Abstract
Purpose:
Ewing sarcoma is a small round blue cell tumor that is highly malignant and predominantly affects the adolescent and young adult population. It has long been suspected that a genetic predisposition exists for this cancer, but the germ-line genetic underpinnings of this disease have not been well established.
Methods:
We performed germline variant analysis of whole-genome or whole-exome sequencing of samples from 175 patients affected by Ewing sarcoma.
Results:
We discovered pathogenic or likely pathogenic germline mutations in 13.1% of our cohort. Pathogenic mutations were highly enriched for genes involved with DNA damage repair and for genes associated with cancer predisposition syndromes.
Conclusion:
Our findings reported here have important clinical implications for patients and families affected by Ewing sarcoma. Genetic counseling should be considered for patients and families affected by this disease to take advantage of existing risk management strategies. Our study also highlights the importance of germline sequencing for patients enrolled in precision-medicine protocols.
Genet Med advance online publication 26 January 2017
Similar content being viewed by others
Main
Ewing sarcoma is a small round blue cell tumor that is highly malignant and predominantly affects adolescent and young adult populations. It has long been suspected that a genetic predisposition exists for this cancer because of the young age of patients involved and wide demographic variations in its incidence.1 Furthermore, it has recently been recognized that there is an excess of cancers among relatives of Ewing sarcoma patients, including a subset of patients that have a pedigree suggestive of a familial cancer syndrome.2 Ewing sarcoma has also been significantly associated with hereditary retinoblastoma based on a meta-analysis that included 10 such cases reported in the literature.3 Germline mutations in cancer predisposition genes have recently been described in 8.5–12% of pediatric cancer cases across a range of cancer types.4,5,6 An excess of pathogenic germline variants has also recently been reported in a large cohort of sarcoma patients encompassing a variety of histologic subtypes.7
Here, we report a germline next-generation sequencing analysis of 175 patients with Ewing sarcoma—the largest and most comprehensive germline genomics analysis to date for this rare tumor type. Our data were derived from three cohorts that have previously been analyzed for somatic mutational spectrum: the National Cancer Institute (56 subjects), the International Cancer Genome Consortium (ICGC; 100 subjects), and the Pediatric Cancer Genome Project (PCGP; 19 subjects).8,9
Materials and Methods
Raw sequencing data from the ICGC and PCGP cohorts were accessed from the European genome–phenome archive, accession numbers EGAS00001000855 (ICGC) and EGA00001000839 (PCGP), respectively. Details of patient selection, informed consent, and clinical characteristics have previously been reported.8,9 Additional data descriptions are provided in the Supplementary Methods online.
Whole-genome sequencing (WGS)/whole-exome sequencing (WES) were processed and mapped and variants were called using methods previously used by our group with very high validation rates.5,8,10 To limit the number of variants for manual review, we implemented a bioinformatics pipeline that included filters for quality, rarity in population databases, and curated knowledge databases such as ClinVar (Supplementary Methods online, Supplementary Figure S1 online). After application of the bioinformatics pipeline, the resultant “tier 1” variants were manually evaluated and classified by a medical oncologist and medical geneticist according to American College of Medical Genetics and Genomics guidelines.11 The college’s classification was the final result used for reporting.
To perform burden testing for genes with pathogenic/likely pathogenic variants, we compared the rate of these classes of variants in these genes in our Ewing sarcoma cohort to that in the Exome Aggregation Consortium (ExAC) population database minus samples contributed from TCGA (The Cancer Genome Atlas). Variants downloaded from the ExAC database were subjected to the same classification methods as used for the study group. A two-sided Fisher’s exact test was used for statistical comparison.
Pathway analysis of pathogenic/likely pathogenic variants was performed using Ingenuity Pathway Analysis software (http://www.ingenuity.com/products/ipa).
Results
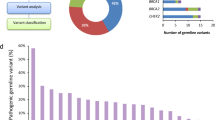
In the cohort of 175 Ewing sarcoma patients analyzed, we identified 52 tier 1 variants for further manual classification. Of these, we classified 23 as pathogenic or likely pathogenic (Supplementary Table S1 online), 12 as variants of unknown significance (Supplementary Table S2 online), 4 as likely benign, and 4 as heterozygous pathogenic variants in MUTYH (Supplementary Table S3 online). We placed the nine remaining variants in a separate category of truncating mutations in a tumor suppressor gene that is not reported to be a germ-line cancer predisposition gene (Supplementary Table S4 online). To help ensure that no potential pathogenic variants were missed as a result of strict initial filtering, we manually reviewed all lower-tier mutations in the 22 genes in which a pathogenic or likely pathogenic mutation was found. No additional pathogenic/likely pathogenic mutations were identified in this expanded review.
The pathogenic/likely pathogenic variants were all mutually exclusive by patient and were therefore found in 13.1% of the population studied. These variants were found in similar percentages among the cohorts studied (National Cancer Institute 14.3% vs. ICGC 12.0% vs. PCGP 15.8%; P = 0.85), between WGS and WES samples (12.0 vs. 16.0%; P = 0.62), and between matched sequencing and tumor-only sequencing (12.3 vs. 16.2%; P = 0.58). Variants of unknown significance were similarly well distributed between groups (Supplementary Table S2 online). Truncating mutations in nonsyndromic tumor suppressor genes, however, were much more commonly observed in tumor-only sequencing samples (19.4 vs. 2.2%; P = 0.003), suggesting that some of these mutations are likely somatic variants that have not been previously reported simply because they are not highly recurrent in this tumor type (Supplementary Table S4 online).
The 23 variants deemed to be pathogenic/likely pathogenic included 22 different genes, with only BLM having multiple pathogenic mutations ( Table 1 ). However, several functional or disease-related clusters of genes were affected. Pathway analysis revealed striking enrichment for hereditary breast cancer signaling, DNA repair pathways, and, notably, DNA double-strand break repair ( Table 2 ). Enrichment in DNA damage response elements such as ATM signaling and GADD45 signaling, embryonic stem cell pluripotency, and molecular mechanisms of cancer were also noted.
To evaluate the possibility that the pathogenic/likely pathogenic variants were detected at a rate similar to that in the general population, we evaluated germline variants from these same 22 genes in the ExAC database (minus TCGA), which included data from 53,105 subjects. We identified 1,367 pathogenic/likely pathogenic variants (2.57%) in this population, which is significantly lower than the 13.1% affected in the Ewing sarcoma cohort (P = 2 × 10–10). We performed a similar analysis for heterozygous mutations in MUTYH. We considered this gene separately because heterozygous pathogenic mutations in this gene are not uncommon in the general population. The rate of MUTYH pathogenic/likely pathogenic mutations was not significantly different between groups (2.29 vs. 1.08%; P = 0.12).
We evaluated the potential association between germline pathogenic mutations and the known recurrent somatic mutations in STAG2, TP53, and CDKN2A in Ewing sarcoma. There were no significant differences between patients with a pathogenic/likely pathogenic germline mutation and those without regarding the rates of somatic mutations in STAG2 (18.1 vs. 17.8%; P = 1.0), CDKN2A (13.6 vs. 12.3%; P = 1.0), or TP53 (9.1 vs. 8.9%; P = 1.0). Interestingly, we noted that the two patients with a somatic mutation in TP53 in the pathogenic/likely pathogenic group had germline mutations in either TP53 itself or the TP53-associated gene WRAP53, suggesting a germline/somatic oncogenic synergy. For matched samples, we evaluated for additional examples of second hits in tumors from patients affected by a pathogenic/likely pathogenic germline mutation, either by loss of heterozygosity or by a truncating somatic mutation; however, we observed no additional cases. We additionally evaluated for potential associations between pathogenic/likely pathogenic mutations and demographic or outcome characteristics in the subset of patients for whom these data were available.8,9 There were no observed differences between groups regarding gender. There was a trend toward younger age among patients with pathogenic/likely pathogenic germline mutations compared with those without (57.1 vs. 42.3% younger than age 12; 42.9 vs. 50.8% age 12–24; 0 vs. 6.9% older than age 24), but this result did not reach statistical significance (P = 0.31). There was no significant difference in the rates of death due to disease. All of these comparisons were limited by small numbers of patients for whom data were available (Supplementary Table S5 online).
Discussion
We present the largest and most comprehensive germline genomics analysis to date involving Ewing sarcoma, utilizing whole-genome or whole-exome sequencing data for 175 patients. We discovered a high rate of pathogenic or likely pathogenic mutations, accounting for 13.1% of the population. This rate is similar but slightly higher than what has recently been reported for pediatric malignancies more generally, including in smaller subsets of Ewing sarcoma patients.4,5,6 Differences in methodology probably explain the differences in incidence between our study and previous studies, particularly regarding the broadness of the gene set used for reporting. For example, because we considered heterozygous deleterious mutations in any member of the Fanconi anemia gene family as being likely to predispose to solid malignancies, we classified them as likely pathogenic in our population. Next-generation sequencing has increasingly identified heterozygous deleterious mutations of members of this gene family outside of the more well-established BRCA1 and BRCA2 as predisposing to solid tumors,12,13 supporting our choice. Furthermore, a strong association has recently been reported between heterozygous germline variants in Fanconi anemia genes and translocation-associated sarcomas.7 We also performed a comparison using a large population database and found that pathogenic/likely pathogenic mutations in the genes reported were much more common in our cohort than in controls.
We observed that pathogenic germline mutations in Ewing sarcoma are not highly recurrent in a single gene; rather, they spread across several genes with potentially similar functional clustering. Mutations affecting DNA double-strand repair, in particular, were highly enriched. We found that second somatic hits in the same genes were uncommon, which is consistent with a previous report in which none of five Ewing sarcoma patients with pathogenic/likely pathogenic germline mutations had a second somatic hit.4 Given that the gain of somatic EWS-ETS fusion is believed to be the seminal event in Ewing sarcoma development,8,9,14 we speculate that the pathogenic germline mutations observed may be permissive for the development of DNA breaks and subsequent translocation. Previous work has noted a similar association between deleterious germline mutations in DNA repair genes and other translocation-associated sarcomas.7 We do caution that similar gene classes of pathogenic germline mutations have been described in the previously aforementioned pediatric malignancy studies that were not Ewing sarcoma–specific. Therefore, further studies are warranted to clarify whether a functional association between this class of mutation and the development of fusion-driven cancer truly exists.
One potential limitation of our study is that we included a portion of patients with tumor-only sequencing. In this smaller subset of patients, we were unable to confirm our findings as germline versus somatic. However, the rate of pathogenic/likely pathogenic mutations discovered in this subgroup is very similar to that of the overall cohort, and removing these patients from the analysis resulted in a virtually identical rate of pathogenic/likely pathogenic mutation overall (13.1 vs. 12.3%). Additionally, three large sequencing efforts for Ewing sarcoma have not reported recurrent somatic mutations in any of the genes identified here.8,9,14
It should be noted that our study was designed to capture only the most clinically relevant germline mutations (i.e., those that would be considered pathogenic or likely pathogenic). The majority of variants of unknown significance mutations, some of which may in time be proven to be clinically important, would have been filtered by design and are outside the scope of this evaluation. Additionally, we did not pursue copy-number evaluation as part of our analysis because we feel that copy-number detection from whole-genome or whole-exome sequencing is more prone to false-positive detection than small-variant detection. We also did not include variants that did not affect either a coding region or a splice site. Finally, because most of our data were derived from online repositories or samples from outside collaborators, we did not have access to family history or familial samples and were therefore unable to determine whether variants were de novo or inherited.
Our findings reported here have important clinical implications for patients and families affected by Ewing sarcoma. Given the high rates of pathogenic germline mutations in this population, we believe that referral to a genetic specialist should be considered for all patients and families affected by this disease. Although screening for Ewing sarcoma itself is unlikely to be undertaken given the rarity of the disease, even in those with a predisposing mutation, patients who survive their cancer and/or potentially family members may benefit from existing risk-management strategies for those with deleterious mutations in genes such as APC or BRCA1 that are associated with cancer syndromes that have a screening or surgical risk reduction management option. For the patients themselves, many of these germline variants may also influence cancer treatment or at least suggest the use of novel therapies, ideally in the setting of clinical trial. As examples, carriers of germline BRCA1 or BRCA2 mutations, even outside of breast or ovarian cancer, may benefit from treatment with poly (ADP-ribose) polymerase (PARP) inhibitors,15,16 whereas Hedgehog pathway inhibitors have activity in tumors associated with germline mutations in PTCH1 or PTCH2.17,18 The high frequency of potentially actionable germline alterations in Ewing sarcoma should also be considered as personalized-medicine approaches are designed and contemplated for patients affected by this disease. This study adds to a growing list highlighting the importance of germline sequencing for patients enrolled in precision-therapy protocols.5,19
Disclaimer
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US government.
Disclosure
The authors declare no conflicts of interest.
References
Worch J, Cyrus J, Goldsby R, Matthay KK, Neuhaus J, DuBois SG. Racial differences in the incidence of mesenchymal tumors associated with EWSR1 translocation. Cancer Epidemiol Biomarkers Prev 2011;20:449–453.
Abbott D, Randall RL, Schiffman J, Lessnick S, Cannon-Albright LA. Abstract 2748: a population-based survey of excess cancers observed in Ewing’s sarcoma and in their first-, second-, and third-degree relatives. Cancer Res 2015;75(15 suppl):2748.
Cope JU, Tsokos M, Miller RW. Ewing sarcoma and sinonasal neuroectodermal tumors as second malignant tumors after retinoblastoma and other neoplasms. Med Pediatr Oncol 2001;36:290–294.
Zhang J, Walsh MF, Wu G, et al. Germline mutations in predisposition genes in pediatric cancer. N Engl J Med 2015;373:2336–2346.
Chang W, Brohl AS, Patidar R, et al. MultiDimensional ClinOmics for precision therapy of children and adolescent young adults with relapsed and refractory cancer: a report from the center for cancer research. Clin Cancer Res 2016;22:3810–3820.
Parsons DW, Roy A, Yang Y, et al. Diagnostic yield of clinical tumor and germline whole-exome sequencing for children with solid tumors. JAMA Oncol; e-pub ahead of print 28 January 2016.
Ballinger ML, Goode DL, Ray-Coquard I, et al.; International Sarcoma Kindred Study. Monogenic and polygenic determinants of sarcoma risk: an international genetic study. Lancet Oncol 2016;17:1261–1271.
Brohl AS, Solomon DA, Chang W, et al. The genomic landscape of the Ewing Sarcoma family of tumors reveals recurrent STAG2 mutation. PLoS Genet 2014;10:e1004475.
Tirode F, Surdez D, Ma X, et al.; St. Jude Children’s Research Hospital–Washington University Pediatric Cancer Genome Project and the International Cancer Genome Consortium. Genomic landscape of Ewing sarcoma defines an aggressive subtype with co-association of STAG2 and TP53 mutations. Cancer Discov 2014;4:1342–1353.
Shern JF, Chen L, Chmielecki J, et al. Comprehensive genomic analysis of rhabdomyosarcoma reveals a landscape of alterations affecting a common genetic axis in fusion-positive and fusion-negative tumors. Cancer Discov 2014;4:216–231.
Richards S, Aziz N, Bale S, et al.; ACMG Laboratory Quality Assurance Committee. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015;17:405–424.
Couch FJ, Hart SN, Sharma P, et al. Inherited mutations in 17 breast cancer susceptibility genes among a large triple-negative breast cancer cohort unselected for family history of breast cancer. J Clin Oncol 2015;33:304–311.
Kiiski JI, Pelttari LM, Khan S, et al. Exome sequencing identifies FANCM as a susceptibility gene for triple-negative breast cancer. Proc Natl Acad Sci USA 2014;111:15172–15177.
Crompton BD, Stewart C, Taylor-Weiner A, et al. The genomic landscape of pediatric Ewing sarcoma. Cancer Discov 2014;4:1326–1341.
Kaufman B, Shapira-Frommer R, Schmutzler RK, et al. Olaparib monotherapy in patients with advanced cancer and a germline BRCA1/2 mutation. J Clin Oncol 2015;33:244–250.
Fong PC, Boss DS, Yap TA, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med 2009;361:123–134.
Rudin CM, Hann CL, Laterra J, et al. Treatment of medulloblastoma with hedgehog pathway inhibitor GDC-0449. N Engl J Med 2009;361:1173–1178.
Von Hoff DD, LoRusso PM, Rudin CM, et al. Inhibition of the hedgehog pathway in advanced basal-cell carcinoma. N Engl J Med 2009;361:1164–1172.
Jones S, Anagnostou V, Lytle K, et al. Personalized genomic analyses for cancer mutation discovery and interpretation. Sci Transl Med 2015;7:283ra53.
Acknowledgements
This study utilized the high-performance computational capabilities of the Biowulf Linux cluster at the National Institutes of Health (hhtp://biowulf.nih.gov).
Author information
Authors and Affiliations
Corresponding authors
Supplementary information
Supplementary Figure and Tables
(PDF 360 kb)
Supplementary Methods
(DOCX 26 kb)
Rights and permissions
About this article
Cite this article
Brohl, A., Patidar, R., Turner, C. et al. Frequent inactivating germline mutations in DNA repair genes in patients with Ewing sarcoma. Genet Med 19, 955–958 (2017). https://doi.org/10.1038/gim.2016.206
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/gim.2016.206
Keywords
This article is cited by
-
Identification of germline cancer predisposition variants in pediatric sarcoma patients from somatic tumor testing
Scientific Reports (2023)
-
An international working group consensus report for the prioritization of molecular biomarkers for Ewing sarcoma
npj Precision Oncology (2022)
-
EWSR1-WT1 gene fusions in neoplasms other than desmoplastic small round cell tumor: a report of three unusual tumors involving the female genital tract and review of the literature
Modern Pathology (2021)
-
Targeting of AKT-Signaling Pathway Potentiates the Anti-cancer Efficacy of Doxorubicin in A673 Ewing Sarcoma Cell Line
BioNanoScience (2021)
-
Review: Ewing Sarcoma Predisposition
Pathology & Oncology Research (2020)