Abstract
Purpose
Monoallelic germ-line mutations in the BRCA1/FANCS, BRCA2/FANCD1 and PALB2/FANCN genes confer high risk of breast cancer. Biallelic mutations in these genes cause Fanconi anemia (FA), characterized by malformations, bone marrow failure, chromosome fragility, and cancer predisposition (BRCA2/FANCD1 and PALB2/FANCN), or an FA-like disease presenting a phenotype similar to FA but without bone marrow failure (BRCA1/FANCS). FANCM monoallelic mutations have been reported as moderate risk factors for breast cancer, but there are no reports of any clinical phenotype observed in carriers of biallelic mutations.
Methods
Breast cancer probands were subjected to mutation analysis by sequencing gene panels or testing DNA damage response genes.
Results
Five cases homozygous for FANCM loss-of-function mutations were identified. They show a heterogeneous phenotype including cancer predisposition, toxicity to chemotherapy, early menopause, and possibly chromosome fragility. Phenotype severity might correlate with mutation position in the gene.
Conclusion
Our data indicate that biallelic FANCM mutations do not cause classical FA, providing proof that FANCM is not a canonical FA gene. Moreover, our observations support previous findings suggesting that FANCM is a breast cancer-predisposing gene. Mutation testing of FANCM might be considered for individuals with the above-described clinical features.
Similar content being viewed by others
Introduction
Breast cancer is the most common female oncological disease worldwide. Up to 15% of all cases have family history for the disease, and the risk of developing breast cancer in individuals with an affected relative is twofold higher compared to the general population.1 BRCA1, BRCA2, and PALB2 are major breast cancer–predisposing genes and monoallelic protein-truncating mutations confer risk of developing the disease by age 70 ranging from 35% to 59%.2, 3 These genes are also designated as FANCS/BRCA1, FANCD1/BRCA2, and FANCN/PALB2, and together with 18 other genes—including the recently identified FANCV/REV74—code for proteins involved in a common pathway responsible for DNA interstrand crosslink (ICL) repair, mediated by homologous recombination. Biallelic mutations in genes of this pathway cause the Fanconi anemia (FA) disease characterized by congenital defects, bone marrow failure, sensitivity to DNA ICLs, and cancer susceptibility. FA patients often show chemotherapy sensitivity, abnormal inflammatory response, reproductive/endocrine defects, and increased risk of hematological tumors and other solid tumors such as squamous cell carcinoma and breast and ovarian cancer. Biallelic mutations in three of the genes involved in the FA pathway, namely FANCS/BRCA1, FANCO/RAD51C, and FANCR/RAD51 cause a FA-like disease that is similar to FA but does not include bone marrow failure (reviewed in ref. 5).
While the FANCM gene is a key component of the FA molecular pathway, its role in the etiology of FA disease has been disputed. In 2005, an individual affected with FA was found to carry biallelic mutations in FAAP250 that were renamed FANCM and proposed as a new FA gene.6 Later, it was discovered that this patient also carried biallelic mutations in the FANCA gene.7 Apart from this case, no other FA patients with FANCM biallelic mutations were described to date. Moreover, seven Finnish individuals were found to be homozygous for FANCM mutations but had no evidence of blood diseases, increased frequency of cancer events or any other chronic disease.8 While these observations raised the question of whether FANCM was a bona fide FA gene, recent case-control studies indicated that monoallelic truncating mutations located in the C-terminus of the gene might be risk factors for breast cancer. The FANCM c.5101C>T mutation (p.Gln1701*, rs147021911) is relatively frequent in Finland, where it was shown to be statistically associated with breast cancer risk, particularly in familial cases (odds ratio, OR=2.11) and individuals with triple-negative tumors (OR=3.56).9 The FANCM c.5791C>T mutation (p.Gly1906Alafs*12; rs144567652), also known as p.Arg1931*, was tested in a large series of familial breast cancer cases and controls from different populations, resulting in a statistically significant association with disease risk (OR = 3.93).10 These observations are supported by a very recent analysis of truncating FANCM mutations found by sequencing German familial cases and controls.11
In this study, we describe the phenotype of five breast cancer probands incidentally found to carry homozygous FANCM loss-of-function mutations. These findings contribute to elucidate the role of this gene in both FA disease and breast cancer predisposition.
Materials and methods
Breast cancer probands and mutation testing strategies
The five female individuals included in this study were originally eligible for mutation testing in breast cancer–predisposing genes based on the facts that they were affected with breast cancer, and they had either early onset or family history for the disease, or were affected with hormone receptor–negative cancer. All probands underwent mutation testing through a diagnostic or a research protocol at four different centers in Italy, Germany, Sweden, and Spain. Mutation testing consisted of next-generation sequencing of custom or commercial gene panels including established and candidate breast cancer genes (Supplementary Table S1 online). In addition, probands from Italy and Germany were subjected to whole-exome sequencing and of all annotated variants we considered only those located within the genes involved in the DNA damage response (Supplementary Table S2). To establish genotype/phenotype correlations, all the variants found in probands’ DNA were annotated and prioritized for causality using different pipelines that classified variants considering (i) their effect on the protein product, (ii) their frequency reported in public databases, and (iii) their clinical classification, if available in public databases or literature. All individuals included in this study and herein described signed an informed consent to the use of their biological samples and clinical data for research projects. This study was approved by local ethics committees.
Results
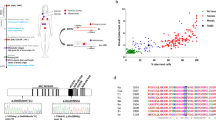
In this collaborative study, we report the clinical phenotype of five female breast cancer probands who were tested for mutations in breast cancer–related genes and found to be homozygous for loss-of-function mutations in the FANCM gene (Table 1 and Figure 1). The presence of homozygous FANCM mutations was confirmed in all probands’ DNA by Sanger sequencing (Supplementary Figure S1). Proband 1 was Italian; she developed early-onset breast cancer at age 29 followed by several other oncological diseases and was found to carry the homozygous mutation FANCM c.1972C>T (p.Arg658*). She was born to first-cousin parents and we speculate they both inherited the same FANCM alleles from a common ancestor. Proband 2 was born in Germany. This woman also developed breast cancer at an early age (31) and inherited one FANCM c.1972C>T (p.Arg658*) allele from each of her heterozygous parents. The two probands from Sweden had a family history for breast or ovarian cancer and were homozygous for FANCM c.5101C>T (p.Gln1701*), which is a relatively common mutation in Finland.9 Finally, the Spanish proband developed a triple-negative breast cancer and was found to be homozygous for the FANCM c.5791C>T (p.Arg1931*) mutation that has been reported as possibly more frequent in southwestern Europe.10 As we could not specifically test for the presence of single-exon rearrangements, we cannot formally exclude that the homozygosity we observed is due to exonic deletions in the FANCM locus.
Family pedigrees of the five probands homozygous for the FANCM c.1972C>T (p.Arg658*), the c.5101C>T (p.Gln1701*), and the c.5791C>T (p.Arg1931*) mutations. Probands are indicated by the arrows. Cancer type, age at diagnosis, and age of death are reported when known. Age of healthy individuals, if known, was at date of genetic counseling. Events occurred after genetic counseling, if known, are annotated. Mutation status is reported as follows:+/+, normal; +/−, heterozygote; −/−, homozygote; (+/−), obligate carrier. Cancer type is reported as follows: An, anal; Br, breast; Br bil, bilateral breast cancer; CR, colorectal; En, endometrial; Gl, glioblastoma; Gy, gynecologic nonspecified cancer; HNSCC, head and neck squamous cells cancer; Ki, kidney; La, larynx; Ly, lymphoma; Lu, lung; Me, melanoma; Ov, ovarian; Pa, pancreas; Pr, prostate; Th, throat; Tr, trichilemmoma.
While these individuals were first diagnosed with breast cancer, the majority of them showed other clinical signs including chemotherapy toxicity and early menopause (Table 1 and Supplementary Table S3). These phenotypes, together with the presence of biallelic mutations in an FA pathway gene, prompted us to measure sensitivity to DNA ICL agents by a chromosome fragility test that was performed as previously described.12 We could only obtain fresh blood cells from probands 2, 3, and 5, and observed sensitivity to DNA ICLs only in proband 2 (Supplementary Table S4).
Apart from the FANCM homozygous mutations, we only found one other known breast cancer–associated variant, CHEK2 c.470C>T (p.Ile157Thr), detected in proband 4. As this variant is reported as a low/moderate risk factor for breast cancer,13 we cannot exclude that it could have contributed to the phenotype observed in this proband. Nor can we exclude that other mutations possibly influencing the clinical phenotypes might remain undetected. However, since our gene panels included known variants associated with a moderate to high risk for developing breast cancer, with the only exception of CHEK2 c.470C>T in proband 4, we believe that the homozygous FANCM mutations were most likely to be considered the only disease-associated mutations identified in the five probands.
Discussion
Our data document for the first time the possible association of biallelic FANCM mutations with a clinical phenotype. The five probands included in this study were all originally recruited as breast cancer probands, and none was clinically diagnosed with FA or FA-like diseases. While none suffered from congenital abnormalities, bone marrow failure, or hemato-oncological diseases, some of them showed clinical signs that are included in the FA phenotype spectrum. In particular, four of the five probands showed chemotherapy-related hematological side effects. Moreover, proband 1 developed three head and neck squamous cell carcinomas and another solid cancer. Probands 2, 4, and 5 showed sign of early menopause, and proband 3 suffered from autoimmune disease (Sjögren syndrome). Finally, proband 2 tested positive for chromosome fragility indicating sensitivity to DNA ICLs (Table 1). These clinical observations indicate that FANCM is not a bona fide FA gene. The small number of carriers of biallelic FANCM mutations that we describe in this study does not allow performing any statistical analysis, or to claim that the FANCM genotypes we detected are causative of the observed clinical phenotypes. However, on a less conservative view, we cannot exclude that homozygosity for truncating mutations in FANCM may have clinical consequences that are more pronounced than for heterozygous mutation carriers. The phenotypes in our five probands are consistent with FANCM being a gene associated with FA-like cancer susceptibility. This appears to be supported by molecular data from human cell lines and by phenotypic observations from Fancm-deficient mice. During the early steps of the physiological repair of DNA ICLs, FANCM and the protein products encoded by the seven bona fide FA genes FANCA, -B, -C, -E, -F, -G, and -L assemble in the “core complex.” This complex monoubiquitinates the ID2 heterodimer, which mediates the recruitment of “effector proteins” responsible for DNA ICLs repair (reviewed in ref. 5). While biallelic mutations in all the core complex genes abolish monoubiquitination of the ID2 complex, lack of FANCM only reduces the efficiency of this mechanism. Moreover, studies in which FANCM was depleted in cell lines showed that this loss neither affects the levels of other FA core complex proteins, such as FANCA and FANCG, nor the ability of FANCL to coimmunoprecipitate with them, suggesting that FANCM is not essential for FA core complex formation and stabilization (reviewed in ref. 14). With respect to FA mice models, mice deficient for Fancm showed increased cancer incidence;15 an excess of ovarian, mammary, and uterine cancers; and sex-independent gonadal defects.16
Monoallelic loss-of-function mutations in FANCM have been suggested to be associated with moderate/low risk for breast cancer.9, 10, 11 The identification of breast cancer probands with biallelic FANCM mutations corroborates the role of this gene as a breast cancer–predisposing factor. Two of these probands showed early-onset breast cancer and one proband developed bilateral breast cancer, suggesting that FANCM biallelic mutations may have a stronger effect on breast cancer risk compared to monoallelic mutations. Such an effect has been previously observed for the moderate breast cancer risk factor CHEK2 c.1100delC. Homozygote carriers of this variant have been estimated to have a twofold and a fourfold higher risk compared to heterozygotes and women of the general population, respectively.17 Moreover, of the nine CHEK2 c.1100delC homozygous familial cases reported to date,17, 18 three developed breast cancer before age 35 and four developed bilateral breast cancer, which is comparable to the clinical phenotypes observed in some of the FANCM homozygous probands.
Our data shows that biallelic FANCM mutations might be associated with a clinical phenotype including breast cancer predisposition, chemotherapy toxicity, and possibly early menopause and chromosome fragility. In the accompanying manuscript,19 Bogliolo et al. describe additional individuals with FANCM homozygous mutations who were affected with different types of early-onset cancer. Altogether, these observations support the hypothesis that biallelic mutations in FANCM may cause a heterogeneous cancer susceptibility phenotype. This heterogeneity and the breast cancer severity in terms of age of onset that we observed in our probands, might be influenced by the position of the mutations in the gene (Supplementary Figure S2). All three carriers of the p.Gln1701* and p.Arg1931* mutations, which are located in the gene C-terminus, developed breast cancer at typical ages and, at least the two who were tested, did not display chromosome fragility. Both carriers of the p.Arg658* mutation, which is expected to produce a protein lacking the domains of interaction with the DNA-binding mediators MHF1 and MHF2, and with the Bloom syndrome complex,20 developed early-onset breast cancer and one showed high chromosome fragility. Similarly, the patients described by Bogliolo et al.19 who suffered from severe early-onset cancers and showed chromosome fragility carried homozygous mutations expected to truncate FANCM at amino acid 503 and 863.
In conclusion, our data point out that biallelic mutations in FANCM do not cause classical FA, providing evidence that FANCM is not a canonical FA gene. Moreover, our observations reinforce the hypothesis that FANCM is a breast cancer–predisposing gene with biallelic mutations possibly conferring higher risk. Finally, we suggest that FANCM biallelic mutations might be associated with an FA-like cancer susceptibility, characterized by increased risk for breast and other types of cancer, chemotherapy toxicity, and possibly early menopause and chromosome fragility. FANCM mutation testing may be offered, in the context of research protocols, to individuals with the above clinical features. Additional genotype/phenotype correlations are necessary to better clarify the clinical impact of FANCM biallelic mutations.
References
Collaborative Group on Hormonal Factors in Breast Cancer. Familial breast cancer: collaborative reanalysis of individual data from 52 epidemiological studies including 58,209 women with breast cancer and 101,986 women without the disease. Lancet 2001;358:1389–1399.
Couch FJ, Nathanson KL, Offit K . Two decades after BRCA: setting paradigms in personalized cancer care and prevention. Science 2014;343:1466–1470.
Antoniou AC, Casadei S, Heikkinen T et al. Breast-cancer risk in families with mutations in PALB2. N Engl J Med 2014;371:497–506.
Bluteau D, Masliah-Planchon J, Clairmont C et al. Biallelic inactivation of REV7 is associated with Fanconi anemia. J Clin Invest 2016;126:3580–3584.
Bogliolo M, Surralles J . Fanconi anemia: a model disease for studies on human genetics and advanced therapeutics. Curr Opin Genet Dev 2015;33:32–40.
Meetei AR, Medhurst AL, Ling C et al. A human ortholog of archaeal DNA repair protein Hef is defective in Fanconi anemia complementation group M. Nat Genet 2005;37:958–963.
Singh TR, Bakker ST, Agarwal S et al. Impaired FANCD2 monoubiquitination and hypersensitivity to camptothecin uniquely characterize Fanconi anemia complementation group M. Blood 2009;114:174–180.
Lim ET, Wurtz P, Havulinna AS et al. Distribution and medical impact of loss-of-function variants in the Finnish founder population. PLoS Genet 2014;10:e1004494.
Kiiski JI, Pelttari LM, Khan S et al. Exome sequencing identifies FANCM as a susceptibility gene for triple-negative breast cancer. Proc Natl Acad Sci USA 2014;111:15172–15177.
Peterlongo P, Catucci I, Colombo M et al. FANCM c.5791C>T nonsense mutation (rs144567652) induces exon skipping, affects DNA repair activity and is a familial breast cancer risk factor. Hum Mol Genet 2015;24:5345–5355.
Neidhardt G, Hauke J, Ramser J et al. Association between loss-of-function mutations within the FANCM gene and early-onset familial breast cancer. JAMA Oncol 2016; advance online publication, 29 December 2016; doi:10.1001/jamaoncol.2016.5592.
Castella M, Pujol R, Callen E et al. Chromosome fragility in patients with Fanconi anaemia: diagnostic implications and clinical impact. J Med Genet 2011;48:242–250.
Kilpivaara O, Vahteristo P, Falck J et al. CHEK2 variant I157T may be associated with increased breast cancer risk. Int J Cancer 2004;111:543–547.
Whitby MC . The FANCM family of DNA helicases/translocases. DNA Repair (Amst) 2010;9:224–236.
Bakker ST, van de Vrugt HJ, Rooimans MA et al. Fancm-deficient mice reveal unique features of Fanconi anemia complementation group M. Hum Mol Genet 2009;18:3484–3495.
Luo Y, Hartford SA, Zeng R, Southard TL, Shima N, Schimenti JC . Hypersensitivity of primordial germ cells to compromised replication-associated DNA repair involves ATM-p53-p21 signaling. PLoS Genet 2014;10:e1004471.
Adank MA, Jonker MA, Kluijt I et al. CHEK2*1100delC homozygosity is associated with a high breast cancer risk in women. J Med Genet 2011;48:860–863.
Zhang S, Phelan CM, Zhang P et al. Frequency of the CHEK2 1100delC mutation among women with breast cancer: an international study. Cancer Res 2008;68:2154–2157.
Bogliolo M, Bluteau D, Lespinasse J et al. Biallelic truncating FANCM mutations cause early-onset cancer but not Fanconi anemia. Genet Med 2017;19:XXX–XXX.
Walden H, Deans AJ . The Fanconi anemia DNA repair pathway: structural and functional insights into a complex disorder. Annu Rev Biophys 2014;43:257–278.
Acknowledgements
The authors are particularly grateful to individuals participating in this study and their families. The authors also thank Valeria Pensotti and Sara Volorio (IFOM, the FIRC Institute of Molecular Oncology, Milan, Italy), Milena Mariani (Fondazione IRCCS Istituto Nazionale dei Tumori, Milan, Italy) for their collaboration in the study. The Swedish BRCA1 and BRCA2 Study Collaborators (SWE-BRCA) are Zakaria Einbeigi (Gothenburg, Sahlgrenska University Hospital); Marie Stenmark-Askmalm (Linköping University Hospital); Hans Ehrencrona, Therese Törngren, Anders Kvist, and Åke Borg (Lund University Hospital); Brita Arver, Annika Lindblom, and Emma Tham (Stockholm, Karolinska University Hospital); Beatrice Melin (Umeå University Hospital); and Ylva Paulsson-Karlsson (Uppsala University Hospital). This work was partially supported by the following: Associazione Italiana per la Ricerca sul Cancro (AIRC) and “Ricerca Finalizzata - Bando 2010” from Ministero della Salute, Italy to P.P.; Fondazione Umberto Veronesi (FUV; postdoctoral fellowships 2016) to I.C.; the Italian citizens who allocated the 5 × 1,000 share of their tax payment in support of the Fondazione IRCCS Istituto Nazionale dei Tumori, according to Italian laws, to S.M.; the German Cancer Aid (grant 109076); the Swedish Cancer Society, to the SWEA study; the Spanish Ministry of Economy and Competitiveness (MINECO) to A.O. (SAF2014-57680-R), FIS PI12/00070, and Spanish Association Against Cancer to J.B.; the ICREA-Academia program, the Spanish Ministry of Health (projects FANCOSTEM and FANCOLEN); the Spanish Ministry of Economy and Competiveness (projects CB06/07/0023, and SAF2015-64152-R); the European Commission (EUROFANCOLEN project HEALTH-F5-2012-305421, FANCODRUG project H2020-703521 and P-SPHERE COFUND project); the Fanconi Anemia Research Fund; the “Fondo Europeo de Desarrollo Regional, una manera de hacer Europa”; and CIBERER, an initiative of the Instituto de Salud Carlos III, Spain to Jordi Surrallés’s laboratory.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
The first four authors contributed equally to this work.
The last five authors contributed equally to this work.
Supplementary material is linked to the online version of the paper
Rights and permissions
About this article
Cite this article
Catucci, I., Osorio, A., Arver, B. et al. Individuals with FANCM biallelic mutations do not develop Fanconi anemia, but show risk for breast cancer, chemotherapy toxicity and may display chromosome fragility. Genet Med 20, 452–457 (2018). https://doi.org/10.1038/gim.2017.123
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/gim.2017.123
Keywords
This article is cited by
-
Leveraging the replication stress response to optimize cancer therapy
Nature Reviews Cancer (2023)
-
Fanconi Anemia Gene Variants in Patients with Gonadal Dysfunction
Reproductive Sciences (2022)
-
Protein truncating variants in FANCM and risk for ER-negative/triple negative breast cancer
npj Breast Cancer (2021)
-
Investigation of monogenic causes of familial breast cancer: data from the BEACCON case-control study
npj Breast Cancer (2021)
-
DONSON and FANCM associate with different replisomes distinguished by replication timing and chromatin domain
Nature Communications (2020)