Abstract
The geographic distribution of genetic diversity can reveal the evolutionary history of a species. For crop plants, phylogeographic patterns also indicate how seed has been exchanged and spread in agrarian communities. Such patterns are, however, easily blurred by the intense seed trade, plant improvement and even genebank conservation during the twentieth century, and discerning fine-scale phylogeographic patterns is thus particularly challenging. Using historical crop specimens, these problems are circumvented and we show here how high-throughput genotyping of historical nineteenth century crop specimens can reveal detailed geographic population structure. Thirty-one historical and nine extant accessions of North European landrace barley (Hordeum vulgare L.), in total 231 individuals, were genotyped on a 384 single nucleotide polymorphism assay. The historical material shows constant high levels of within-accession diversity, whereas the extant accessions show more varying levels of diversity and a higher degree of total genotype sharing. Structure, discriminant analysis of principal components and principal component analysis cluster the accessions in latitudinal groups across country borders in Finland, Norway and Sweden. FST statistics indicate strong differentiation between accessions from southern Fennoscandia and accessions from central or northern Fennoscandia, and less differentiation between central and northern accessions. These findings are discussed in the context of contrasting historical records on intense within-country south to north seed movement. Our results suggest that although seeds were traded long distances, long-term cultivation has instead been of locally available, possibly better adapted, genotypes.
Similar content being viewed by others
Introduction
Population genetics and phylogeography are important tools that provide insight into the evolutionary history of species. Geographic patterns in the distribution of genetic diversity can give information about the geographic origin of lineages or the effects of migration routes (Avise, 2009) and have, among other species, been applied to crop plants (for example, Olsson and Schaal, 1999; Londo et al., 2006; Saisho and Purugganan, 2007). Insights into the genetic structure of crop species provide not only a better understanding of their evolutionary history but can also increase the knowledge about cultural exchange in agrarian communities (Van Heerwaarden et al., 2011; Oliveira et al., 2012; Roullier et al., 2013).
Barley (Hordeum vulgare L. ssp. vulgare) was domesticated about 10 000 years ago (Badr et al., 2000), most likely from multiple domestication centers (Morrell and Clegg, 2007; Saisho and Purugganan, 2007; Fuller et al., 2011; Ren et al., 2013). An adaptable species, barley has been of great importance also in regions where climate or soil is sub-optimal for agriculture. As a result of crop improvement during the twentieth century, present-day cultivars primarily show phylogeographic structuring on a continental scale. Agronomic traits, such as winter or spring growth habit and two-row or six-row type, have been found to be as relevant as geographic origin in determining population clustering (Malysheva-Otto et al., 2006).
To detect traces of evolutionary history, landraces are a preferable choice to modern cultivars. The definition of a landrace is not without controversy (Zeven, 1998; Camacho Villa et al., 2005), but they can be described as locally adapted populations, genetically diverse and with a historical origin, lacking formal crop improvement. During centuries of continuous cultivation in their respective area, their genetic composition has shifted because of gene flow and seed trade as well as local adaptation and genetic drift, but landraces are generally considered to have been relatively stable over time (Brown, 1999; Jones et al., 2008). Importantly, landraces show phylogeographic patterns unencumbered by the overwriting effects of the intense plant breeding during the last century.
Extant landrace materials preserved in genebanks, however, also suffer from limitations to their usefulness in phylogeographic studies. In some areas, such as northern Europe, the number of available extant landraces is very low (Jones et al., 2008). The passport data of these accessions are often limited to a country of origin and it is not always possible to determine whether or not older accessions are actual landraces or early cultivars. Extant accessions are, furthermore, maintained ex situ and at small population sizes, which unavoidably leads to genetic drift, in addition to the risk of contamination during propagation (Steiner et al., 1997; Börner et al., 2000; Parzies et al., 2000; Chebotar et al., 2002; Hagenblad et al., 2012). Consequently, extant material, at least in certain geographic areas, may inadequately represent the genetic diversity once present in landraces, thereby obscuring phylogeographic patterns.
Few studies have reported fine-scale geographic structure of landrace crops (but see Pandey et al., 2006; Yahiaoui et al., 2007; Rodriguez et al., 2012). This may, in part, be due to the lack of suitable landrace material. Fennoscandia (Norway, Sweden, Finland and Denmark), however, provides unique opportunities to explore fine-scale geographic structure in crops and how it relates to known agrarian history. In Sweden, Finland and Norway, seed collections, compiled by agronomists during the late nineteenth century, mainly to be used as display objects, provide an alternative material to extant landraces (Leino et al., 2009; Leino, 2010). Sampling was carried out ‘on farm’, and sampling location is frequently detailed down to the specific farmstead. Although documentation on the sampling method is not available, it is reasonable to assume that samples are representative for the farms where they were collected as the seed volumes are large and show no signs of seed sorting. The historical seeds are original samples and have, in contrast to extant material, not been subject to any change in genetic diversity since the sampling.
Fennoscandia holds the northernmost expansion of barley in the world, and encompasses large variation in climate, soil and light regimes. Historically, the Baltic Sea and the Bay of Bothnia between Sweden and Finland have facilitated trade in the region, while the Scandes mountain range between Norway and Sweden has been a natural barrier for both trade and agriculture (Flygare, 2011). Historical documents describe both trade regulations limiting seed import and crop failures leading to the need for long-range seed trade. A study of fine-scale geographic structure would allow the long-term genetic effects of such historical events to be assessed. Studying Swedish barley accessions from the historical collections, genotyped for 14 microsatellite markers, Leino and Hagenblad (2010) detected a north-south separation of genetic diversity and suggested two separate colonization routes into the country. The number of markers used did not, however, allow any detailed genetic structuring to be detected and the sampling limited to Sweden prevented broader conclusions concerning Fennoscandian barley from being drawn.
Here we report a study of the genetic structure of landrace barley sampled with a high geographic resolution across all of Fennoscandia. We have used high-throughput single nucleotide polymorphism (SNP genotyping to screen multiple individual seeds from a large number of primarily historical specimens. This allows us a population genomic approach to explore the details of the species’ genetic structure at the northernmost limit of its distribution range.
Materials and Methods
Plant material
A total of 40 six-row barley accessions were studied (Table 1). Of these the majority, 31 accessions, were taken from the nineteenth century historical seed collections; Tromsø University Museum in Norway (TR, three accessions), Mustiala Agricultural College in Finland (MU, six accessions) and the Swedish Museum of Cultural History in Sweden (NM, 21 accessions) (Leino et al., 2009; Leino, 2010). The seeds, which are no longer viable, were collected at harvest on farm in 1869 (TR), 1890s (MU) and 1896 (NM), respectively, except for NM264 that was collected in 1882. Accessions were chosen for best possible coverage of Fennoscandia (Figure 1, Table 1), and where geographic coverage was lacking in the seed collections, the historical accessions were complemented with nine extant accessions provided by the Nordic Genetic Resource Center (NGB) and the N.I. Vavilov Research Institute of Plant Industry (VIR). On the basis of passport data, seed jar labels and visual inspection, only hulled six-row spring barley landraces were chosen. Plant improvement of six-row barley for Fennoscandia did not begin until the 1920s (Osvald, 1959) and the historical material can therefore be considered to be genuine landraces.
DNA analysis
DNA was extracted from six individual seeds from each accession using FastDNA Spin Kits and the FastPrep Instrument (MP Biochemicals, Solon, OH, USA). Extractions were performed at a laboratory separate from that where SNP genotyping was carried out to reduce the risk of contamination. A negative control was included in each extraction series. Two present day cultivars (cv. ‘Morex’ and cv. ‘Rolfi’) were used as positive controls and three negative extraction controls were included in the SNP genotyping. SNP data were generated using an Illumina Golden Gate assay (Illumina Inc., San Diego, CA, USA), with the C-384 SNP set designed for optimal diversity for European barley cultivars, as developed by Moragues et al. (2010). The resulting data were processed and studied with the Bead Studio 3.1.3.0 software packager (Illumina Inc., San Diego, CA, USA). To verify that repeatable and authentic SNP calling could be performed on historical material by the assay, four DNA extracts from kernels of the same 100-year-old ear (NM76) were genotyped. Additionally, DNA extracts from historical samples of the cultivars ‘Gull’ (NM52) and ‘Princess’ (NM60) and extant material of the same cultivars (NGB1480 and NGB9424) were compared. To evaluate ascertainment bias, folded minor allele frequency spectra were generated for the full data and for three regional subsets of the data. Accessions with an origin north of the 65th parallel were categorized as ‘North’, accessions with an origin between the 60th and 65th parallel as ‘Mid’ and accessions with an origin south of the 60th parallel categorized as ‘South’.
Linkage disequilibrium
Linkage disequilibrium (LD) was calculated as r2 (Hill and Robertson, 1968) using a purpose-written Perl script. Intrachromosomal LD was calculated for pairs of polymorphic loci residing on the same chromosome and interchromosomal LD was calculated for pairs of polymorphic loci located on different chromosomes. LD was calculated both across all individuals and for the individuals of each accession.
Statistical analysis
Principal component analysis (PCA) was performed using the prcomp function in the statistical software R (R Development Core Team, 2013, version 3.0.2) to visualize both within accession diversity and structure between accessions. The SNP data were analyzed both as individuals and on an accession level. For the individual level, each homozygous SNP was treated as either 1 or 0 and missing data were replaced with the allele frequency in the full dataset of the allele designated as ‘1’. For the accession level PCA, allele frequencies of each accession for each of the SNPs were calculated and treated as independent variables. A measure of genetic relatedness between individuals within accessions, based on principal components, was calculated using R. This measure, called PC dispersion, was the mean pairwise distance in PC-space between individuals within accessions. Data of all principal components for each individual in an accession were used as coordinates in a multidimensional space and the average distance between individuals belonging to the same accession in this multidimensional space was calculated.
The software structure (Pritchard et al., 2000; Falush et al., 2003, version 2.3.3) was used to cluster accessions into populations. As suggested by Nordborg et al. (2005) for selfing species, and applied in other studies (Pandey et al., 2006; Leino and Hagenblad, 2010; Leino et al., 2013), we analyzed data as haploid, treating heterozygous loci as missing data. Structure simulations were carried out using an admixture model. Burn-in period was set to 25 000 iterations and estimations were based on 50 000 iterations. The simulations were repeated 20 times for K-values of 1–10. The choice of relevant numbers of clusters was guided by calculating ΔK using the method presented in Evanno et al. (2005) and the change in H’ from CLUMPP. To properly evaluate multimodality of the structure output, the 20 repeats for each K from the structure simulations were merged using the CLUMPP software (Jakobsson and Rosenberg, 2007). CLUMPP was used with the Greedy Algorithm method and the results were visualized using the Distruct v1.1 software (Rosenberg, 2004). The same procedure and settings were used for all analyses of genetic structure.
To verify that our results were not influenced by the structure assumption of Hardy-Weinberg equilibrium, discriminant analysis of principal components (DAPC) was used as an alternative method of evaluating population clustering. DAPC is a multivariate method included in the Adegenet R package (Jombart et al., 2010) and requires no prior assumption of the underlying population genetic model. The method was applied both to the full data and to a subset consisting of only the historical accessions. All principal components were utilized for prior group clustering and DAPC analysis used a subset of 50 PCs to prevent over-fitting.
Within-accession genetic diversity was assessed by calculating Nei’s h (h=1–Σ pi2, where pi is the frequency of the ith allele) for each accession (Nei, 1973). This was carried out both for individual SNPs and for haplotypes (length 2–10 SNPs) consisting of merged neighboring SNPs. The distribution of genetic diversity was further explored by calculating pairwise FST values (Weir and Cockerham, 1984) between the accessions. Pairwise FST values were also analyzed between pairs of accessions in the three previously defined latitudinal groups. Average genetic diversity (Nei’s h) for the genotyped SNPs and pairwise FST between accessions and groups of accessions were calculated using the Arlequin 3.5 software (Excoffier and Lischer, 2010). The significances of FST values were estimated with permutation tests (1000 permutations). The relative effect of gene flow and drift was also analyzed by plotting pairwise FST values against geographic distances (Hutchison and Templeton, 1999). Geographic distances between accessions were calculated in R using the haversine formula. Total genotype sharing, that is when two individuals had identical genotypes at all loci, excluding missing data, was also determined using Arlequin 3.5.
Geographic visualization
Maps for geographic visualization of genetic structure were created using ArcGIS (ESRI, 2011), with geographic data available through the ‘ESRI data and maps v. 9.3’ database (2008).
Results
SNP calling in historical and extant barley accessions
We have measured genetic diversity in more than 115-year-old historical and extant barley using an Illumina Golden Gate SNP assay. DNA from barley seeds in the historical collections is degraded to fragments of typically 100–200 bp and yield 200 ng mg−1 seed, compared with extant seeds where DNA yield is typically four times higher and DNA length is above 10 000 bp (Leino et al., 2009). DNA concentrations in the extracts used for SNP genotyping were in the range of 20–65 ng μl−1. Although DNA concentration is low, it proved to be of sufficient quality for successful genotyping and whole genome amplification using Illustra GenomiPhi (GE Healthcare Life Sciences, Buckinghamshire, UK), of eight test individuals did not improve genotyping success.
To verify that repeatable and accurate signals could be obtained from the historical material, four replicate DNAs extracted from seeds from the same 100-year-old ear were analyzed. As barley is highly inbreeding, the seeds can be expected to have very high genetic similarity and SNP calling from the four extracts were indeed identical, demonstrating repeatability. To further verify that extant and historical material could produce comparable genotype scores, historical and extant accessions of two cultivars, ‘Gull’ and ‘Princess’ were tested. Even though it is possible that the cultivars have changed slightly over the past 100 years, we nonetheless expect, if our genotyping method yields accurate results, historical and extant samples from the same cultivars to be more similar than when comparing the historical and the extant accessions, respectively, with each other. Comparing historical and extant accessions of the same cultivar resulted on average in 91.8% identical scores (100.0% and 85.4% for ‘Gull’ and ‘Princess’ respectively), whereas comparing extant material of different cultivars resulted in 63.2% identical scores and comparing historical material resulted in 71.7% identical scores. We thus concluded that SNP calling of the historical material was sufficiently accurate and not influenced by the nature of aged DNA.
A total of 231 individuals from 40 different accessions originating across Fennoscandia were obtained from historical seed collections (31 accessions) and genebanks (9 accessions) (Table 1, Figure 1). These, together with three negative extraction controls were assayed for 384 SNPs. Neither of the extraction controls yielded detectable SNP signals verifying the absence of contamination. Loci scored as heterozygous within an individual were extremely rare in the dataset, 0.076% averaged across all loci and individuals. Because of difficulties of separating unclear marker distinction from actual heterozygotes, such loci were scored as missing data. Of the 384 SNPs assayed, 63 were excluded because of high levels (>15%) of missing data and one because of being monomorphic, leaving 320 SNPs to be used for analyses. Additionally, nine individuals with more than 40% missing data were excluded from further analysis leaving an average 1.6% missing SNP data per individual (on average, 2.01% for the historical material and 0.09% for the extant material). Success rates of individual SNP markers are reported as supporting information (Supplementary Table 1).
Minor allele frequency spectra for the total dataset and the predefined latitudinal groups showed that the spectra for the subgroups differ somewhat from each other and that of the total dataset (Supplementary Figure 1). This is likely due to stochastic effects as the underlying data for the subgroups are based on fewer individuals. In all spectra, the frequency of the minor allele was in most cases 5% or less. The number of SNPs within each category of minor allele frequency was becoming increasingly lower as the minor allele frequency approached 50% (Supplementary Figure 1), as expected under the basic model (see Nielsen et al., 2004).
LD in Fennoscandian barley does not vary with latitude
LD between pairs of polymorphic loci was calculated as r2. Average interchromosomal LD was for the complete dataset 0.0664 but with a skewed distribution (median 0.012). Intrachromosomal LD declined with the distance between markers, but r2 values of 0.4 and more could still be observed between markers 100 cM of more apart (Supplementary Figure 2). We also calculated interchromosomal LD for each accession separately (Table 1). Average interchromosomal LD ranged from 0.2 for NM671 to 1 for NGB468, NM633 and NGB9529 (average 0.364) but was not significantly correlated with latitude of the accession (P=0.247). We also compared LD values between accessions from each of three predetermined latitudinal groups: South, Mid and North. None of the three geographical groups South, Mid and North differed significantly in their LD from each other (two-sided t-test, all P>0.5). LD was, however, higher in extant material (average 0.612) than in historical (average 0.293) accessions (two-sided t-test, P<0.001).
Both genetic diversity and total genotype sharing is higher within extant landraces compared with historical accessions
To assess the genetic composition of the accessions, both Nei’s h and the amount of total genotype sharing within accessions were calculated. Within-accession genetic diversity ranged from 0.005 in the extant Norwegian landrace NGB2072 to 0.331 in the extant Russian landrace VIR2174 with an average within-accession diversity of 0.101 (Table 1, Figure 2). A genetic bottleneck leading to loss of genetic diversity could be expected in populations migrating northwards. However, within-accession genetic diversity was not found to be significantly correlated with latitude (r2=0.11672, P=0.205). The average within-accession genetic diversity of the historical accessions (0.087) was significantly lower than extant accessions (0.152) (two-tailed unpaired t-test, P<0.001).
Ascertainment bias can affect comparisons of genetic diversity between unascertained and ascertained populations, or populations similar to ascertained populations. The effect of ascertainment bias can be alleviated by combining SNPs into haplotypes (Conrad et al., 2006; Oliveira et al., 2014). For this reason, Nei’s h was also calculated for haplotypes of length 2–10. The haplotype diversities suggested little effect of ascertainment bias. The accessions with the highest and lowest diversity remained the same for all lengths of haplotypes and most differences in relative rank were minor (Supplementary Table 2). The few accessions that showed a marked change in relative diversity could be explained by an increase of missing data as the haplotypes were merged.
All but one extant accession (88.9%) included individuals sharing identical total genotypes (that is, two or more individuals within the same accession having all genotyped SNPs scored as identical), something that was only found in 6 out of 31 accessions (19.4%) among the historical material (Table 1). It is likely that the amount of total genotype sharing in the historical accessions is even lower, as missing data are more common, increasing the likelihood of individuals being classified as identical. It is worth noting that the three accessions with the highest genetic diversity, VIR2143, VIR2174 and VIR3221, also have a high degree of total genotype sharing, meaning they consist of a few, but very different, lines (Table 1). Although most of the historical accessions are more variable than the extant material, it should be noted that some of the individuals share total genotypes not only within accessions but also between accessions. This occurs in the accessions MU13, MU69, NM633 and NM668, all of which originate from relatively nearby places in northern Finland and northern Sweden.
Genetic diversity in extant landraces is inflated by a few individuals with distinctly different genotypes
We explored the distribution of genetic diversity by PCA. In the accession level PCA, the Estonian accession, VIR2143, clustered separately from all other accessions along PC2, whereas the Karelian accessions appeared closer to the Fennoscandian material (Figure 3a). There was, however, little indication of accessions clustering according to country of origin (Figure 3a). Classifying accessions according to latitudinal groups described above revealed a certain amount of clustering according to latitude along the first PC (Figure 3b), a pattern that was made clearer when omitting extant accessions (Figure 3c).
PCA of accessions and individuals. (a) PCA of 40 accessions of landrace barley on accession level. Accessions are divided by color depending on country of origin according to present day national borders and by symbol according to seed source. PC1 and PC2 explain 22.88% and 11.43% of the total variation, respectively. (b) PCA of the same data as in (a) presented according to latitude. Accessions with an origin north of 65° N (‘North’) are shown in blue, accessions from between 60° N and 65° N (‘Mid’) in red and accessions south of 60° N (‘South’) in black. Symbols are as in (a). (c) PCA of historical accessions only. PC1 and PC2 explain 25.00% and 17.57% of the total variation, respectively. Color coding is as in (b). (d) PCA of 321 individuals from 40 accessions of landrace barley, performed on an individual level. PC1 and PC2 explain 12.25% and 7.46% of the total variation, respectively. Color coding is as in (b). Individuals from the two Karelian accessions are marked with filled circles to visualize their within-accession differentiation.
The geographic distances between accessions were well explained by the distances in PC space (r2=0.265, P<0.001). Analyzing latitude and longitude separately as explanatory variables showed that latitude explained the distances between accessions in PC space much better (r2=0.224, P<0.001) than longitude (r2=0.0366, P<0.001). This is noteworthy as the geographical range of the study area is approximately equal in both directions.
The individual level PCA showed that specimens of the Karelian accessions (VIR2174 and VIR3221) were widely distributed along both PC1 and PC2 with individuals being either very similar to the Fennoscandian material or highly divergent (Figure 3d). To explore this further, a proxy of the genetic variation within accessions, PC dispersion, was quantified from the PCA results using mean pairwise distances in PC-space (Table 1). These values ranged from 1.063 for the Norwegian accession NGB2072 to 9.319 for the Karelian accession VIR2174 and were highly correlated with the within-accession genetic diversity (r=0.867, P<0.001). The variance of the PC dispersion statistic provided additional information to the mean PC dispersion as it is strongly inflated where the genetic variation between individuals within an accession is uneven, such as when a sample contains seed from mixed sources. Thus, a high PC dispersion variance indicates accessions that may have been subject to seed mixing during rejuvenation. For example, the accession NGB468 shows a high variance in PC dispersion (24.048) because of a single individual which differs greatly, whereas the remainder of the material contains multiple identical total genotypes (NUGen/NInd=0.33); this could not be detected from the genetic diversity index (h=0.101), which is in fact below average. High PC dispersion variance was found in several extant accessions but none of the historical accessions (Table 1).
Differences in the genetic diversity of extant and historical landraces were also evaluated by comparing extant and historical accessions from the same geographic origin to rule out any geographical effects on diversity. The two pairs of extant and historical accessions with the shortest geographic distance are NGB15103 (extant)—NM727 (historical) and NGB27 (extant)—MU52 (historical) where the accessions NGB15103 and NM727 originate from nearly the same site. The extant NGB15103 had significantly higher genetic diversity (0.136 vs 0.077, two-tailed unpaired t-test, P<0.001), fewer unique total genotypes (four vs six) and a higher PC dispersion (6.680 vs 0.217) compared with the historical NM727, which could indicate that it is a mixture of seeds with different origins rather than a genuine landrace. In contrast, the genetic diversity of the accessions NGB27 and MU52 were not significantly different (Nei’s h=0.102 vs 0.107 for NGB27 and MU52, respectively, two-tailed unpaired t-test, P=0.76) and each accession had two individuals with total genotype sharing among six individuals. The variances of the PC dispersion of NGB27 and MU52 were also quite similar, being 2.528 in NGB27 and 1.611 in MU52. FST values further indicated differences between NGB15103 and NM727 with a high and significant FST (FST=0.422, P<0.001). NGB27 and MU52 in contrast had a low and non-significant FST value (FST=0.077, P=0.144).
A latitudinal structure of genetic diversity
We explored geographic structuring of the accessions with the software structure. We initially analyzed the complete dataset where Evanno’s ΔK suggested that a two-cluster model best described the data and the H’ values from CLUMPP indicated either two or three populations with equal support (Supplementary Table 3). Although ΔK cannot be calculated for k=1, log-likelihood values calculated by structure were consistently much higher for k>1 than k=1, which, together with the distinct and reproducible clustering patterns, indicates that population structure is best described by more than one cluster. The two-cluster model separated the entire VIR2143 accession and individuals from the accessions VIR2174, VIR3221, NGB321 and NGB468 (Figure 4a). Increasing the number of clusters to three (data not shown) divided the Fennoscandian accessions in a north and south cluster.
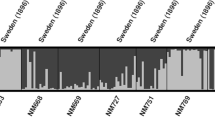
(a–c) Genetic structure from 20 individual structure simulations merged with the CLUMPP software. (a) K=2, complete dataset. (b) K=2, subset with the minor (black) cluster from (b) removed. (c) K=3, historical accessions only. (d) Output from DAPC analysis, visualized with the Distruct software, K=5, historical accessions only. A full color version of this figure is available at the Heredity journal online.
To properly evaluate any substructure that may have been obscured by large difference between the main clusters, and based on the information from the PCA and the structure analysis, we created two subsets of accessions. The first subset excluded the divergent cluster from the first structure analysis by removing individuals that clustered to the minor cluster in the k=2 model to an extent of 90% or more (shown as black in Figure 4a). For this subset, ΔK again supported k=2, while H’ indicated k=2, but with nearly as high support for k=4 (Supplementary Table 3). The two-cluster model of the first subset divides Fennoscandia into a northern and southern cluster, across national boundaries (Figure 4b). The four-cluster model adds two additional, latitudinal, clusters (data not shown).
The second subset consisted of only historical accessions to assess whether the inclusion of the extant material in general has an effect on the genetic structure. Indeed, the clusters in Fennoscandia proved to be much more distinct with the omission of extant accessions (Figure 4c). For the historical seed subset, the ΔK indicated k=2 while H’ was highest for k=3 and nearly as high for k=2 and k=6 (Supplementary Table 3). The three-cluster model divided the accessions into three latitudinal groups, the southernmost encompassing the southern third of Sweden and the southern tip of Finland, a middle group of accessions stretching eastward across central Sweden and Finland and a northernmost group with accessions from northern Norway, northern Finland and Sweden along the border to Finland (Figures 4c and 5).
Geographic visualization of structure results from 20 simulations with K=3, joined together with CLUMPP and visualized with ArcMap 10. Clusters are colored as in Figure 4. A full color version of this figure is available at the Heredity journal online.
We also evaluated the data with discriminant analysis of principal components (DAPC), an alternative method not assuming Hardy-Weinberg equilibrium. These DAPC analyses supported k≈9–10 (Supplementary table S2) for the complete dataset and clustering for k=2 to k=10 were nearly identical to those of the structure analyses; for all levels of k, the structuring observed was primarily latitudinally distributed. For the historical subset, DAPC clearly supported k=5 (Figure 4d) and the overall structure was again very similar to that of the k=3 model from structure, with clear latitudinal structuring, albeit separating the NM625 into its own cluster and most Swedish accessions between the 60th and 65th parallel into yet another cluster (Figure 4d).
FST values were calculated between all pairs of accessions and 89.6% of the pairwise FST values were significant at the level P<0.05. Pairwise FST values and geographic distances were moderately but significantly correlated (r=0.497, P<0.001). A scatter plot visualized this correlation and also indicates increasingly varying FST values as distances increase (Figure 6). The resulting correlation suggests a lack of regional equilibrium between gene flow and drift (see Hutchison and Templeton, 1999). For the landrace barley, it would appear that gene flow is more effective at shorter distances and drift is more influential at greater distances of geographic separation.
After excluding the extant material, we averaged pairwise FST for comparisons within and between the previously defined latitudinal regions ‘North’, ‘Mid’ and ‘South’ (Table 2). In the three groups, the highest average pairwise FST was found between accessions in the ‘South’ group, which could be an indication of a higher genetic diversity within this latitudinal group. The corresponding statistic was lower for the ‘North’ and ‘Mid’ group where average FST values were approximately the same. The pairwise comparisons between groups revealed that the differences between accessions in the ‘North’ and ‘Mid’ group were less than the difference of either group from the ‘South’ group. The FST averages for the different comparisons between regions were all significantly different from each other (two-tailed unpaired t-test, P<0.001).
Discussion
Resolving the historical spread of agricultural crops is a challenging task requiring combined archaeological, botanical and historical evidence. Recently, phylogeographic studies have added substantially to the understanding of crop history (Van Heerwaarden et al., 2011; Oliveira et al., 2012; Roullier et al., 2013). However, if a study is performed on a finer geographical scale, more power in terms of genetic markers and number of individuals is needed. Additionally, the use of landraces with strong genetic integrity becomes even more important. When these requirements are fulfilled, as shown in the present study, patterns of historical crop spread can become visible.
Phylogeographic studies of landrace crops have often been restricted to single individuals (for example, Saisho and Purugganan, 2007; Isaac et al., 2010; Van Heerwaarden et al., 2011) or aggregate samples from accessions, using pooling schemes (for example, Hunt et al., 2011; Jones et al., 2011; Oliveira et al., 2012). This allows a much higher number of accessions to be studied, but comes at the cost of ignoring within-accession genetic diversity. This is unfortunate, both because knowledge on within-accession diversity is valuable per se, but even more so as within-population diversity aids in the proper identification of population structure (Fogelqvist et al., 2010; Lascoux and Petit, 2010).
The trade-off between the number of individuals per accession studied and number of accessions that can be included in a study means a careful balance must be struck. Although a higher number of individuals aids in the identification of population structure, the overall scatter obtained here when plotting FST vs geographic distance (Figure 6) illustrates the need also for a large number of accessions to determine genetic structure of the species. This is particularly the case when the distance between sampling locations is high. The increasing variance in FST values as distance increase suggests that a dense sampling of populations will facilitate the evaluation of genetic structure. Ideally, a minimum number of individuals, such as the 5–10 individuals suggested by Fogelqvist et al. (2010), should be sampled while ensuring that the necessary number of populations can still be studied.
Our analysis of the between-individual distribution of within-accession diversity suggests that NGB468, VIR3221 and VIR2174 may be of mixed origin. Although the genetic diversity of these accessions is extremely high, individuals sharing the same total genotype are common, that is, there are several distinct total genotypes each shared by more than one individual, as would be expected in seed mixtures or lineages descending from seed mixtures. Additionally, although some individuals share origin with the Nordic group, others appear to have a different origin (Figure 3d, Karelia). It is not possible to determine whether this mixture is a result of contamination during genebank maintenance or if the mixture was present at the site at the time of collection. If the original collection occurred after a recent crop failure event, a part of the seed could have been recently imported from a different area. Further testing of material from areas suggested as potential trade partners in historical records will be needed to elucidate this.
Although the benefit of genebank conservation for plant breeders is unquestionable (reviewed by De Carvalho et al., 2013), concerns have been raised regarding the use of extant landrace material for studying questions regarding crop evolution (Lister et al., 2009; Hagenblad et al., 2012, Leino et al., 2013; Roullier et al., 2013). Genetic drift, selection and contamination are all processes that can lead to changes in the genetic identity of landraces during ex situ regenerations. Recently collected in situ preserved landraces, when available, can instead have been affected by twentieth century large-scale seed trade and movement. Our results suggest that some of these processes may well have had an effect on the genetic composition of our extant material. The extant material contains accessions that are both markedly less diverse (such as NGB2072 and NGB9529), possibly a consequence of strong genetic drift, and clearly more variable (for example, NGB321, VIR3221, VIR2143 and VIR2174), which could be due to contamination, than all or most of the historical accessions (Figure 2). However, it must be noted that the extant material in this study was chosen to cover geographic areas from which historical material was not available. This means that both age and geography may play a part in any comparisons between extant and historical material. From the historical samples, we draw the conclusion that the within-accession genetic diversity of Fennoscandian landrace barley before the introduction of modern plant improvement was, for these SNP markers, typically somewhere between 0.05 and 0.1.
The SNP markers used were originally ascertained on smaller discovery panels of barley cultivars (Rostoks et al., 2005, 2006; Close et al., 2009), and hence ascertainment bias may influence the results. Moragues et al. (2010) tested a set of 1536 SNP markers on a large set of 500 cultivars and 169 landraces, evaluating ascertainment bias and its effect on diversity and proposed two reduced sets of 384 SNPs optimized for European cultivars and Syrian and Jordanian landraces, respectively. Of these, we have used the one optimized for European cultivars. Our material is restricted to landraces and although ascertainment bias could render inferences of genetic variation outside the area of the study somewhat inaccurate, it is unlikely to have an effect on the genetic structure or comparisons of population parameters within the area. We also note that neither the minor allele frequency spectra (Supplementary Figure 1) nor the genetic diversity of the haplotype groups (Supplementary Table 2) indicate ascertainment bias as a major issue in this dataset.
Genetic analyses of historical specimens are a desirable alternative to steer clear of some of the issues associated with extant landraces (Jones et al., 2008; Lister et al., 2010, Hagenblad et al., 2012). However, the difficulties involved in obtaining sufficient numbers of individuals and quality of DNA for such studies means the number of investigations reporting within-accession diversity in historical material is even lower than that of extant landraces (but see Leino and Hagenblad, 2010; Hagenblad et al., 2012; Leino et al., 2013). Additionally, the degraded quality of DNA in most historical samples means that genetic analysis can be difficult and in many cases a relatively low number of genetic markers, usually chloroplast or mitochondrial and sometimes microsatellite markers have been analyzed (reviewed by Palmer et al., 2012).
Here we show that seeds more than 100 years old, when stored under favorable conditions, contain sufficient DNA to be analyzed with high-throughput SNP genotyping. SNP genotyping of historical specimens has previously been reported from animal samples, such as cattle (for example, Svensson et al., 2007) and salmon (for example, Johnston et al., 2013), and in small-scale studies of plants (for example, Lister et al., 2013). To our knowledge, this is the first report where historical plant samples have been analyzed by large-scale SNP genotyping, with more than one individual per accession and a high number of markers. Should this prove to be possible also for other material, it will expand the field of population genetics of historical plant specimens into that of population genomics. Historical seed collections such as those at the Swedish Museum of Cultural History, the Mustiala Agricultural College and Tromsø University Museum, are likely to prove unusually suited for these types of studies as the number of seeds in collections allows genomic analysis on a population scale.
Our genomic scale analysis allowed us to investigate issues such as LD. We found limited evidence of breakdown of intrachromosomal LD (Supplementary Figure 2), contrasting with previous findings in both wild barley (Morrell et al., 2005; Caldwell et al., 2006) and barley cultivars (Caldwell et al., 2006) but showing striking similarities to the landrace dataset from Syria and Jordan studied by Caldwell et al. (2006). Average intrachromosomal LD was also similar to that reported for landrace barley from Syria and Jordan by Russell et al. (2011) but higher than in wild barley from the same area and lower than in a worldwide set of both two- and six-row barley cultivars (Malysheva-Otto et al., 2006). Our results thus suggest that a similar genetic structure is maintained between landraces from the area of domestication and those from Fennoscandia. Additionally, landraces in general seem to show more intrachromosomal LD than wild barley, but less than improved cultivars. Our interchromosomal LD was, however, an order of magnitude higher than the one found in populations of Sardinian landrace barley (Rodriguez et al., 2012). Future studies will reveal whether this is an effect of the sampling size or the result of differences in population structure and gene flow between different areas. The similar levels of within-population LD in the different geographical groups suggest that levels of outcrossing and gene flow between accessions are of similar magnitude across Fennoscandia.
The spatial genetic structure of extant barley has been described earlier both on a worldwide scale (Malysheva-Otto et al., 2006; Saisho and Purugganan, 2007) and across Europe (Jones et al., 2011, 2012) using microsatellite markers. With only 14 microsatellite markers genotyped in landrace barley from historical seed collections, Leino and Hagenblad (2010) were able to show geographic structuring in barley on a much finer scale—within Sweden. Six-row barley landraces from the far north were genetically distinct from those of the rest of Sweden. This tentatively suggested two separate routes of migration into Sweden, but raised questions regarding the phylogeographic patterns of barley in the neighboring Fennoscandian countries.
Our analysis of 384 SNPs allowed us to verify the far northern Swedish population detected by Leino and Hagenblad (2010). We could further show that the cluster is distributed across the whole far-north Fennoscandia. Additionally, although Leino and Hagenblad (2010) could only detect a single far-north cluster, the increased power from the higher number of markers in this study presents a more detailed picture. In addition to the far-north cluster, two or possibly three more latitudinal clusters can be detected, all being shared across the longitudinal breadth of Fennoscandia. The far northern group detected by Leino and Hagenblad (2010) thus seemed not to be a Finnish group migrating into northern Sweden but the northernmost of a set of several latitudinally structured clusters shared across Fennoscandia. These results contrast with several historical records on seed movement within the area.
When analyzing the subset with only historical material the most conspicuous detail of the three-cluster structure model and the five-cluster is the far-northern group (depicted in turquoise/light grey in Figures 4 and 5). It stretches from Tromsø in the far north of Norway reaching east through the northernmost Sweden and Finland. This connection between northern Norway, northern Sweden and Finland is particularly fascinating as barley cultivation is not continuous in the area (Flygare, 2011), but interrupted by the Scandes mountain range. The genetic similarity instead appears to be a result of international seed trade over the mountain range. Such a trade in the far north of Fennoscandia is noteworthy because mercantilist policies during the period 1593–1788 restricted foreign seed import into Norway, at that time under Danish rule (Herstad, 2000). This culminated with a state monopoly on seed trade in the period 1735–1788 for northern and western Norway (Lunden, 2004). It is, however, known from historical records that settlers in inland northern Sweden brought harvested seed to millers across the border (Kjellström, 2012). To what extent the traded seed was grown in Norway, or whether the Swedish farmers brought back seed from the other side of the border is not clear; but from the results in this study, it seems likely that seed trade across the borders of northern Fennoscandia had an impact on the genetic composition of the cultivated barley.
Historical sources mention that during times of crop failure, seed was transported in large quantities from Estonia to Sweden (Heckscher, 1935), but traces of such trade is not evident in this study. The extant accession from Estonia, VIR2143, clearly and consistently separates from the historical Fennoscandian accessions (Figures 3d and 4a). More samples with origins around the eastern Baltic, preferably from historical collections, are needed to clarify the relationship between the Estonian and Fennoscandian landrace barley.
A striking feature from the genetic structure is its apparent independence from national borders within the region, contrary to what historical records suggest. Not only in the north but also along the whole north–south range of Fennoscandia, we see genetic clustering shared between countries along the approximately same latitude. For example, in 1867–1868, just a few decades before the sampling of the accessions studied here, a severe crop failure in northern Sweden resulted in large quantities of barley for food and seed being imported from southern Sweden (Häger et al., 1978). In spite of this, no mixing of south Swedish barley in the landraces from northern Sweden is evident. Although seeds were traded and documented human migrants presumably brought seeds with them, it is evident that long-term cultivation has been of the genotypes locally available, rather than imported seed. Possibly, adaptation to different climatic conditions has favored cultivation of local genotypes over imported ones. We are currently investigating the distribution of genetic diversity in candidate genes for climate adaptation to explore their role in barley cultivation.
Data archiving
Genotype data available from the Dryad Digital Repository: doi:10.5061/dryad.293hd.
References
Avise JC (2009). Phylogeography: retrospect and prospect. J Biogeogr 36: 3–15.
Badr A, Muller K, Schafer-Pregl R, El Rabey H, Effgen S, Ibrahim HH et al. (2000). On the origin and domestication history of barley (Hordeum vulgare). Mol Biol Evol 17: 499–510.
Brown TA (1999). How ancient DNA may help in understanding the origin and spread of agriculture. Philos Trans R Soc Lond B Biol Sci 354: 89–98.
Börner A, Chebotar S, Korzun V (2000). Molecular characterization of the genetic integrity of wheat (Triticum aestivum L.) germplasm after long-term maintenance. Theor Appl Genet 100: 494–497.
Caldwell KS, Russell J, Langridge P, Powell W (2006). Extreme population-dependent linkage disequilibrium in an inbreeding plant species, Hordeum vulgare. Genetics 172: 557–567.
Camacho Villa TC, Maxted N, Scholten M, Ford-Lloyd B (2005). Defining and identifying crop landraces. Plant Genet Res 3: 373–384.
Chebotar S, Roder MS, Korzun V, Borner A (2002). Genetic integrity of ex situ genebank collections. Cell Mol Biol Lett 7: 437–444.
Close TJ, Bhat PR, Lonardi S, Wu Y, Rostoks N, Ramsay L et al. (2009). Development and implementation of high-throughput SNP genotyping in barley. BMC Genomics 10: 582.
Conrad DF, Jakobsson M, Coop G, Wen X, Wall JD, Rosenberg NA et al. (2006). A worldwide survey of haplotype variation and linkage disequilibrium in the human genome. Nat Genet 38: 1251–1260.
De Carvalho MAP, Bebeli PJ, Bettencourt E, Costa G, Dias S, Dos Santos TM et al. (2013). Cereal landraces genetic resources in worldwide GeneBanks. A review. Agron Sustain Dev 33: 177–203.
ESRI (2011). ArcGIS Desktop: Release 10. Environmental Systems Research Institute: Redlands, CA.
Evanno G, Regnaut S, Goudet J (2005). Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14: 2611–2620.
Excoffier L, Lischer HEL (2010). Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Res 10: 564–567.
Falush D, Stephens M, Pritchard JK (2003). Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164: 1567–1587.
Flygare I (2011). The structure of agriculture. In: Jansson U, Wastenson L, Aspenberg P (eds.) National atlas of Sweden. Agriculture and forestry in Sweden since 1900—a cartographic description. Norstedt: Stockholm. p 58–70.
Fogelqvist J, Niittyvuopio A, Agren J, Savolainen O, Lascoux M (2010). Cryptic population genetic structure: the number of inferred clusters depends on sample size. Mol Ecol Res 10: 314–323.
Fuller DQ, Willcox G, Allaby RG (2011). Cultivation and domestication had multiple origins: arguments against the core area hypothesis for the origins of agriculture in the Near East. World Archaeol 43: 628–652.
Hagenblad J, Zie J, Leino MW (2012). Exploring the population genetics of genebank and historical landrace varieties. Genet Res Crop Evol 59: 1185–1199.
Heckscher EF (1935-1949). Sveriges ekonomiska historia från Gustav Vasa. Bonnier: Stockholm.
Herstad J (2000). I helstatens grep: kornmonopolet. Tano Aschehoug: Oslo, 1735–1788.
Hill WG, Robertson A (1968). Linkage disequilibrium in finite populations. Theor Appl Genet 38: 226–231.
Hunt HV, Campana MG, Lawes MC, PARK YJ, Bower MA, Howe CJ et al. (2011). Genetic diversity and phylogeography of broomcorn millet (Panicum miliaceum L.) across Eurasia. Mol Ecol 20: 4756–4771.
Hutchison DW, Templeton AR (1999). Correlation of pairwise genetic and geographic distance measures: Inferring the relative influences of gene flow and drift on the distribution of genetic variability. Evolution 53: 1898–1914.
Häger O, Torell C, Villius H (1978). Ett satans år: Norrland 1867. Sveriges Radio: Stockholm.
Isaac AD, Muldoon M, Brown KA, Brown TA (2010). Genetic analysis of wheat landraces enables the location of the first agricultural sites in Italy to be identified. J Archaeol Sci 37: 950–956.
Jakobsson M, Rosenberg NA (2007). CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23: 1801–1806.
Johnston SE, Lindqvist M, Niemela E, Orell P, Erkinaro J, Kent MP et al. (2013). Fish scales and SNP chips: SNP genotyping and allele frequency estimation in individual and pooled DNA from historical samples of Atlantic salmon (Salmo salar). BMC Genom 14: 439.
Jombart T, Devillard S, Balloux F (2010). Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet 11: 1–15.
Jones G, Jones H, Charles MP, Jones MK, Colledge S, Leigh FJ et al. (2012). Phylogeographic analysis of barley DNA as evidence for the spread of Neolithic agriculture through Europe. J Archaeol Sci 39: 3230–3238.
Jones H, Civan P, Cockram J, Leigh FJ, Smith LMJ, Jones MK et al. (2011). Evolutionary history of barley cultivation in Europe revealed by genetic analysis of extant landraces. BMC Evol Biol 11: 320.
Jones H, Lister DL, Bower MA, Leigh FJ, Smith LM, Jones MK (2008). Approaches and constraints of using existing landrace and extant plant material to understand agricultural spread in prehistory. Plant Gen Res 6: 98–112.
Kjellström R (2012). Nybyggarliv i Vilhelmina, Uppsala, Kungl. Gustav Adolfs Akademien för svensk folkkultur.
Lascoux M, Petit RJ (2010). The ‘New Wave’in plant demographic inference: more loci and more individuals. Mol Ecol 9: 1075–1078.
Leino MW (2010). Frösamlingar på museum. Nord Mus 2010: 96–108.
Leino MW, Boström E, Hagenblad J (2013). Twentieth-century changes in the genetic composition of Swedish field pea metapopulations. Heredity 110: 338–346.
Leino MW, Hagenblad J (2010). Nineteenth century seeds reveal the population genetics of landrace barley (Hordeum vulgare). Mol Biol Evol 27: 964–973.
Leino MW, Hagenblad J, Edqvist J, Strese EMK (2009). DNA preservation and utility of a historic seed collection. Seed Sci Res 19: 125–135.
Lister DL, Bower MA, Jones MK (2010). Herbarium specimens expand the geographical and temporal range of germplasm data in phylogeographic studies. Taxon 59: 1321–1323.
Lister DL, Jones H, Jones MK, O'Sullivan DM, Cockram J (2013). Analysis of DNA polymorphism in ancient barley herbarium material: Validation of the KASP SNP genotyping platform. Taxon 62: 779–789.
Lister DL, Thaw S, Bower MA, Jones H, Charles MP, Jones G et al. (2009). Latitudinal variation in a photoperiod response gene in European barley: insight into the dynamics of agricultural spread from ‘historic’ specimens. J Archaeol Sci 36: 1092–1098.
Londo JP, Chiang Y-C, Hung K-H, Chiang T-Y, Schaal BA (2006). Phylogeography of Asian wild rice, Oryza rufipogon, reveals multiple independent domestications of cultivated rice, Oryza sativa. Proc Natl Acad Sci USA 103: 9578–9583.
Lunden K (2004). Recession and new Expansion. In: Almås R (Ed) Norwegian Agricultural History. Tapir Academic Press: Trondheim, 141–232.
Malysheva-Otto LV, Ganal MW, Roder MS (2006). Analysis of molecular diversity, population structure and linkage disequilibrium in a worldwide survey of cultivated barley germplasm (Hordeum vulgare L.). BMC Genet 7: 6.
Moragues M, Comadran J, Waugh R, Milne I, Flavell AJ, Russell JR (2010). Effects of ascertainment bias and marker number on estimations of barley diversity from high-throughput SNP genotype data. Theor Appl Genet 120: 1525–1534.
Morrell PL, Clegg MT (2007). Genetic evidence for a second domestication of barley (Hordeum vulgare) east of the Fertile Crescent. Proc Natl Acad Sci USA 104: 3289–3294.
Morrell PL, Toleno DM, Lundy KE, Clegg MT (2005). Low levels of linkage disequilibrium in wild barley (Hordeum vulgare ssp. spontaneum) despite high rates of self-fertilization. Proc Natl Acad Sci USA 102: 2442–2447.
Nei M (1973). Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci USA 70: 3321–3323.
Nielsen R, Hubisz MJ, Clark AG (2004). Reconstituting the frequency spectrum of ascertained single-nucleotide polymorphism data. Genetics 168: 2373–2382.
Nordborg M, Hu TT, Ishino Y, Jhaveri J, Toomajian C, Zheng H et al. (2005). The pattern of polymorphism in Arabidopsis thaliana. PLoS Biol 3: e196.
Oliveira HR, Campana M, Jones H, Hunt H, Leigh F, Lister DL et al. (2012). Tetraploid wheat landraces in the Mediterranean basin: taxonomy, evolution and genetic diversity. PLoS One 7: e37063.
Oliveira HR, Hagenblad J, Leino MW, Leigh FJ, Lister DL, Penã-Chocarro L et al. (2014). Wheat in the Mediterranean revisited—tetraploid wheat landraces assessed with elite bread wheat single-nucleotide polymorphism markers. BMC Genetics 15: 54.
Olsen KM, Schaal BA (1999). Evidence on the origin of cassava: phylogeography of Manihot esculenta. Proc Natl Acad Sci USA 96: 5586–5591.
Osvald H (1959). Åkerns nyttoväxter. Stockholm: Sv. litteratur.
Palmer SA, Smith O, Allaby RG (2012). The blossoming of plant archaeogenetics. Ann Anat 194: 146–156.
Pandey M, Wagner C, Friedt W, Ordon F (2006). Genetic relatedness and population differentiation of Himalayan hulless barley (Hordeum vulgare L.) landraces inferred with SSRs. Theor Appl Genet 113: 715–729.
Parzies HK, Spoor W, Ennos RA (2000). ‘Genetic diversity of barley landrace accessions (Hordeum vulgare ssp vulgare) conserved for different lengths of time in ex situ gene banks.’. Heredity 84: 476–486.
Pritchard JK, Stephens M, Donnelly P (2000). Inference of population structure using multilocus genotype data. Genetics 155: 945–959.
R Development Core Team (2013). R: A language and environment for statistical computing. R Foundation for Statistical Computing: Vienna, Austria.
Ren X, Nevo E, Sun D, Sun G (2013). ‘Tibet as a potential domestication center of cultivated barley of China.’. PLoS ONE 8: 1.
Rodriguez M, Rau D, O’Sullivan D, Brown AHD, Papa R, Attene G (2012). Genetic structure and linkage disequilibrium in landrace populations of barley in Sardinia. Theor Appl Genet 125: 171–184.
Rosenberg NA (2004). DISTRUCT: a program for the graphical display of population structure. Mol Ecol Notes 4: 137–138.
Rostoks N, Mudie S, Cardle L, Russell J, Ramsay L, Booth A et al. (2005). Genome-wide SNP discovery and linkage analysis in barley based on genes responsive to abiotic stress. Mol Genet Genomics 274: 515–527.
Rostoks N, Ramsay L, MacKenzie K, Cardle L, Bhat PR, Roose ML et al. (2006). Recent history of artificial outcrossing facilitates whole-genome association mapping in elite inbred crop varieties. Proc Natl Acad Sci USA 103: 18656–18661.
Roullier C, Benoit L, McKey DB, Lebot V (2013). Historical collections reveal patterns of diffusion of sweet potato in Oceania obscured by modern plant movements and recombination. Proc Natl Acad Sci USA 110: 2205–2210.
Russell J, Dawson IK, Flavell AJ, Steffenson B, Weltzien E, Booth A et al. (2011). Analysis of >1000 single nucleotide polymorphisms in geographically matched samples of landrace and wild barley indicates secondary contact and chromosome-level differences in diversity around domestication genes. New Phytol 191: 564–578.
Saisho D, Purugganan MD (2007). Molecular phylogeography of domesticated barley traces expansion of agriculture in the Old World. Genetics 177: 1765–1776.
Steiner AM, Ruckenbauer P, Goecke E (1997). Maintenance in genebanks, a case study: contaminations observed in the Nurnberg oats of 1831. Gen Res Crop Evol 44: 533–538.
Svensson EM, Anderung C, Baubliene J, Persson P, Malmström H, Smith C et al. (2007). Tracing genetic change over time using nuclear SNPs in ancient and modern cattle. Anim Genet 38: 378–383.
Van Heerwaarden J, Doebley J, Briggs WH, Glaubitz JC, Goodman MM, Gonzalez JJS et al. (2011). Genetic signals of origin, spread and introgression in a large sample of maize landraces. Proc Natl Acad Sci USA 108: 1088–1092.
Weir BS, Cockerham CC (1984). Estimating F-statistics for the analysis of population structure. Evolution 38: 1358–1370.
Yahiaoui S, Igartua E, Moralejo M, Ramsay L, Molina-Cano JL, Ciudad FJ et al. (2007). Patterns of genetic and eco-geographical diversity in Spanish barleys. Theor Appl Genet 116: 271–282.
Zeven AC (1998). Landraces: A review of definitions and classifications. Euphytica 104: 127–139.
Acknowledgements
This work was funded by the Swedish Research Council for Environment, Agricultural Sciences and Spatial Planning (FORMAS) and the Lagersberg Foundation. Seed material was kindly provided by Fredrik Ottosson at NordGen, Külli Annamaa at the Jögevaa Plant Breeding Institute, Igor Loskutov at the N.I. Vavilov Institute of Plant Industry, Annika Michelsson at Mustiala Agricultural College, Hannu Ahokas at MTT and Torbjørn Alm at Tromsø University Museum. Maria Lundström is acknowledged for skillful technical assistance.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on Heredity website
Rights and permissions
About this article
Cite this article
Forsberg, N., Russell, J., Macaulay, M. et al. Farmers without borders—genetic structuring in century old barley (Hordeum vulgare). Heredity 114, 195–206 (2015). https://doi.org/10.1038/hdy.2014.83
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/hdy.2014.83
This article is cited by
-
Genetic diversity and population structure of masson pine (Pinus massoniana Lamb.) superior clones in South China as revealed by EST-SSR markers
Genetic Resources and Crop Evolution (2021)
-
Genetic diversity and structure of Elymus tangutorum accessions from western China as unraveled by AFLP markers
Hereditas (2019)
-
Population structure in landrace barley (Hordeum vulgare L.) during the late 19th century crop failures in Fennoscandia
Heredity (2019)
-
Population genetic structure in Fennoscandian landrace rye (Secale cereale L.) spanning 350 years
Genetic Resources and Crop Evolution (2019)
-
Morphological and genetic characterization of barley (Hordeum vulgare L.) landraces in the Canary Islands
Genetic Resources and Crop Evolution (2019)