil (288) samples were screened via PCR for the presence of the hrcR gene. Overall, 19/151 different isolates from the mycosphere and 8/288 from the bulk soil were shown to be hrcR-positive. Thus, a significant increase (Mann–Whitney U-test, P<0.05) was seen in the average fractions of hrcR-positive isolates in the mycosphere soil (12.6%±6.8%) in comparison to that in the bulk soil (2.8%±2.3%). The diversity of bacterial species within the hrcR-positive strains was, however, low. Among the hrcR-positive strains from mycosphere soil, an organism affiliated with P. fluorescens (16S ribosomal RNA gene sequencing, 99% similarity) was found to dominate (17 isolates, estimated at 8.0 × 105 CFU g−1 dry soil). This P. fluorescens BS053 strain encompassed 89% of all hrcR-positive isolates obtained from the mycosphere. Two other hrcR-positive organisms found to occur in the mycosphere (both estimated at 5.1 × 104 CFU g−1 dry soil) were identified as B. terrae (16S ribosomal RNA gene sequencing, 99% similarity) and B. cepacia (99%). All hrcR-positive strains obtained from bulk soil belonged to just two species of the genus Burkholderia, namely B. cepacia (3 × 104 CFU g−1 dry soil; 99%) and B. phenazinium (6 × 104 CFU g−1 dry soil; 99%).
Occurrence of hrcR in mycosphere and bulk soil via molecular detection
In total, 50 hrcR sequences were obtained each from the 2004 mycosphere and bulk soil samples via analysis of the hrcR clone library. All sequences indeed were identified as hrcR-like genes by BLAST-N comparison with entries in the NCBI database.
Clustering analysis (Mega 4, Kumar et al., 2004) showed the existence of three distinct groups (Figure 5). Two groups, denoted A and B, were apparently specific for the mycosphere, although a small bulk soil subcluster was found within group B. The third group, C, consisted of a mixture of both mycosphere and bulk soil derived sequences. The group A sequences were related (81% similarity) to a hrcR-like gene of P. aeruginosa UCBPP-PA14, the group B sequences were related (87% similarity) to a hrcR-like gene of P. syringae pv. tomato DC3000 and those of group C were most related (80% similarity) to the hrcR-like gene SctR of B. thailandensis E264.
Clustering of hrcR-like genes from total bacterial communities of the Laccaria proxima mycosphere (codes with M) and bulk soil (codes with S). Bootstrap values below 50 are not shown. Strains indicated by name/code (accession number). Pseudomonas fluorescens BS053 and Burkholderia terrae BS110: own isolates described in this study.
A comparison of sequences of isolates and the library showed that the hrcR-like gene of the mycosphere-specific P. fluorescens BS053 strain clustered in group B, whereas that of the B. terrae BS110 strain clustered within group A.
HrcR specific PCR-DGGE analyses
The hrcR specific PCR-DGGE profiles (Figure 6) were visually consistent across, on the one hand, the mycosphere soil samples and, on the other hand, the bulk soil samples. Two clusters, in line with habitat, thus emerged. Analyses of the profiles with GelcomparII confirmed this observation, primarily showing strong clustering of the mycosphere soil derived profiles in relation to the bulk soil derived ones.
Differences between the profiles were mainly seen in band prevalence, as a larger degree of relative dominance was observed in the mycosphere than in the bulk soil samples. Moreover, absence of certain bands was observed in the mycosphere soil compared to the bulk soil samples. These observations indicate the occurrence of selection/deselection processes in bacterial communities in the mycosphere.
Discussion
L. proxima is a common ectomycorrhizal symbiont in deciduous and coniferous tree species in temperate climate ecosystems. In spite of the fact that bacterial–fungal interactions play an important role in, for example, mycorrhization processes (Frey-Klett et al., 2005), the interaction of this fungal partner with soil bacteria is virtually unknown. We hypothesized that the process of fruiting body formation of ectomycorrhizal fungi may result in changes of local conditions in soil, which may lead to enhanced ecological opportunities for local bacterial inhabitants. We thus set out to sample soil at the feet of fruiting bodies—here called the ‘mycosphere’—of L. proxima in the field in three consecutive years in a plot close to hazel trees. The samples consisted of a dense network of hyphae throughout the top soil. We opted for this type of sample, as it allowed a robust definition of the mycosphere as the soil zone under strong influence of dense fungal hyphae, and a clear-cut comparison with corresponding bulk soil. Another common sampling method of ectomycorrhizal fungal tissue in soil is the collection of plant root tips colonized by fungal tissue. However, in that strategy the direct influence of roots—via root exudates—on the bacterial community must also be taken into account and, hence, it is difficult to discern a fungus-specific effect (Frey-Klett et al., 1999). Taking fungal hyphae out of soil with tweezers under a binocular offers another possibility, but this method is time-consuming and is only useful for a few fungal species, as it depends on the width and strength of the hyphal structures.
In this study, the mycosphere of naturally growing L. proxima was shown to indeed exert a selective effect, in three consecutive years, on the bacterial populations that are locally present. This evidence was primarily based on a comparison of bacterial PCR-DGGE profiles between mycosphere and corresponding bulk soil samples. Specifically, and in spite of the fact that the apparent diversity of the bacterial communities was not strongly shifted, the clustering of the DGGE profiles of these two habitats showed the emergence of two major groups—defined by habitat—with consistently only low similarity between them. Thus, the emergence of the L. proxima hyphal network did not result in a clear change of the apparent diversity of the dominant bacterial phylotypes in soil, but it did induce a strong shift in the composition of these communities. The influence of the hyphal network at the base of fungal fruiting bodies on the local bacterial community structure in natural soil has, as far as we know, never been described in detail by a fingerprinting technique such as DGGE. Most research in this area so far has focused on specific groups of culturable bacteria selected under the influence of a certain fungus and not on bacterial community analyses (Frey et al., 1997; Bending et al., 2002; Rangel-Castro et al. 2002; Tornberg et al. 2003). Hence, for the first time we reveal a glimpse of an ecological effect on soil bacterial communities brought about by emerging fungal structures and presumably related to upshifts in growth-enhancing ecological opportunities for bacterial associates.
The shift in the bacterial community structure in the mycosphere was also seen in the analysis of the 2004 culturable bacterial community by plating on R2A. In total, 14 mycosphere-specific groups identified by GTG-5 genomic fingerprinting were found. Among these, three encompassed bacteria easily recognizable on plates by their yellow color and identified as belonging to the genus Chryseobacterium (Table 3). These organisms were apparently quite specific for the mycosphere, as they were never found in any of the corresponding bulk soil samples in any of the three sampling years. They consistently formed a large part of the bacterial communities of the L. proxima mycosphere, with 14% or more of all culturable bacteria in this mycosphere belonging to this genus. The putative role of this bacterial group in the mycosphere is unknown and has never been described in the literature. Moreover, a bacterial strain that turned out to be most strongly selected in the 2004 mycosphere samples was identified as a P. fluorescens.
Our hrcR based detection system was presumed to allow the detection of host-interactive bacteria containing a full TTSS (Table 2), with a basis in a range of plant-interactive organisms, possibly exempting some rhizobial types (Figure 1). We propose the use of this detection system to broadly screen for the presumptive presence of a TTSS in bacteria in soil that interact with eukaryotic organisms. Thus, the use of the hrcR gene based molecular detection system provided evidence for the presence of TTSS in this P. fluorescens BS053 population selected by the mycosphere. The data, therefore, hinted at a role of this secretion system in the presumed interactions between this soil-dwelling prokaryote and the fungal host. The prevalence of this organism at the cultivation-independent level was corroborated by Pseudomonas-specific DGGE profiles, revealing its absolute dominance among the pseudomonads that were locally present.
In a generic sense, the analyses of TTSS prevalence showed an enrichment of hrcR-positive bacterial species in the culturable fraction of the mycosphere soil of L. proxima, as compared to the bulk soil, which provides a hint of the putative role of TTSS in the interaction of soil bacteria with fungal hosts. Specifically, this effect in 2004 was dominated by the strongly selected P. fluorescens BS053 species, although preliminary evidence also hinted at the selection of a B. terrae type.
Selection of hrcR-like genes in the mycosphere was also evidenced by analyses of the hrcR-specific PCR-DGGE profiles (Figure 6). Clustering of the profiles generated from the mycosphere and bulk soil showed a strong separation of both habitats. This separation was based on the selection of dominant hrcR gene types. Several types showed higher abundance in mycosphere as compared to corresponding bulk soil samples. Differences were also noted in the less intense bands. Hence, selection of different hrcR genes was indicated in the mycosphere, suggesting a role for TTSS in this habitat. This correlates with the results shown in the culturable fraction analyses. Furthermore, grouping of the hrcR gene sequences obtained with clone library analyses of the mycosphere and bulk soil samples was quite consistent with the clustering of the hrcR-specific PCR-DGGE profiles, in which the dissection between mycosphere and bulk was mainly based on relative intensities of bands. Specifically, three main groups (A, B and C; Figure 5) were identified, of which one (A) was specific for the mycosphere, another one (B) nearly so, whereas the third (largest) one (C) encompassed sequences from both mycosphere and bulk soil. Hence, although specificity for the mycosphere was detected, we also found the large group of mixed sequences. Sequence groups A and B were remotely related (about 80% similarity) to hrcR-like genes of Pseudomonas spp. As TTSS is mainly found on (mobile) pathogenicity islands (and may be in the horizontal gene pool), we cannot affirm the identity of the gene hosts. Concerning this issue, the hrcR gene of the mycosphere-isolated P. fluorescens BS053 strain clustered within a mycosphere hrcR group, although not being identical to the sequences in this group. The failure to find the exact hrcR gene sequence of the P. fluorescens BS053 isolate in the mycosphere clone library may relate to the great plate count anomaly, which indicates that, in most of the cases, only roughly 1% to 5% of all soil bacteria can be brought into culture using common cultivation techniques.
The important role in the mycosphere of the hrcR-positive P. fluorescens BS053 strain was corroborated by the isolation of a similar hrcR-positive P. fluorescens strain (based on 16S rRNA gene identity and Biolog assays) from the mycospheres of three other fungal species, that is, Nolanea conferenda, Lactarius hepaticus and Mycena galericulata (Warmink et al., in preparation). Hence, we assume that this organism has the capacity to associate with fungal tissue that emerges in its habitat, which possibly goes beyond the mere interaction with L. proxima. TTSS may play a role in this interaction, although we have not yet obtained mechanistic evidence for this contention.
Recent published work provides support for our hypothesis about a key role of TTSS in pseudomonad–lower eukaryote interactions in soil. Thus, a recent study (Rezzonico et al., 2005) postulated a role for the TTSS present in P. fluorescens KD in protecting cucumber against the phytopathogenic oomycete Pythium ultimum. This pathogenic eukaryote physically resembles fungi, but is genetically linked to the Chromista/Stramenophila, a group that also contains brown algae and diatoms. This study, thus, provides a glimpse of a potentially broader role of bacterial TTSS, that is, beyond the interactions of prokaryotes with plants and animals, extending the effective interactive range to members of the Chromista/Stramenophila. Furthermore, Preston et al. (2001) described an experiment with P. fluorescens SBW25, in which expression of the hrcC gene of the TTSS machinery in the rhizosphere was detected, without any evidence for a role of TTSS in the interaction of the organism with plant tissue. In the light of our findings, it is possible that a rhizosphere-inhabiting fungus was actually responsible for triggering the expression of the hrcC gene in the organism used.
Although only preliminary data were obtained to support this (trehalose utilization in Biolog assays, Warmink et al., in preparation), we hypothesize that the TTSS positive P. fluorescens BS053 might be thriving in the mycosphere as a result of the enhanced availability of nutrients, for example sugars such as trehalose or even oxalate, made available to it by the fungus (De Boer et al., 2005; Van Elsas et al., 2006b). If such an analysis holds true, this makes cultivation-based approaches, next to direct molecular work, quite useful, as the bacterium-L. proxima interaction can be readily assessed in soil via such analyses.
On the other hand, the great plate count anomaly (Staley and Konopka, 1985) should be taken into account when cultivation-dependent bacterial analyses are interpreted, as only up to 5% of all bacteria in soil can be cultivated. Thus, conclusions obtained from data obtained as regards the culturable fraction do not automatically hold for the whole bacterial community, yet they might give a good indication of the mechanisms behind selection in the mycosphere. This conclusion is supported by the cultivation-independent data of the mycosphere, in which hrcR-like genes were found to be selected in comparison to the bulk soil samples.
In summary, our data showed:
-
1)
the occurrence of a clear and consistent bacterial community shift in the mycosphere soil of L. proxima in relation to corresponding bulk soil, as evidenced by both cultivation-based and cultivation-independent methods,
-
2)
the selection of bacteria that produce the flexirubin pigment (assigned to three Chryseobacterium spp) and
-
3)
the positive selection of TTSS-harboring bacteria, in particular different Pseudomonas sp. and to a lesser extent Burkholderia sp.
Future work should attempt to determine whether the selected TTSS serves a fitness-enhancing ecological role for pseudomonads in the mycosphere, for instance in the liberation of nutrients from fungal tissue to the bacterial host. More research on this putative mechanism is, thus, required to provide a more conclusive answer to this question.
Accession codes
References
Bending GD, Poole EJ, Whipps JM, Read DJ . (2002). Characterisation of bacteria from Pinus sylvestris-Suillus luteus mycorrhizas and their effects on root-fungus interactions and plant growth. FEMS Microbiol Ecol 39: 219–227.
Buell CR, Joardar V, Lindeberg M, Selengut J, Paulsen IT, Gwinn ML . (2003). The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. Proc Natl Acad Sci USA 100: 10181–10186.
Costa R, Gomes NCM, Peixoto RS, Rumjanek N, Berg G, Mendonca-Hagler LCS et al. (2006). Diversity and antagonistic potential of Pseudomonas spp. associated to the rhizosphere of maize grown in a subtropical organic farm. Soil Biol Biochem 38: 2434–2447.
De Boer W, Folman LB, Summerbell RC, Boddy L . (2005). Living in a fungal world: impact of fungi on soil bacterial niche development. FEMS Microbiol Rev 29: 795–811.
De Boer W, Leveau JHJ, Kowalchuk GA, Gunnewiek PJAK, Abeln ECA, Figge MJ et al. (2004). Collimonas fungivorans gen. nov., sp nov., a chitinolytic soil bacterium with the ability to grow on living fungal hyphae. Int J Syst Evol Microbiol 54: 857–864.
Delsal G, Manfioletti G, Schneider C . (1989). The CTAB-DNA precipitation method—a common mini-scale preparation of template DNA from phagemids, phages or plasmids suitable for sequencing. Biotechniques 7: 514–520.
Feil H, Feil WS, Chain P, Larimer F, DiBartolo G, Copeland A . (2005). Comparison of the complete genome sequences of Pseudomonas syringae pv. syringae B728a and pv. tomato DC3000. Proc Natl Acad Sci USA 102: 11064–11069.
Frederick RD, Ahmad M, Majerczak DR, rroyo-Rodriguez AS, Manulis S, Coplin DL . (2001). Genetic organization of the Pantoea stewartii subsp stewartii hrp gene cluster and sequence analysis of the hrpA, hrpC, hrpN, and wtsE operons. Mol Plant Microbe Interact 14: 1213–1222.
Frey P, Frey-Klett P, Garbaye J, Berge O, Heulin T . (1997). Metabolic and genotypic fingerprinting of fluorescent pseudomonads associated with the Douglas fir Laccaria bicolor mycorrhizosphere. Appl Environ Microbiol 63: 1852–1860.
Frey-Klett P, Chavatte M, Clausse ML, Courrier S, Le Roux C, Raaijmakers J et al. (2005). Ectomycorrhizal symbiosis affects functional diversity of rhizosphere fluorescent pseudomonads. New Phytologist 165: 317–328.
Frey-Klett P, Churin JL, Pierrat JC, Garbaye J . (1999). Dose effect in the dual inoculation of an ectomycorrhizal fungus and a mycorrhiza helper bacterium in two forest nurseries. Soil Biol Biochem 31: 1555–1562.
Garbaye J . (1994). Helper Bacteria—a new dimension to the mycorrhizal symbiosis. New Phytologist 128: 197–210.
Gardes M, Bruns TD . (1993). ITS primers with enhanced specificity for basidiomycetes—application to the identification of mycorrhizae and rusts. Mol Ecol 2: 113–118.
Gude H . (1980). Occurrence of cytophagas in sewage plants. Appl Environ Microbiol 39: 756–763.
He SY, Nomura K, Whittam TS . (2004). Type III protein secretion mechanism in mammalian and plant pathogens. Biochimica et Biophysica Acta-Molecular Cell Research 1694: 181–206.
Heuer H, Smalla K . (1997). Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) for studying soil microbial communities. In: Van Elsas JD, Trevors JT, Wellington EMH (eds). Modern Soil Microbiology. Marcel Dekker, Inc.: New York, pp 353–373.
Heuer H, Wieland J, Schönfeld J, Schönwälder A, Gomes NCM, Smalla K . (2001). Bacterial community profiling using DGGE or TGGE analysis. In: Rouchelle P (ed). Environmental Molecular Microbiology: Protocols and Applications. Horizon Scientific Press: Wymondham, UK, pp 177–190.
Holden MTG, Titball RW, Peacock SJ, Cerdeno-Tarraga AM, Atkins T, Crossman LC . (2004). From the cover: genomic plasticity of the causative agent of melioidosis, Burkholderia pseudomallei. Proc Natl Acad Sci USA 101: 14240–14245.
Hueck CJ . (1998). Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol Mol Biol Rev 62: 379–433.
Johansson JF, Paul LR, Finlay RD . (2004). Microbial interactions in the mycorrhizosphere and their significance for sustainable agriculture. FEMS Microbiol Ecol 48: 1–13.
Kaneko T, Nakamura Y, Sato S, Asamizu E, Kato T, Sasamoto S . (2000). Complete genome structure of the nitrogen-fixing symbiotic bacterium Mesorhizobium loti. DNA Res 7: 331–338.
Klimyuk VI, Carroll BJ, Thomas CM, Jones JDG . (1993). Alkali treatment for rapid preparation of plant material for reliable PCR analysis. Plant Journal 3: 493–494.
Kumar S, Tamura K, Nei M . (2004). MEGA 3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinformatics 5: 150–163.
Lee BM, Park YJ, Park DS, Kang HW, Kim JG, Song ES . (2005). The genome sequence of Xanthomonas oryzae pathovar oryzae KACC10331, the bacterial blight pathogen of rice. Nucleic Acids Res 33: 577–586.
Marie C, Broughton WJ, Deakin WJ . (2001). Rhizobium type III secretion systems: legume charmers or alarmers? Curr Opin Plant Biol 4: 336–342.
Mazurier S, Lemunier M, Hartmann A, Siblot S, Lemanceau P . (2006). Conservation of type III secretion system genes in Bradyrhizobium isolated from soybean. FEMS Microbiol Lett 259: 317–325.
Mazurier S, Lemunier M, Siblot S, Mougel C, Lemanceau P . (2004). Distribution and diversity of type III secretion system-like genes in saprophytic and phytopathogenic fluorescent pseudomonads. FEMS Microbiol Ecol 49: 455–467.
Muyzer G, Teske A, Wirsen CO, Jannasch HW . (1995). Phylogenetic relationships of Thiomicrospira species and their identification in deep-dea hydrothermal vent samples by denaturing gradient gel electrophoresis of 16S rDNA fragments. Arch Microbiol 164: 165–172.
Nübel U, Engelen B, Felske A, Snaidr J, Wieshuber A, Amann RI et al. (1996). Sequence heterogeneities of genes encoding 16S rRNAs in Paenibacillus polymyxa detected by temperature gradient gel electrophoresis. J Bacteriol 178: 5636–5643.
Preston GM, Bertrand N, Rainey PB . (2001). Type III secretion in plant growth-promoting Pseudomonas fluorescens SBW25. Mol Microbiol 41: 999–1014.
Rangel-Castro JI, Levenfors JJ, Danell E . (2002). Physiological and genetic characterization of fluorescent Pseudomonas associated with Cantharellus cibarius. Can J Microbiol 48: 739–748.
Rezzonico F, Binder C, Defago G, Moenne-Loccoz Y . (2005). The type III secretion system of biocontrol Pseudomonas fluorescens KD targets the phytopathogenic chromista Pythium ultimum and promotes cucumber protection. Mol Plant Microbe Interact 18: 991–1001.
Salanoubat M, Genin S, Artiguenave F, Gouzy J, Mangenot S, Arlat M et al. (2002). Genome sequence of the plant pathogen Ralstonia solanacearum. Nature 415: 497–502.
Sneath PH, Sokal RR . (1973). Numerical Taxonomy: The Principles and Practice of Numerical Classification. Freeman WH: San Francisco.
Staley JT, Konopka A . (1985). Measurement of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Annu Rev Microbiol 39: 321–346.
Toljander JF, Artursson V, Paul LR, Jansson JK, Finlay RD . (2006). Attachment of different soil bacteria to arbuscular mycorrhizal fungal extraradical hyphae is determined by hyphal vitality and fungal species. FEMS Microbiol Lett 254: 34–40.
Tornberg K, Baath E, Olsson S . (2003). Fungal growth and effects of different wood decomposing fungi on the indigenous bacterial community of polluted and unpolluted soils. Biol Fertil Soils 37: 190–197.
Van Breemen N, Finlay R, Lundstrom U, Jongmans AG, Giesler R, Olsson M . (2000). Mycorrhizal weathering: a true case of mineral plant nutrition? Biogeochemistry 49: 53–67.
Van Elsas JD, Jansson JK, Trevors J . (2006a). Modern Soil Microbiology, 2nd edn, CRC Press: Boca Raton, FL, USA.
Van Elsas JD, Tam L, Finlay RD, Killham K, Trevors J . (2006b). Microbial interactions in soil. In: Van Elsas JD, Jansson JK, Trevors J (eds). Modern Soil Microbiology, 2nd edn, CRC Press: London, pp 177–210.
Versalovic J, Schneider M, De Bruijn FJ, Lupski JR . (1994). Genomic fingerprinting of bacteria using repetitive sequence-based polymerase chain reaction. Methods Mol Cell Biol 5: 25–40.
Acknowledgements
We thank Jolanda Brons for all the sequencing work she has done for this study and Bert Venema for the chemical analyses of the soil samples. We also thank Jacques Hille and Wietse de Boer for the helpful discussions.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Warmink, J., van Elsas, J. Selection of bacterial populations in the mycosphere of Laccaria proxima: is type III secretion involved?. ISME J 2, 887–900 (2008). https://doi.org/10.1038/ismej.2008.41
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2008.41
Keywords
This article is cited by
-
Microfluidic chips provide visual access to in situ soil ecology
Communications Biology (2021)
-
Root-associated bacteria influencing mycelial growth of Tricholoma matsutake (pine mushroom)
Journal of Microbiology (2018)
-
Draft genome sequences of three fungal-interactive Paraburkholderia terrae strains, BS007, BS110 and BS437
Standards in Genomic Sciences (2017)
-
Pseudomonas fluorescens C7R12 type III secretion system impacts mycorrhization of Medicago truncatula and associated microbial communities
Mycorrhiza (2017)
-
Ericoid Roots and Mycospheres Govern Plant-Specific Bacterial Communities in Boreal Forest Humus
Microbial Ecology (2017)