Abstract
Allopolyploidy is an evolutionary and mechanistically intriguing process, in that it entails the reconciliation of two or more sets of diverged genomes and regulatory interactions. In this study, we explored gene expression patterns in interspecific hybrid F1, and synthetic and natural allopolyploid cotton using RNA-Seq reads from leaf transcriptomes. We determined how the extent and direction of expression level dominance (total level of expression for both homoeologs) and homoeolog expression bias (relative contribution of homoeologs to the transcriptome) changed from hybridization through evolution at the polyploid level and following cotton domestication. Genome-wide expression level dominance was biased toward the A-genome in the diploid hybrid and natural allopolyploids, whereas the direction was reversed in the synthetic allopolyploid. This biased expression level dominance was mainly caused by up- or downregulation of the homoeolog from the ‘non-dominant’ parent. Extensive alterations in homoeolog expression bias and expression level dominance accompany the initial merger of two diverged diploid genomes, suggesting a combination of regulatory (cis or trans) and epigenetic interactions that may arise and propagate through the transcriptome network. The extent of homoeolog expression bias and expression level dominance increases over time, from genome merger through evolution at the polyploid level. Higher rates of transgressive and novel gene expression patterns as well as homoeolog silencing were observed in natural allopolyploids than in F1 hybrid and synthetic allopolyploid cottons. These observations suggest that natural selection reconciles the regulatory mismatches caused by initial genomic merger, while new gene expression conditions are generated for evaluation by selection.
Similar content being viewed by others
Introduction
Polyploidy is a fundamental process in plant evolution, leading to the merger of two or more genomes into the same nucleus. All flowering plant lineages are known to have experienced at least one round of polyploidization in their evolutionary history, as indicated by genome analysis in diverse lineages of plants (Jiao et al., 2011). The additional chromosome set(s) can be derived from intraspecific genome duplication (autopolyploidy) or hybridization and genome duplication of two different species (allopolyploidy) (Doyle et al., 2008).
Allopolyploidy is evolutionary and mechanistically intriguing, in that the process entails the reconciliation of two or more sets of diverged genomes and regulatory interactions. This coexistence of diverged genomes is associated with major changes at the genetic and epigenetic levels, with important consequences for the expression patterns of genes from both genomes and the phenotype (Doyle et al., 2008; Jackson and Chen, 2010). Recent findings show that duplicate gene pairs may display homoeolog expression bias, where the two homoeologs are expressed unequally, often varying among tissues and from a priori expectations based on progenitor diploid expression levels (Flagel et al., 2008; Chaudhary et al., 2009; Buggs et al., 2011; Dong and Adams, 2011; Combes et al., 2012). In addition to expression bias, a second phenomenon was more recently described, based on the relative expression levels of the two parents compared with the total expression of a gene pair in their allopolyploid derivative. Specifically, the total expression level of a homoeolog pair in an allopolyploid may be similar to that exhibited by only one of the two diploid parents, irrespective of whether that parent displays up- or downregulation with respect to the other parent. This phenomenon has been termed genomic dominance (Rapp et al., 2009). Originally described in cotton (Rapp et al., 2009; Flagel and Wendel, 2010), genomic dominance appears to be a common and perhaps ubiquitous property in allopolyploids, having recently been reported in Spartina (Chelaifa et al., 2010), wheat (Chagué et al., 2010) and coffee (Bardil et al., 2011). Because this is a RNA- rather than DNA-level process, and to avoid confusion caused by alternative usages of the expression genomic dominance, we recently have proposed that a more appropriate term for the phenomenon is ‘expression level dominance’ (Grover et al., 2012).
Here, we use the terms ‘homoeolog expression bias’ and ‘expression level dominance’ for expression patterns observed within individual homoeologous genes, and for overall expression of both homoeologs, respectively. Importantly, both homoeolog expression bias and expression level dominance may be balanced among loci in a genome with respect to diploid parent-of-origin or, alternatively, they may exhibit a preference toward one parental genome (referred to as unbalanced) (Grover et al., 2012).
Mechanism(s) explaining the more recently described phenomenon of expression level dominance remain unclear, and the phenomenon itself needs additional illumination. In particular, the individual contributions of homoeologous transcripts to the pattern of expression level dominance have not been explored. In principle, two homoeologs of a gene may contribute equally or unequally to total gene expression for each gene pair in allopolyploids, the latter reflecting homoeolog expression bias, a phenomenon sometimes misinterpreted in the literature as genomic dominance (here described as expression level dominance) (Grover et al., 2012).
In cotton, two studies reported expression level dominance (previously, genomic dominance) in synthetic and natural allopolyploids (Rapp et al., 2009; Flagel and Wendel, 2010). The magnitude of expression level dominance was reported to be high, with respect to the number of genes, in resynthesized allopolyploid cotton, with as many as 11 700 out of 40 100 genes displaying expression levels that were statistically equal to one of the two diploid parents (Rapp et al., 2009); Flagel and Wendel (2010) also showed that about 34% of genes investigated exhibited this phenomenon. Interestingly, this study showed that the direction of expression level dominance in cotton became more balanced on an evolutionary timescale, with the bias in direction of expression level dominance being most severe in the interspecific diploid hybrid F1, and attenuated in natural allopolyploids, even though the overall magnitude of gene pairs displaying expression level dominance remained high (Flagel and Wendel, 2010).
In general, deciphering the contribution of homoeologs to the transcript pool in allopolyploids has been hampered by the lack of genomic data and low divergence between progenitor diploid genomes. For these reasons, most studies involve small-scale experiments, for example, 13 genes in coffee (Combes et al., 2012) and 10 to 144 genes in Tragopogon (Koh et al., 2010; Buggs et al., 2011). By leveraging EST resources from both progenitor diploids of allopolyploid cotton, Flagel and Wendel (2010) were able to extend analysis to include a larger number of homoeolog gene pairs (1383), providing an overview of a homoeolog expression bias on a genomically more widespread scale. However, they did not examine how homoeolog expression bias at individual genes was related to the phenomenon of expression level dominance. Because of the plausible mechanistic connections between bias and dominance and to further our understanding of these complex transcriptomic dynamics in allopolyploids, here we used total transcriptome profiling to explore expression level dominance in polyploid cotton and its relationship to homoeologous gene expression.
Cotton (Gossypium) is a useful model for studying polyploid evolution because of its well-documented evolutionary history and extensive genetic and genomic resources. Approximately 1–2 million years ago (mya), allopolyploidization between Old World, A-genome (resembling modern Gossypium arboreum, designated A2, and G. herbaceum) and New World, D-genome (similar to modern G. raimondii (D5)) taxa resulted in a new allopolyploid lineage, which now includes five species, with genome designations AD1 to AD5 (Wendel and Cronn, 2003). In addition, because several species have been independently domesticated for their cotton ‘fiber’ (single-celled, epidermal trichomes), comparative analyses of wild and domesticated cottons may provide insight into how selection under domestication affects gene expression in allopolyploids.
To date, all studies of expression level dominance have used microarrays to investigate global gene expression patterns in polyploids and their progenitors. These techniques rarely are ideal for large-scale analyses of homoeolog expression due to overlap in hybridization among homoeologs (see, however, Udall et al., 2006). RNA-Seq has several key advantages for transcriptome profiling, including the lack of a priori information on genome sequences, no upper limit for quantification, higher accuracy for distinguishing and quantifying expression levels of homoeologous copies, and a high level of reproducibility (Wang et al., 2009). Using RNA-Seq, we extend earlier findings of homoeolog expression bias and expression level dominance by parsing expression patterns of leaf transcriptomes in hybrid and polyploid cottons, and their parents, into the 12 categories by which three entities (two parents and either a hybrid or a polyploid) may statistically differ in expression (as described by Rapp et al. (2009)). We address the magnitude and directionality of expression level dominance and homoeolog expression bias, the relationships between dominance and bias, and how hybridization, polyploidy and domestication have contributed to these phenomena.
Materials and methods
Plant material
For this study, we used seven accessions, including two natural allopolyploids (a wild form of G. hirsutum accession TX2094 and the domesticated cultivar ‘Maxxa’) with models of their A- and D-genome diploid progenitors, G. arboreum (A2) and G. raimondii (D5) (Wendel and Cronn, 2003), as well as the corresponding interspecific diploid F1 hybrid (A2 × D5) (Table 1). The latter accession represents genome merger without chromosome doubling, while the inclusion of both TX2094 and Maxxa permits an initial exploration of the possible effects of human selection. The synthetic allopolyploid corresponding to this hybrid has not been successfully created because of sterility of the F1 hybrid; therefore, a substitute synthetic allopolyploid was used, 2(A2D1), generated from a different D-genome (G. thurberi, D1) but the same female species (G. arboretum, A2). Despite the different paternal parent (D1 instead of D5), the phylogenetic closeness of these two D-genomes (Wendel and Cronn, 2003) justifies using this synthetic allopolyploid for inferring the effect of polyploidization on gene expression evolution in allopolyploids.
Seeds were scarified and germinated under high humidity in a 1:1 mix of sand and soil. As the F1 hybrid is not fertile, we used vegetatively propagated clones using cuttings. A young branch from a mature plant was cut and dipped in a solution containing a growth hormone. Then, it was placed in a 1:1 mix of soil and sand for 6 weeks. To minimize developmental or environmental effects, we collected newly emerged young leaf tissue from plants grown under the same conditions as other accessions had. Three biological replicates of each taxon were grown in a randomized complete block design under controlled greenhouse conditions in the Bessey Hall at Iowa State University (Ames, IA, USA).
RNA extraction and library construction for sequencing
The seventh fully expanded true leaf (3 cm in length) was collected and frozen in liquid nitrogen and stored at −80 °C until extraction. RNA was extracted using a CTAB extraction protocol (Doyle and Doyle, 1987), and then purified with the RNeasy Plant Mini Kit (Qiagen, Stanford, CA, USA). The quality and concentration of extracted RNAs were determined using the Agilent 2100 Bioanalyzer (Agilent, Santa Clara, CA, USA). mRNA purification was performed using the MicroPoly(A) Purist kit (Ambion, Austin, TX, USA), and then processed for RNA-Seq library construction following a suggested protocol (Nagalakshmi et al., 2010). The 21 constructed libraries, indexed (bar-coded) with six nucleotide sequences, were pooled together in equimolar amounts and were sequenced on the Illumina Genome Analyzer IIx sequencer (Illumina, San Diego, CA, USA) with 80 base reads at the Genomics Core Facility at the University of Oregon.
Analysis of RNA-Seq data: mapping and differential expression
Raw reads were classified into the correct accession according to their individual barcodes using fastx (http://hannonlab.cshl.edu/fastx_toolkit/index.html). After trimming the indexed sequences, fastq formatted reads from each library were mapped to the reference genome using Burrows Wheeler Alignment (Li and Durbin, 2009). We utilized the Cotton46A EST assembly (http://www.cottonevolution.info/) as the reference genome (with 56 372 genes) because this database was built using transcripts from both diploid and allopolyploid species (A2, D5 and AD1) and various tissues (floral, vegetative and fiber tissues). Most relevant to this study, this resource identifies single-nucleotide polymorphisms (SNPs) between the A- and D-genome progenitors for 25 317 genes, identified based on several sequencing techniques, including Sanger sequencing, 454 and Illumina (Solexa) (Flagel et al., 2012). We also mapped our reads using the recently available Cotton D5- (G. raimondii) genome sequence (DOE Joint Genome Institute: Cotton D V.1.0; http://www.phytozome.net/cotton.php) to compare the mapping results.
Aligned sequences were filtered by MAPQ (MAPping Quality) equal to or larger than either 24 or 25 (depending on accession) using Samtools (Li et al., 2009). Reads with SNPs that differed between the A- and D-genome progenitors were parsed into A or D homoeolog-specific bins using custom python scripts. We use the term homoeolog in place of parental alleles in the F1 hybrid to facilitate ease of comparison across ploidy levels. Also, we designate homoeolog-specific reads as A or D reads in the diploid progenitors, and At or Dt reads in the allopolyploids.
Mapped, filtered and sorted reads were analyzed with the edgeR package (ver. 2.0.5) in R software version 2.13 (R Foundation for Statistical Computing, Vienna, Austria). First, replicates were normalized by the TMM (trimmed mean of M values) method, and then using overall expression level of both homoeologs for a gene, differential expression was assessed in the F1 hybrid and allopolyploids relative to their diploid parents using Fisher’s exact tests (Robinson et al., 2010). The distribution of P-values was controlled for a false discovery rate by the BH method (Benjamini and Hochberg, 1995) at α=0.05. To remove the negative effect of background expression noise on differential expression calls, we restricted analysis to genes having read counts (≧1) in all accessions and biological replicates in each contrast.
Analyses of expression level dominance and homoeolog expression bias
We first explored the data for novel expression (new expression of a gene for this tissue) and homoeolog silencing patterns (no expression of one homoeolog) in the hybrid and polyploids. Novel expression was inferred when both parental species had no reads for a gene, yet hybrids and/or allopolyploids displayed more than 10 read counts per gene per million reads in all three biological replicates. If both parental species had more than 10 read counts per homoeolog per million reads, but hybrids and/or allopolyploids had zero counts for the same homoeolog, this was considered silencing. These two cases were eliminated from further analysis, focusing on genes that are expressed in at least one parent and where both homoeologs are expressed in the polyploids and F1 hybrid.
Genes identified as differentially expressed in the hybrid and allopolyploid relative to their diploid parents were binned into 12 possible expression classes of differential expression (see Figure 2), that is, expression level dominance, additivity and transgressive (outside the range of either parent), according to Rapp et al. (2009). Briefly, genes were parsed into these 12 categories (using Roman numerals; see Figure2), depending on relative expression levels between the two parents and that of the hybrid or polyploid. Examined in this manner, genes may display additivity (I and XII), D-expression level dominance (II and XI), A-expression level dominance (IV and IX), transgressive expression lower than either parent (III, VII and X) or transgressive expression higher than either parent (V, VI and VIII).
For each of the 12 categories above (which are based on joint expression levels for both homoeologs), we tabulated homoeolog-specific reads (available for 25 317 genes) to examine how homoeolog usage for each gene pair was related to total gene expression for each homoeolog pair for each of the 12 categories. For example, a gene displaying additivity (for example, category I) may, in principle, do so because of equal expression of both homoeologs or because of some highly unbalanced ratio of the two homoeologs. For this analysis, the relative contribution of homoeologs to gene expression in the F1 and allopolyploids (At or Dt up- or downregulation based on comparison of At vs Dt) was assessed via Fisher’s exact tests. In addition, we examined if individual homoeologs display a conserved level of expression relative to the diploid progenitors. To accomplish this, we compared the expression level of each homoeolog to that of the parental gene in the diploid species using Fisher’s exact tests (that is, At vs A or Dt vs D).
A temporal dimension to expression level dominance and homoeolog expression bias
To determine how the extent and direction of homoeolog expression bias and expression level dominance have changed in response to hybridization, evolution at the polyploid level and domestication, we compared gene expression categories and homoeolog expression bias across the F1 diploid hybrid, the synthetic and natural polyploids, and the in silico mid-parent expression values (MPV). Values from the three biological replicates of each parent were averaged for MPV and then analyzed in the same manner as described above.
For homoeolog expression bias, comparisons were made in two ways, including and excluding the synthetic allopolyploid, because it has a different D-genome parent (D1) than the D-genome diploid (D5) used for comparisons with natural allopolyploids. For the analyses involving the synthetic allopolyploid, we therefore included only those genes where the two different D-genome diploid parents had equivalent expression.
Results
Mapping efficiency of RNA-Seq data
To investigate and compare transcript accumulation in synthetic diploid hybrid and synthetic and natural allopolyploids relative to their diploid progenitors, 56 372 genes were surveyed for their expression using RNA-Seq. A total of 148 million (M) reads from 21 libraries were obtained and an average of 80% of those reads mapped to the reference transcriptome assembly Cotton46A (Flagel et al., 2012) (Supplementary Table S1). After excluding reads with low mapping quality scores, 43.7% on average of the total reads (approximately 3 M reads per library) were mapped, and these mapped reads were statistically evaluated for differential expression between progenies and their diploid progenitors. Among the 56 372 genes in the cotton assembly, an average of 47.5% were expressed in leaf tissue (Table 1). Notably, percentages were similar among the two D-genome diploids, as expected from their high genetic similarity (>99% nucleotide identity in exons) (Alvarez et al., 2005). We also mapped the reads from D5 onto the recently released D5-genome draft sequence to compare mapping efficiency; this resulted in 75% of reads mapping onto the 13 scaffolds of the draft assembly (Supplementary Table S2). As this genome was not annotated at the time of this writing, we used the Cotton46A assembly for further analysis.
Differential gene expression, novel expression and silencing
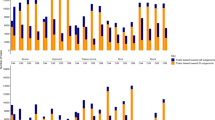
As a prerequisite to study gene expression patterns in allopolyploid or hybrid plants, we performed pairwise comparisons between the diploid parents to assess pre-existing differential gene expression (Figure 1). About 42.0–50.0% of genes were differentially expressed between the diploid parents, with equivalent proportions of upregulated orthologous gene copies relative to the other parent for A2 vs D5 (P>0.05 in all comparisons; Fisher’s exact test). For example, in panel a of Figure 1, 5288 genes (21.4% of the genes analyzed) were upregulated in A2 relative to D5, whereas an additional 5154 genes (20.8%) were upregulated in the latter species relative to the former. In contrast, slightly more genes were upregulated in A2 vs D1 (26.4% vs 23.8%, respectively, P<0.001; Fisher’s exact test).
Differentially expressed genes in each contrast between progenies and their diploid progenitors. Bold text indicates the total number and fraction of genes differentially expressed in each contrast. Also shown for each contrast is the partitioning of the total number of differentially expressed genes into the direction of upregulation. For example, in (a), 10 442 genes are indicated as being differentially expressed between G. arboreum and G. raimondii. Of these, 5288 are upregulated in G. arboreum, and 5154 genes are upregulated in G. raimondii. About 42.2% of 24 733 genes commonly expressed in the F1 hybrid and diploid parents were differentially expressed between diploids, with a range of 22.2–30.1% between the F1 hybrid and its diploid parents. The asymmetry between differential expression between the progeny and its diploid parents corresponds to genome-wide expression level dominance toward one parental genome. (a) An interspecific hybrid F1 A2 × D5 generated from the diploid parents G. arboreum (A2) and G. raimondii (D5). (b) G. hirsutum var. yucatanense accession TX2094 is a wild form of G. hirsutum, based on morphological evidence and restriction fragment length polymorphism (RFLP) data (Brubaker and Wendel, 1994). (c) G. hirsutum cv. Acala Maxxa represents elite modern domesticated cotton. (d) Synthetic allopolyploid 2(A2D1) generated from the diploid parents G. arboreum (A2) and G. thurberi (D1).
In all comparisons, the percentage of genes showing differential expression between the F1 hybrid or allopolyploids and their two parents was asymmetric (P<0.05; Fisher’s exact tests). That is, there was a bias in the direction of differential expression relative to the parents. For three out of the four plants (F1, TX2094 and Maxxa), global expression patterns were closer to the A progenitor than to the D progenitor. For example, 22.2% of genes were differentially expressed between the F1 and the A2 parent, whereas this number was 30.1% relative to D5 (Figure 1, P<0.001; Fisher’s exact test). Conversely, for the fourth case involving the synthetic allopolyploid, the global pattern of gene expression was biased toward the D-genome parent (D1).
In the synthetic F1 hybrid and synthetic allopolyploid, only 28 (0.05%) and 24 genes (0.04%) exhibited novel expression, respectively. In the natural allopolyploids, however, novel expression was greatly increased, with 611 (1.08%) and 688 (1.22%) genes in TX2094 and Maxxa, respectively (Table 2).
We evaluated homoeolog silencing for the 25 317 genes that have SNP information that diagnoses A and D homoeologs. There were 14 cases of homoeolog silencing in the F1 hybrid and 14 cases in the synthetic allopolyploid, while natural allopolyploids showed more silencing events. Specifically, TX2094 and Maxxa displayed 180 (0.71%) and 191 (0.75%) cases (out of 25 317 genes), respectively (P<0.001 in Fisher’s exact test), distributed equally between A and D silencing events (P>0.05 in Fisher’s exact tests; Table 2).
Expression level dominance in the leaf transcriptome
To detect additivity, transgressive expression and expression level dominance, we binned genes in the F1 hybrid and allopolyploids into 12 categories, as described (see Materials and methods). The categories of additivity (I and XII in Figure 2) and equivalent expression (‘no change’ in Figure 2) were relatively static; 16.6% (derived as (2180+1934)/24 733) of the gene pairs in the F1 hybrid, representing the initial stage of genome merger, displayed additivity, whereas the two natural allopolyploids, TX2094 (wild G. hirsutum) and Maxxa (domesticated G. hirsutum), showed 14.1% and 13.8%, respectively (P<0.001, Fisher’s exact tests; Figure 2). As one might expect, there was an appreciable increase in genes that were transgressively upregulated (categories V, VI and VIII in Figure 2) and downregulated (categories III, VII and X in Figure 2) in the natural allopolyploids, which originated 1- 2 mya (Wendel and Cronn, 2003), relative to the numbers observed in the F1 hybrid (P<0.001, Fisher’s exact tests). Notably, more transgressive expression was observed in domesticated relative to wild cotton (P<0.001, Fisher’s exact test). Because the synthetic allopolyploid has a different parentage, comparisons may not be entirely appropriate, but we do note that their patterns are more like those of the synthetic diploid hybrid than the natural allopolyploids, with respect to the proportion of genes exhibiting additivity and transgressive expression (Figure 2).
The 12 possible differential expression states in the F1 hybrid, natural (TX2094 and Maxxa) and synthetic (2(A2D1) allopolyploids relative to their diploid parents. Roman numerals indicate the same categorization as used in Rapp et al. (2009), with figures schematizing their respective gene expression pattern for the A-genome, diploid maternal parent (♀), the F1 or allopolyploids (P) and the D-genome diploid paternal parent (♂).
We examined all four progenies for evidence of expression level dominance. Overall, approximately 25% of genes fell into an expression level dominance category (Figure 2). Paralleling the depictions in Figure 1, the F1 hybrid and two natural allopolyploids exhibited genome-wide expression level dominance favoring the A-genome, while the direction of expression level dominance was reversed in the synthetic allopolyploid. The degree of biased expression level dominance was the most severe in the F1 hybrid, where 3738 (15.2% of all genes, categories IV and IX) had expression levels statistically equivalent to the A-genome parent, compared with 2248 (9.0%, categories II and XI) for the D-genome parent (P<0.001, Fisher’s exact test). Thus, 62.4% more gene pairs (3738 vs 2248) exhibited expression level dominance toward the A-parent than the D-parent. This percentage increase in the bias of expression level dominance was similar for domesticated allopolyploid cotton (61.5%), but significantly smaller (54.7%) for the wild allopolyploid (P<0.001, Fisher’s exact test).
The relationship of homoeolog-specific expression to expression level dominance
To evaluate how expression of individual homoeologs relates to joint homoeolog expression, we examined homoeolog bias in each of the 12 categories of differential expression. The results showed that the number of genes showing homoeolog expression bias was balanced with respect to parent of origin in the F1 hybrid and natural allopolyploids (for example, A-bias vs D-bias=1180 vs 1219 in the F1), whereas the synthetic allopolyploid showed unbalanced biased expression with a preference toward the A-genome (A-bias vs D-bias=1671 vs 1486).
Notably, for most genes homoeologs contributed equally to the transcriptome of the F1 hybrid and the allopolyploids for all 12 categories, even when they exhibited unequal expression between the parents. For example, in TX2094, 277 genes in category II displayed similar levels of A and D homoeolog expression, even though by definition these genes were upregulated in the D-genome parent relative to the A-genome parent (Supplementary Table S3). When we examined the mapped reads for these genes, A homoeologs were upregulated in TX2094 relative to the A-genome parent (a generalized example shown in Figure 3a). In contrast, 310 genes in category XI, where the A-genome parent has a higher expression level than D-genome parent, display equal expression of both homoeologs in TX2094, demonstrating downregulation of A homoeologs in TX2094 relative to its A-genome parent (cf., Figure 3b). The same phenomenon was observed for genes showing expression level dominance toward the A-genome (cf., Figures 3c and d). Investigation of homoeolog expression levels relative to those of their progenitors showed that a significant number of genes support this explanation for the homoeolog basis of expression level dominance, namely, that it reflects up- or downregulation of the alternative homoeolog, or far less often, up- or downregulation of both homoeologs (Figure 4). This phenomenon is variable among the four progenies; in the F1, for example, there is more modification of the D homoeolog of gene pairs (534 genes total, based on 371 up- and 163 downregulated) than of the A homoeolog (170 genes=88 up- and 82 downregulation; P<0.001, Fisher’s exact test) (Figure 4). In contrast, we observed slightly more A homoeolog modification in the synthetic allopolyploid, where genome-wide expression level dominance favoring the D-genome was present (419 vs 347=A vs D; P>0.05, Fisher’s exact test). This regulation pattern was also observed for the transcriptome overall; for example, there was more novel D homoeolog regulation in the F1 hybrid and natural allopolyploids than for the A homoeolog, while the reverse was observed for the synthetic allopolyploid (P<0.01 in Fisher's exact tests; Supplementary Table 6).
Partial explanation for expression level dominance. If the progeny AB maintains the parental expression pattern, its homoeolog expression pattern will be additive (blue text). If, however, the parent B has higher expression than the parent A, expression level dominance in the direction of parent B can be explained by upregulation of the A homoeolog (a). If the parent A shows higher expression than the parent B, expression level dominance in the direction of parent B can be explained by downregulation of the A-homoeolog (b). Up- or downregulation of the A homoeolog can thus result in equal expression of both homoeologs in the progeny AB (red text). This same logic may be applied to explain expression level dominance in the direction of the parent A by up- or downregulation of the B homoeologs (c and d), respectively.
Homoeolog expression level relative to parental expression levels, and expression level dominance. Shown at the top are figures that parallel those from Figure 2, representing the four categories of expression level dominance in the hybrid and allopolyploids (II and XI: D-genome expression level dominance; IV and IX: A-genome expression level dominance). The total number of genes falling into each category was tabulated based on comparisons of homoeolog expression levels to those of the diploid parents.
To address whether the patterns of homoeolog expression bias observed above simply reflect the vertical transmission of pre-existing expression differences, or if instead they reflect novel regulatory and/or evolutionary interactions, we compared genes on a case-by-case basis for the parental diploids and their derived hybrid and polyploids. As shown in Table 3, the patterns observed in the diploid parents were often conserved in the hybrid and allopolyploid derivatives. For example, the first three lines demonstrate that the parental expression patterns were maintained for greater than half of all genes, reflecting 59.4% (in the synthetic) to 70.9% (in the F1) of all genes included in this analysis (P<0.001, Fisher’s exact tests). Lines 4 and 5 represent the second most common class of genes, representing 18.3–25.7% of all genes studied. In these cases pre-existing expression bias reverted to non-differential expression of homoeologous copies in the hybrid and allopolyploids. For smaller numbers of genes, other novel patterns have arisen in the hybrid and allopolyploids, including a number of cases (rows 8 and 9 of Table 3) where the bias in homoeolog expression is in the opposite direction to that observed between the parents. We also observed differences in comparisons of the F1 to the allopolyploids; most notable in this respect is the approximately 2.6-fold increase in genes showing novel patterns of biased expression in natural allopolyploids relative to the F1 hybrid (P<0.001, Fisher’s exact tests).
The temporal dimension of homoeolog expression bias and expression level dominance
For the 25 359 genes jointly expressed in the F1 diploid, the synthetic allopolyploid and both natural allopolyploids, we tabulated gene expression patterns, with respect to 12 possible expression categories, using an in silico MPV as a reference point for comparisons to expression patterns in the F1 hybrid and synthetic and natural allopolyploids. This four-entity comparison showed that only 6.1% of genes (1547 out of 25 359 genes) changed expression patterns in response to genome merger (gene expression changes in Table 4). Among these, almost twice as many genes altered their expression categories to A-expression level dominance (828 genes) than to D-expression level dominance (444 genes) in response to hybridization (Supplementary Table S4). During the 1–2 mya since polyploid formation, 9.1% of genes changed expression patterns, while domestication itself had an almost equally large effect on gene expression (an additional 8.6% of genes; P<0.05, Fisher’s exact test). More of these genes displayed changes from A- than to D-expression level dominance, although the number of genes in this category decreased over time. In contrast, transgressively up- or downregulated genes increased in number over time. For example, only 222 genes were transgressively expressed as a result of genome merger, while there were more than 700 and 1200 genes in the comparisons of MPV-F1 to TX2094-Maxxa and MPV-F1-TX2094 to Maxxa, respectively (P<0.001, Fisher’s exact tests; Supplementary Table S4).
We also analyzed how homoeolog expression bias changed during the course of evolution. For this analysis, we included all seven accessions studied and 7861 gene pairs commonly expressed, while 12 523 gene pairs were examined in an analysis without the synthetic allopolyploid and its paternal parent (D1). Both analyses showed that genome merger itself had the greatest effect on homoeolog expression bias changes, compared with other effects (P<0.01, Fisher’s exact tests; homoeolog expression bias in Table 4). It is noteworthy that 543 gene pairs (out of 7861) were equally expressed in response to genome merger but differentially expressed between parental genomes, while there were only 113 or 79 gene pairs for this category due to polyploidy (Supplementary Table S5). The analysis shows that there was more novel biased expression in response to polyploidy (397 genes) and domestication (290 genes) than from genome merger (79 genes) and genome doubling (143 genes) (P<0.01, Fisher’s exact tests; Supplementary Table S5).
Discussion
In this article, we present a novel approach to study expression level dominance (previously referred to as genomic dominance; Rapp et al., 2009) and homoeolog expression bias in polyploids using next-generation sequencing. Our results demonstrate that polyploidy is accompanied by different phases of gene expression evolution, with a decrease in unbalanced expression level dominance over evolutionary time, but increasing levels of transgressive and novel expression, as well as more homoeolog expression bias. Expression level dominance seems phenomenologically connected to homoeolog-specific gene regulation established at the time of genome merger, suggesting differential epigenetic markings between diploids as a prominent underlying mechanism. This, however, is supplemented by evolutionary processes that we will discuss below.
Unbalanced expression level dominance in the leaf transcriptome
As shown in previous studies, genomes of allopolyploids are subject to various forms of altered duplicate gene expression (Doyle et al., 2008; Jackson and Chen, 2010). Apparently, one of the most prominent forms of altered expression is expression level dominance, where the total gene expression level resembles that of one of the two parents.
In this study, we generated millions of short reads to investigate the leaf transcriptome in a diploid F1 hybrid, synthetic and natural allopolyploids and models of their two progenitors. Like other studies, which have shown a high correlation between microarray and RNA-Seq results (Marioni et al., 2008), the comparison between our work and the microarray study of Rapp et al. (2009), using a synthetic allopolyploid 2(A2D1), also showed general agreement, such as similar proportion of genes showing non-differential expression, additivity and more transgressively downregulated than upregulated genes. However, there also were differences in the results obtained using the two techniques. Although Rapp et al.’s study (2009) and our study demonstrated D-biased expression level dominance in synthetic allopolyploids, the magnitude of the imbalance was different (93% vs 64% of D-biased expression level dominance in the microarray and RNA-Seq results, respectively). This difference may be explained by technical considerations (for example, an improved assembly with more sensitivity in this study) and environmental factors. Despite utilizing the same tissue of the same species in both studies, Rapp et al. (2009) used plants grown under controlled conditions in a growth chamber (26 °C for 12/12 (day/night)), whereas we grew plants in a greenhouse under natural light. A recent study using diploid and allopolyploid Coffea reported that temperature during plant growth can modulate the ‘global’ transcription divergence in allopolyploids in comparison with their diploid parents (Bardil et al., 2011). In particular, the magnitude of expression level dominance was similar under two different conditions, but the bias in expression level dominance was severely affected by growth temperature (E genome (C. eugenioides) vs C genome (C. canephora)=2713 vs 1279 at 26–22 °C, while 35 vs 3997 at 30–26 °C; Bardil et al., 2011). Like the coffee study, the higher growth temperature in Rapp et al.’s study (2009) may be related to the higher level of biased expression level dominance observed compared with this study. This intriguing suggestion of a genome-wide ‘stress response’ in the transcriptome is experimentally testable, requiring further study using plants grown under different temperature regimens and using the same technology (for example, RNA-Seq).
We also compared our results with those of Flagel and Wendel (2010), where microarrays were used to examine gene expression in petals from a synthetic F1 hybrid and five natural allopolyploids. Both studies retrieved the same results for natural allopolyploid G. hirsutum, that is, A-biased expression level dominance. However, there was a discrepancy in the direction of expression level dominance for the F1 hybrid, which was D-biased in Flagel and Wendel (2010), but A-biased in the present analysis. We note that Flagel and Wendel (2010) studied petal tissue, raising the possibility that there are tissue-specific differences in intergenomic transcriptome dynamics. Alternatively, sequencing and mapping differences, and perhaps even temperature regimens, may have a role in the differences between the two studies.
The homoeolog basis of expression level dominance
To distinguish how homoeolog expression itself contributes to duplicate gene expression patterns, we examined homoeolog expression levels for the 12 categories possible for differential expression. In contrast to the hypothesis that expression level dominance might be explained by up- or downregulation of the homoeolog derived from the ‘dominant’ genome, for most genes both homoeologs were equally represented in the hybrid and allopolyploids despite differential gene expression between the diploid parents (Supplementary Table S3). In particular, investigation of each homoeolog expression level relative to its parental genome revealed that counterpart homoeologs were frequently up- or downregulated in each expression level category, indicating that D-expression level dominance often reflected upregulation of the A homoeolog (Figure 3a) or vice versa (Figure 3b). Therefore, up- or downregulation of the homoeolog from one parent (the ‘non-dominant’ parent; cf., Figure 3) is the most common cause of expression level dominance in the hybrid and allopolyploids (Figure 4). In addition, lack of parental equivalence with respect to expression level dominance seems to be caused by bias in this type of homoeolog regulation. For example, if there is more D homoeolog than A homoeolog modification in expression level dominance categories, it may lead to A-biased expression level dominance, as we can see in the F1 and natural allopolyploids.
Considering all the categories, our data demonstrate that there was no overall bias in the interspecific diploid hybrid and natural allopolyploids, whereas there was an overall preferential transcription of A-genome homoeologs in the synthetic allopolyploid (bottom two rows in Table 3). Although the majority of genes (59–71%) demonstrate simple vertical inheritance of pre-existing patterns (first three rows in Table 3), approximately 3800–5000 gene pairs have experienced alterations in homoeolog usage relative to the parental gene (Supplementary Table S6).
The temporal dimension of homoeolog expression bias and expression level dominance
The question arises as to whether hybridization or genome doubling has a greater impact on the genome-wide alterations in gene expression that characterize allopolyploids. A number of examples suggest that genome merger (hybridization) alters the transcriptome more than does genome doubling itself (for example, in cotton (Flagel et al., 2008; Chaudhary et al., 2009), Senecio (Hegarty et al., 2006), Brassica napus (Gaeta et al., 2009) and Spartina (Chelaifa et al., 2010)). For 1383 gene pairs in cotton petals (Flagel and Wendel, 2010), the total number of genes exhibiting statistically significant homoeolog expression bias was higher in natural polyploids compared with the synthetic diploid and allopolyploid accessions. These results suggest the reasonable conclusion that homoeolog usage and expression are responsive to long-term evolutionary alteration, which increases with time.
To address this temporal dimension of expression evolution further, we compared in silico MPV expression levels to those actually observed in F1 hybrid and natural allopolyploid cottons. Our analyses show that time since polyploid formation is associated with more gene expression change than that caused by genome merger, in contrast to the results for homoeolog expression bias (Table 4). Nearly twice as many genes evolved A-expression level dominance rather than D-expression level dominance in response to genome merger, and this preference for A-expression level dominance is inferred to have been retained during evolution at the polyploid level and during domestication, although the number of gene pairs decreased over time (Supplementary Table S4). Phrased alternatively, genome merger appears to have set in motion a suite of regulatory interactions that were enhanced and further shaped during the 1–2 mya since polyploids formed. This indication that genome merger per se radically alters gene expression patterns is further supported by the data of Figure 2, which shows that 8630 gene pairs fall into non-additive categories or expression level dominance in the F1, whereas 9869 do so in TX2094; the difference between these two values (=1239) represents that additional increment associated with genome doubling and subsequent evolution at the polyploid level, although this is a minimal estimate because some gene pairs (about 3000) that were non-additive in the F1 became additive at the polyploid level.
In addition, the number of transgressively expressed genes increased over evolutionary time (gene expression changes in Table 4). Some transgressively expressed genes in the allopolyploids exhibited ‘no change’ in the F1, supporting a previous indication that these expression changes evolve de novo in allopolyploids (Flagel and Wendel, 2010). Other genes showing ‘no change’ in the MPV were transgressively expressed in the F1 hybrid, and thus these novel expression patterns are not unique to allopolyploids, showing that regulatory networks may be disturbed upon hybridization even for genes showing equal expression levels in the parents. Natural allopolyploids have been evolving for 1–2 mya, while domesticated cottons have experienced strong artificial selection for only 5000–7000 years (Brubaker et al., 1999). Yet Maxxa, the domesticated form, has more transgressively expressed genes than does its wild counterpart, TX2094. Considering the recency of cotton domestication relative to polyploid formation, one is tempted to suggest that selection under domestication has greatly altered the leaf transcriptome of cultivated cotton, even without leaves being obvious, direct phenotypic targets. This selection may operate at the level of individual genes, but given the scope of transcriptome alterations, we suspect that most gene expression changes reflect the downstream consequences of entire regulatory networks that have been subtly or not so subtly affected by the domestication process. Further research is required to elucidate the extent to which network modulation is related to global patterns of expression alteration in polyploids, including on transgressive regulation, novel expression, level of biased homoeolog expression and expression level dominance.
Possible mechanisms underlying homoeolog expression bias and expression level dominance
Given the ubiquity of expression level dominance and homoeolog expression bias in allopolyploid cotton and in other allopolyploids examined to date, it is of interest to understand the possible underlying causes. These likely reflect multiple underlying mechanisms, but they collectively may be considered a manifestation or form of ‘transcriptomic shock’ (Hegarty et al., 2006; Buggs et al., 2011). Merging two divergent genomes in a nucleus may lead to trans-activation and repression due to the divergence in parental regulatory machineries that become reunited in the hybrid and allopolyploids. According to this model, genome merger combines diverged cis- and trans-acting regulatory elements, resulting in both novel patterns of homoeolog activation and repression. This is illustrated here, for example, in Figures 3 and 4 for the case of up- and downregulation of A homoeologs resulting from the new encounter with D-genome-derived elements. The net effects of these regulatory interactions are reflected as expression level dominance and transgressive expression. This suggests a form of transactivation or repression, entailing the joint up- or downregulation of homoeologs due to the novel regulatory environment of the hybrid or polyploid nucleus. One can imagine that perturbations operate both at the level of single gene pairs and entire networks of coexpression. Although no study yet reveals how duplicated networks affect gene expression in allopolyploids, one can envision that the combinatorial complexity of suddenly doubled networks of interconnected genes and regulatory elements have myriad subtle and perhaps dramatic effects on individual gene expression levels and expression cascades.
The biased homoeolog-specific gene regulation observed here also suggests a role for epigenetic modulation, which has been shown to be prevalent in response to hybridization and genome doubling (Madlung et al., 2002, 2005; Levy and Feldman, 2004; Salmon et al., 2005; Paun et al., 2007; Ha et al., 2009; He et al., 2010; Li et al., 2010). For example, Ha et al. (2009) showed that miRNAs lead to non-additive expression patterns in allopolyploid Arabidopsis by unequal degradation of parental target genes. Similarly, He et al. (2010) showed that allelic biases in rice hybrids were highly positively correlated with differences in gene expression or epigenetic modifications between parents, suggesting that differential epigenetic markings and possibly small RNA populations inherited from parents could be responsible for biased expression. Further insights into the proximate and ultimate causes of gene expression alteration in polyploids are likely to emerge from integrated analyses of gene expression and epigenetic and chromatin level modifications.
Additional clues as to the underlying mechanism may derive from the observation that there exists a great deal of variation in the bias and direction of expression level dominance. This may be an even bigger mystery than expression level dominance per se, as comparison to other results on cotton or to those from Coffea (Bardil et al., 2011) shows that biased expression level dominance may be contingent upon tissue type and environmental conditions. Given the scale of the phenomenon, it is evident that the global transcriptome and its relative levels of homoeolog usage may be highly sensitive to environmental perturbation or ‘stress’. To the extent that polyploids have the potential for broader niches than their diploid progenitors, we hypothesize that environmental conditions influence parental genome usage in response to changing environments. It may be, for example, that certain environmental conditions favor one homoeolog, while the reverse is true under other conditions. In this respect, we note that Dong and Adams (2011) demonstrated that abiotic stress can profoundly alter duplicate gene expression ratios in Gossypium hirsutum; among the 60 pairs of genes they examined, 70% exhibited changes in homoeolog expression levels with varying stress conditions. Possible mechanisms include organ-specific or stress-activated small RNAs or epigenetic pathways that mediate cascades, which affect homoeologs either differentially or jointly.
Conclusions
Homoeolog expression bias and especially expression level dominance represent underexplored dimensions of the transcriptomic dynamics and evolution of allopolyploids, with only a handful of species (cotton, Spartina, coffee and wheat) evaluated for the latter phenomenon and only under one or two experimental conditions for few tissue types. Here we explored those two phenomena in detail using RNA-Seq, which enables genome-wide analysis for both homoeolog expression bias and expression level dominance at the same time. Our results showed that the extent of homoeolog expression bias and expression level dominance increases over time from hybridization through evolution at the polyploid level. In addition, we observed higher rates of transgressive and novel gene expression patterns, as well as homoeolog silencing in natural allopolyploids than in F1 hybrid and synthetic allopolyploid cottons. Additional studies using various species, tissues and environmental conditions are needed to further describe the various expression phenomena involved and their interconnections. In addition, more integrated studies are needed involving sequenced genomes and simultaneous analyses of epigenetic features and gene expression. At present, we are even further from an understanding of how these transcriptomic dynamics affect function and mediate phenotypes that are responsive to selection. Achieving progress in this direction may facilitate an understanding of the evolutionary significance of allopolyploidy and how two differentiated genomes become reconciled, stabilized and utilized.
Data archiving
Short read sequences were deposited in the NCBI Sequence Read Archive with study number SRA056385.
References
Alvarez I, Cronn R, Wendel JF (2005). Phylogeny of the New World diploid cottons (Gossypium L., Malvaceae) based on sequences of three low-copy nuclear genes. Plant Syst Evol 252: 199–214.
Bardil A, de Almeida JD, Combes MC, Lashermes P, Bertrand B (2011). Genomic expression dominance in the natural allopolyploid Coffea arabica is massively affected by growth temperature. New Phytol 192: 760–774.
Benjamini Y, Hochberg Y (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B 57: 289–300.
Brubaker CL, Bourland FM, Wendel JF (1999). The origin and domestication of cotton. In: Smith CW, Cothren JT (eds). Cotton: Origin, History, Technology and Production. Wiley: New York, pp 3–31.
Brubaker CL, Wendel JF (1994). Reevaluating the origin of domesticated cotton (Gossypium hirsutum; Malvaceae) using nuclear restriction fragment length polymorphisms (RFLPs). Am J Bot 81: 1309–1326.
Buggs RJ, Zhang L, Miles N, Tate JA, Gao L, Wei W et al. (2011). Transcriptomic shock generates evolutionary novelty in a newly formed, natural allopolyploid plant. Curr Biol 21: 551–556.
Chagué V, Just J, Mestiri I, Balzergue S, Tanguy AM, Huneau C et al. (2010). Genome-wide gene expression changes in genetically stable synthetic and natural wheat allohexaploids. New Phytol 187: 1181–1194.
Chaudhary B, Flagel L, Stupar RM, Udall JA, Verma N, Springer NM et al. (2009). Reciprocal silencing, transcriptional bias and functional divergence of homeologs in polyploid cotton (Gossypium). Genetics 182: 503–517.
Chelaifa H, Monnier A, Ainouche M (2010). Transcriptomic changes following recent natural hybridization and allopolyploidy in the salt marsh species Spartina × townsendii and Spartina anglica (Poaceae). New Phytol 186: 161–174.
Combes MC, Cenci A, Baraille H, Bertrand B, Lashermes P (2012). Homeologous gene expression in response to growing temperature in a recent allopolyploid (Coffea arabica L.). J Hered 103: 36–46.
Dong S, Adams KL (2011). Differential contributions to the transcriptome of duplicated genes in response to abiotic stresses in natural and synthetic polyploids. New Phytol 190: 1045–1057.
Doyle JJ, Doyle JL (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19: 11–15.
Doyle JJ, Flagel L, Paterson AH, Rapp RA, Soltis DE, Soltis PS et al. (2008). Evolutionary genetics of genome merger and doubling in plants. Annu Rev Genet 42: 443–461.
Flagel L, Udall J, Nettleton D, Wendel JF (2008). Duplicate gene expression in allopolyploid Gossypium reveals two temporally distinct phases of expression evolution. BMC Biol 6: 16.
Flagel L, Wendel JF (2010). Evolutionary rate variation, genomic dominance and duplicate gene expression evolution during allotetraploid cotton speciation. New Phytol 186: 184–193.
Flagel L, Wendel JF, Udall J (2012). Duplicate gene evolution, homoeologous recombination, and transcriptome characterization in allopolyploid cotton. BMC Genomics 13: 302.
Gaeta RT, Yoo SY, Pires JC, Doerge RW, Chen ZJ, Osborn TC (2009). Analysis of gene expression in resynthesized Brassica napus allopolyploids using arabidopsis 70mer oligo microarrays. PLoS One 4: e4760.
Grover CE, Gallagher JP, Szadkowski E, Yoo M-J, Flagel LE, Wendel JF (2012). Homoeolog expression bias and expression level dominance in allopolyploids. New Phytol 196: 966–971.
Ha M, Lu J, Tian L, Ramachandran V, Kasschau KD, Chapman EJ et al. (2009). Small RNAs serve as a genetic buffer against genomic shock in Arabidopsis interspecific hybrids and allopolyploids. Proc Natl Acad Sci USA 106: 17835–17840.
He G, Zhu X, Elling AA, Chen L, Wang X, Guo L et al. (2010). Global epigenetic and transcriptional trends among two rice subspecies and their reciprocal hybrids. Plant Cell 22: 17–33.
Hegarty MJ, Barker GL, Wilson ID, Abbott RJ, Edwards KJ, Hiscock SJ (2006). Transcriptome shock after interspecific hybridization in Senecio is ameliorated by genome duplication. Curr Biol 16: 1652–1659.
Jackson S, Chen ZJ (2010). Genomic and expression plasticity of polyploidy. Curr Opin Plant Biol 13: 153–159.
Jiao Y, Wickett NJ, Ayyampalayam S, Chanderbali AS, Landherr L, Ralph PE et al. (2011). Ancestral polyploidy in seed plants and angiosperms. Nature 473: 97–100.
Koh J, Soltis PS, Soltis DE (2010). Homeolog loss and expression changes in natural populations of the recently and repeatedly formed allotetraploid Tragopogon mirus (Asteraceae). BMC Genom 11: 97.
Levy AA, Feldman M (2004). Genetic and epigenetic reprogramming of the wheat genome upon allopolyploidization. Biol J Linn Soc 82: 607–613.
Li H, Durbin R (2009). Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25: 1754–1760.
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N et al. (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079.
Li X, Guo W, Wang B, Chen H, Wei L, Wang Y et al. (2010). Instability of chromosome number and DNA methylation variation induced by hybridization and amphidiploid formation between Raphanus sativus L. and Brassica alboglabra Bailey. BMC Plant biol 10: 207.
Madlung A, Masuelli RW, Watson B, Reynolds SH, Davison J, Comai L (2002). Remodeling of DNA methylation and phenotypic and transcriptional changes in synthetic Arabidopsis allotetraploids. Plant Physiol 129: 733–746.
Madlung A, Tyagi AP, Watson B, Jiang H, Kagochi T, Doerge RW et al. (2005). Genomic changes in synthetic Arabidopsis polyploids. Plant J 41: 221–230.
Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y (2008). RNA-Seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res 18: 1509–1517.
Nagalakshmi U, Waern K, Snyder M (2010). RNA-Seq: a method for comprehensive transcriptome analysis. Curr Protoc Mol Biol 11, (Unit 4) 1–13.
Paun O, Fay MF, Soltis DE, Chase MW (2007). Genetic and epigenetic alterations after hybridization and genome doubling. Taxon 56: 649–656.
Rapp RA, Udall J, Wendel JF (2009). Genomic expression dominance in allopolyploids. BMC Biol 7: 18.
Robinson MD, McCarthy DJ, Smyth GK (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140.
Salmon A, Ainouche ML, Wendel JF (2005). Genetic and epigenetic consequences of recent hybridization and polyploidy in Spartina (Poaceae). Mol Ecol 14: 1163–1175.
Udall JA, Swanson JM, Nettleton D, Percifield RJ, Wendel JF (2006). A novel approach for characterizing expression levels of genes duplicated by polyploidy. Genetics 173: 1823–1827.
Wang Z, Gerstein M, Snyder M (2009). RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10: 57–63.
Wendel JF, Cronn RC (2003). Polyploidy and the evolutionary history of cotton. Adv Agron 78: 139–186.
Acknowledgements
We thank Marta Matvienko and Michael Baker for technical assistance, and Lex Flagel for bioinformatics assistance. We also express our gratitude to Barbara Mable for her many suggestions to improve the manuscript. This work was supported by National Science Foundation Plant Genome Program to JFW.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Heredity website
Rights and permissions
About this article
Cite this article
Yoo, MJ., Szadkowski, E. & Wendel, J. Homoeolog expression bias and expression level dominance in allopolyploid cotton. Heredity 110, 171–180 (2013). https://doi.org/10.1038/hdy.2012.94
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/hdy.2012.94
Keywords
This article is cited by
-
Identifying the genetic and epigenetic basis for asymmetric bZIP expression in temperature-stressed bread wheat
Plant Growth Regulation (2024)
-
Dynamic patterns of gene expression and regulatory variation in the maize seed coat
BMC Plant Biology (2023)
-
Transposon signatures of allopolyploid genome evolution
Nature Communications (2023)
-
Comparative transcriptomic analysis reveals the molecular mechanism underlying seedling biomass heterosis in Brassica napus
BMC Plant Biology (2022)
-
Proteome evaluation of homolog abundance patterns in Arachis hypogaea cv. Tifrunner
Plant Methods (2022)