Abstract
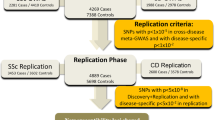
A recent meta-analysis identified seven single-nucleotide polymorphisms (SNPs) with suggestive evidence of association with multiple sclerosis (MS). We report an analysis of these polymorphisms in a replication study that includes 8,085 cases and 7,777 controls. A meta-analysis across the replication collections and a joint analysis with the discovery data set were performed. The possible functional consequences of the validated susceptibility loci were explored using RNA expression data. For all of the tested SNPs, the effect observed in the replication phase involved the same allele and the same direction of effect observed in the discovery phase. Three loci exceeded genome-wide significance in the joint analysis: RGS1 (P value=3.55 × 10−9), IL12A (P=3.08 × 10−8) and MPHOSPH9/CDK2AP1 (P=3.96 × 10−8). The RGS1 risk allele is shared with celiac disease (CD), and the IL12A risk allele seems to be protective for celiac disease. Within the MPHOSPH9/CDK2AP1 locus, the risk allele correlates with diminished RNA expression of the cell cycle regulator CDK2AP1; this effect is seen in both lymphoblastic cell lines (P=1.18 × 10−5) and in peripheral blood mononuclear cells from subjects with MS (P=0.01). Thus, we report three new MS susceptibility loci, including a novel inflammatory disease locus that could affect autoreactive cell proliferation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 6 digital issues and online access to articles
$119.00 per year
only $19.83 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
International Multiple Sclerosis Genetics Consortium (IMSGC). The expanding genetic overlap between multiple sclerosis and type I diabetes. Genes Immun 2009; 10: 11–14.
International Multiple Sclerosis Genetics Consortium (IMSGC). Refining genetic associations in multiple sclerosis. Lancet Neurol 2008; 7: 567–569.
De Jager P, Baecher-Allan C, Maier LM, Arthur AT, Ottoboni L, Barcellos L et al. The role of the CD58 locus in multiple sclerosis. Proc Natl Acad Sci USA 2009; 106: 5264–5269.
International Multiple Sclerosis Genetics Consortium (IMSGC). Risk alleles for multiple sclerosis identified by a genomewide study. N Engl J Med 2007; 357: 851–862.
Australia and New Zealand Multiple Sclerosis Genetics Consortium (ANZgene). Genome-wide association study identifies new multiple sclerosis susceptibility loci on chromosomes 12 and 20. Nat Genet 2009; 41: 824–828.
Aulchenko YS, Hoppenbrouwers IA, Ramagopalan SV, Broer L, Jafari N, Hillert J et al. Genetic variation in the KIF1B locus influences susceptibility to multiple sclerosis. Nat Genet 2008; 40: 1402–1403.
De Jager PL, Jia X, Wang J, de Bakker PI, Ottoboni L, Aggarwal NT et al. Meta-analysis of genome scans and replication identify CD6, IRF8 and TNFRSF1A as new multiple sclerosis susceptibility loci. Nat Genet 2009; 41: 776–782.
Hunt KA, Zhernakova A, Turner G, Heap GA, Franke L, Bruinenberg M et al. Newly identified genetic risk variants for celiac disease related to the immune response. Nat Genet 2008; 40: 395–402.
Hirschfield GM, Liu X, Xu C, Lu Y, Xie G, Lu Y et al. Primary biliary cirrhosis associated with HLA, IL12A, and IL12RB2 variants. N Engl J Med 2009; 360: 2544–2555.
Barrett JC, Hansoul S, Nicolae DL, Cho JH, Duerr RH, Rioux JD et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn′s disease. Nat Genet 2008; 40: 955–962.
Musone SL, Taylor KE, Lu TT, Nititham J, Ferreira RC, Ortmann W et al. Multiple polymorphisms in the TNFAIP3 region are independently associated with systemic lupus erythematosus. Nat Genet 2008; 40: 1062–1064.
Trynka G, Zhernakova A, Romanos J, Franke L, Hunt KA, Turner G et al. Coeliac disease-associated risk variants in TNFAIP3 and REL implicate altered NF-kappaB signalling. Gut 2009; 58: 1078–1083.
Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA et al. A second generation human haplotype map of over 3.1 million SNPs. Nature 2007; 449: 851–861.
Dixon AL, Liang L, Moffatt MF, Chen W, Heath S, Wong KC et al. A genome-wide association study of global gene expression. Nat Genet 2007; 39: 1202–1207.
Moratz C, Harrison K, Kehrl JH . Regulation of chemokine-induced lymphocyte migration by RGS proteins. Methods Enzymol 2004; 389: 15–32.
Han SB, Moratz C, Huang NN, Kelsall B, Cho H, Shi CS et al. Rgs1 and Gnai2 regulate the entrance of B lymphocytes into lymph nodes and B cell motility within lymph node follicles. Immunity 2005; 22: 343–354.
Trinchieri G . Interleukin-12 and the regulation of innate resistance and adaptive immunity. Nat Rev Immunol 2003; 3: 133–146.
Segal BM, Dwyer BK, Shevach EM . An interleukin (IL)-10/IL-12 immunoregulatory circuit controls susceptibility to autoimmune disease. J Exp Med 1998; 187: 537–546.
Brok HP, van Meurs M, Blezer E, Schantz A, Peritt D, Treacy G et al. Prevention of experimental autoimmune encephalomyelitis in common marmosets using an anti-IL-12p40 monoclonal antibody. J Immunol 2002; 169: 6554–6563.
Segal BM, Constantinescu CS, Raychaudhuri A, Kim L, Fidelus-Gort R, Kasper LH . Repeated subcutaneous injections of IL12/23 p40 neutralising antibody, ustekinumab, in patients with relapsing-remitting multiple sclerosis: a phase II, double-blind, placebo-controlled, randomised, dose-ranging study. Lancet Neurol 2008; 7: 796–804.
Shintani S, Ohyama H, Zhang X, McBride J, Matsuo K, Tsuji T et al. p12(DOC-1) is a novel cyclin-dependent kinase 2-associated protein. Mol Cell Biol 2000; 20: 6300–6307.
Kohno Y, Patel V, Kim Y, Tsuji T, Chin BR, Sun M et al. Apoptosis, proliferation and p12(doc-1) profiles in normal, dysplastic and malignant squamous epithelium of the Syrian hamster cheek pouch model. Oral Oncol 2002; 38: 274–280.
Jakkula E, Leppa V, Sulonen AM, Varilo T, Kallio S, Kemppinen A et al. Genome-wide association study in a high-risk isolate for multiple sclerosis reveals associated variants in STAT3 gene. Am J Hum Genet 2010; 86: 285–291.
Cenit MC, Alcina A, Marquez A, Mendoza JL, Diaz-Rubio M, de Las Heras V et al. STAT3 locus in inflammatory bowel disease and multiple sclerosis susceptibility. Genes Immun 2010; 11: 264–268.
Mero IL, Lorentzen AR, Ban M, Smestad C, Celius EG, Aarseth JH et al. A rare variant of the TYK2 gene is confirmed to be associated with multiple sclerosis. Eur J Hum Genet 2009; 18: 502–504.
De Jager PL, Chibnik LB, Cui J, Reischl J, Lehr S, Simon KC et al. Integration of genetic risk factors into a clinical algorithm for multiple sclerosis susceptibility: a weighted genetic risk score. Lancet Neurol 2009; 8: 1111–1119.
Baranzini SE, Wang J, Gibson RA, Galwey N, Naegelin Y, Barkhof F et al. Genome-wide association analysis of susceptibility and clinical phenotype in multiple sclerosis. Hum Mol Genet 2009; 18: 767–778.
Comabella M, Craig DW, Camina-Tato M, Morcillo C, Lopez C, Navarro A et al. Identification of a novel risk locus for multiple sclerosis at 13q31.3 by a pooled genome-wide scan of 500,000 single nucleotide polymorphisms. PLoS One 2008; 3: e3490.
Hafler JP, Maier LM, Cooper JD, Plagnol V, Hinks A, Simmonds MJ et al. CD226 Gly307Ser association with multiple autoimmune diseases. Genes Immun 2009; 10: 5–10.
D’Alfonso S, Bolognesi E, Guerini FR, Barizzone N, Bocca S, Ferrante D et al. A sequence variation in the MOG gene is involved in multiple sclerosis susceptibility in Italy. Genes Immun 2008; 9: 7–15.
Kallio SP, Jakkula E, Purcell S, Suvela M, Koivisto K, Tienari PJ et al. Use of a genetic isolate to identify rare disease variants: C7 on 5p associated with MS. Hum Mol Genet 2009; 18: 1670–1683.
Lundmark F, Duvefelt K, Iacobaeus E, Kockum I, Wallstrom E, Khademi M et al. Variation in interleukin 7 receptor alpha chain (IL7R) influences risk of multiple sclerosis. Nat Genet 2007; 39: 1108–1113.
McDonald WI, Compston A, Edan G, Goodkin D, Hartung HP, Lublin FD et al. Recommended diagnostic criteria for multiple sclerosis: guidelines from the International Panel on the diagnosis of multiple sclerosis. Ann Neurol 2001; 50: 121–127.
Polman CH, Reingold SC, Edan G, Filippi M, Hartung HP, Kappos L et al. Diagnostic criteria for multiple sclerosis: 2005 revisions to the ‘McDonald Criteria’. Ann Neurol 2005; 58: 840–846.
Dalton CM, Brex PA, Miszkiel KA, Hickman SJ, MacManus DG, Plant GT et al. Application of the new McDonald criteria to patients with clinically isolated syndromes suggestive of multiple sclerosis. Ann Neurol 2002; 52: 47–53.
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81: 559–575.
Mantel N, Haenszel W . Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 1959; 22: 719–748.
DerSimonian R, Laird N . Meta-analysis in clinical trials. Control Clin Trials 1986; 7: 177–188.
Cochran W . The combination of estimates from different experiments. Biometrics 1954; 10: 101–129.
Higgins JP, Thompson SG . Quantifying heterogeneity in a meta-analysis. Stat Med 2002; 21: 1539–1558.
de Bakker PI, Ferreira MA, Jia X, Neale BM, Raychaudhuri S, Voight BF . Practical aspects of imputation-driven meta-analysis of genome-wide association studies. Hum Mol Genet 2008; 17: R122–R128.
Stranger BE, Forrest MS, Dunning M, Ingle CE, Beazley C, Thorne N et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science 2007; 315: 848–853.
Gauthier SA, Mandel M, Guttmann CR, Glanz BI, Khoury SJ, Betensky RA et al. Predicting short-term disability in multiple sclerosis. Neurology 2007; 68: 2059–2065.
Acknowledgements
We are grateful to the patients and the healthy controls for their participation in this study. PLD is a Harry Weaver Neuroscience Scholar Award Recipient of the National MS Society (NMSS). DAH is a Jacob Javits Scholar of NIH. SD is supported by a FISM grant (2008/R/11). BF and IR are authors on behalf of REFGENSEP, a national French clinical and research network funded by INSERM, ARSEP and AFM. The International MS Genetics Consortium is supported by R01NS049477 and by the National Multiple Sclerosis Society. This work was also supported by the Medical Research Council (G0700061) and by the Cambridge NIHR Biomedical Research Centre. The sampling and analysis of the Swedish cohorts have received grant support from The Swedish Research Council, the EU fp6 program neuropromise (LSHM-CT-2005-018637), as well as from the Bibbi and Niels Jensens Foundation, The Montel Williams Foundation and the Söderberg Foundation.
Author information
Authors and Affiliations
Consortia
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Web Resources
The URLs for data presented herein are as follows:
International HapMap Project, http://hapmap.ncbi.nlm.nih.gov/
GENe Expression VARiation, http://www.sanger.ac.uk/humgen/genevar/
meta: Meta-Analysis, R package http://CRAN.R-project.org/package=meta
rmeta: Meta-Analysis, R package http://CRAN.R-project.org/package=rmeta
‘mRNA-by-SNP-browser’ v1.0.1, http://www.sph.umich.edu/csg/liang/asthma/
The Gene Expression Omnibus accession number for the RNA data obtained from PBMCs and reported in this paper is GSE16214.
Supplementary Information accompanies the paper on Genes and Immunity website
Supplementary information
Appendix
Appendix
Members of the International Multiple Sclerosis Genetics Consortium
F Esposito1,2, NA Patsopoulos3,4,5, S Cepok6, I Kockum7,8, V Leppä9,10, DR Booth11, RN Heard11, GJ Stewart11, M Cox12, RJ Scott12, J Lechner-Scott12, A Goris13, R Dobosi13, B Dubois13, JD Rioux14,15, AB Oturai16, HB Søndergaard16, F Sellebjerg16, PS Sørensen16, M Reunanen17, K Koivisto18, I Cournu-Rebeix19,20,21, B Fontaine19,20,21, J Winkelmann22,23,24, C Gieger25, C Infante-Duarte26, F Zipp27, L Bergamaschi28, M Leone29, R Bergamaschi30, P Cavalla31, ÅR Lorentzen32,33, I-L Mero33,34, EG Celius34, HF Harbo32,34, A Spurkland35, M Comabella36, B Brynedal7, L Alfredsson37, L Bernardinelli38,39, NP Robertson40, CP Hawkins41,42, LF Barcellos43, G Beecham44, W Bush45, BAC Cree46, MJ Daly4,47,48, AJ Ivinson49, C Aubin48, A Compston50, S D’Alfonso28, JL Haines45, SL Hauser46, B Hemmer6, J Hillert8,37, JL McCauley44, J Oksenberg46, T Olsson7,8, A Palotie9,10, L Peltonen9,10,51, MA Pericak-Vance44, J Saarela9,10, SJ Sawcer50, B Stranger3,4,48, FM Boneschi1,2, G Comi1,2, DA Hafler4,48,52, PIW de Bakker3,4,48 and PL De Jager4,5,48, International Multiple Sclerosis Genetics Consortium members.
1Department of Neurology, Scientific Institute San Raffaele, Milan, Italy; 2Institute of Experimental Neurology, Scientific Institute San Raffaele, Milan, Italy; 3Division of Genetics, Department of Medicine, Brigham & Women's Hospital, Boston, MA, USA; 4Harvard Medical School, Boston, MA, USA; 5Program in Translational NeuroPsychiatric Genomics, Department of Neurology, Brigham & Women's Hospital, Boston, MA, USA; 6Klinikum rechts der Isar, Technische Universität, Münch, Germany; 7Neuroimmunology Unit, Department of clinical neuroscience, Karolinska Institutet at Karolinska University Hospital, Solna, Sweden; 8Center for Molecular Medicine, Karolinska Institutet at Karolinska University Hospital, Solna, Sweden; 9Public Health Genomics Unit, National Institute for Health and Welfare, Helsinki, Finland; 10Institute for Molecular Medicine Finland (FIMM), University of Helsinki, Helsinki, Finland; 11University of Sydney, Institute for Immunology and Allergy Research, Westmead Millennium Institute, Westmead Hospital, NSW, Australia; 12University of Newcastle, Callaghan, NSW Australia; 13Section for Experimental Neurology, Katholieke Universiteit Leuven, Leuven, Belgium; 14Université de Montréal, Montréal, Québec, Canada; 15Montreal Heart Institute, H1 T 1C8, Montréal, Québec, Canada; 16The Danish Multiple Sclerosis Research Center, Copenhagen University Hospital, Rigshospitalet, DK-2100, Copenhagen, Denmark; 17Department of Neurology, Oulu University Hospital, Oulu, Finland; 18Central Hospital of Seinäjoki, Seinäjoki, Finland; 19INSERM, UMR_S975, Paris, France; 20UPMC Univ Paris 06, UMR_S975, Centre de Recherche Institut du Cerveau et de la Moelle, CNRS 7225, Paris, France; 21Département de Neurologie, Pitié –Salpêtrière Hospital, AP-HP, 75651 Paris, France; 22Klinik für Neurologie, Technische Universität München, München, Deutsch; 23Institut für Humangenetik, Technische Universität München, München, Deutsch; 24Institut für Humangenetik, Helmholtz Zentrum München, München, Deutsch; 25Institute of Epidemiology, Helmholtz Zentrum München-German Research Center for Environmental Health, Munich, Germany; 26Charite-Universitaetsmedizin Berlin, Cecilie Vogt Clinic for Neurology, Berlin, Germany; 27Department of Neurology, University Medicine Mainz, Johannes Gutenberg University, Mainz, Germany; 28Department of Medical Sciences and Interdisciplinary Research Center of Autoimmune Diseases, University of Eastern Piedmont, Novara, Italy; 29Clinica Neurologica, AOU Maggiore della Carità, Novara, Italy; 30Neurological Institute C. Mondino, IRCCS, Pavia, Italy; 31Department of Neurology, Ospedale San Giovanni Battista, Torino, Italy; 32Department of Neurology, Faculty Division Ullevål University Hospital, University of Oslo, Oslo, Norway; 33Institute of Immunology, Rikshospitalet, Oslo University Hospital, Oslo, Norway; 34Department of Neurology, Oslo University Hospital, Ullevål, Oslo, Norway; 35Institute of Basal Medical Sciences, University of Oslo, Blindern, Oslo, Norway; 36Unitat de Neuroimmunologia Clínica, CEM-Cat, Hospital Universitari Vall d’Hebron, Barcelona, Spain; 37Institute of Environmental Medicine, Karolinska Institutet, Stockholm, Sweden; 38University of Pavia, Department of Applied Sciences, Pavia, Italy; 39University of Cambridge, Statistical Laboratory, Centre for Mathematical Sciences, Wilberforce Road, Cambridge, UK; 40Department of Neurology, University Hospital of Wales, Heath Park, Cardiff, UK; 41Human Genomics research group, Keele University, Stoke-on-Trent, UK; 42Department of Neurology, University Hospital North Staffordshire, Stoke-on-Trent, UK; 43Division of Epidemiology, School of Public Health, University of California at Berkeley, Berkeley, CA, USA; 44John P. Hussman Institute for Human Genomics, The University of Miami Miller School of Medicine, Miami, FL, USA; 45Center for Human Genetics Research, Vanderbilt University Medical Center, Nashville, TN, USA; 46Department of Neurology, University of California San Francisco, San Francisco, CA, USA; 47Massachusetts General Hospital, Boston, MA, USA; 48Program in Medical & Population Genetics, Broad Institute of Harvard University and Massachusetts Institute of Technology, Cambridge, MA, USA; 49Harvard NeuroDiscovery Center, Harvard Medical School, Boston, MA, USA; 50University of Cambridge, Department of Clinical Neuroscience, Addenbrooke's Hospital, Cambridge, UK; 51The Wellcome Trust Sanger Institute, Hinxton, Cambridge, UK and 52Department of Neurology, Yale University Medical School, New Haven, CT, USA.
Rights and permissions
About this article
Cite this article
The International Multiple Sclerosis Genetics Conssortium (IMSGC). IL12A, MPHOSPH9/CDK2AP1 and RGS1 are novel multiple sclerosis susceptibility loci. Genes Immun 11, 397–405 (2010). https://doi.org/10.1038/gene.2010.28
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/gene.2010.28
Keywords
This article is cited by
-
Transcriptome-wide association study reveals increased neuronal FLT3 expression is associated with Tourette’s syndrome
Communications Biology (2022)
-
Replication study of GWAS risk loci in Greek multiple sclerosis patients
Neurological Sciences (2019)
-
Genome-wide DNA methylation changes in CD19+ B cells from relapsing-remitting multiple sclerosis patients
Scientific Reports (2018)
-
Differential methylation at MHC in CD4+ T cells is associated with multiple sclerosis independently of HLA-DRB1
Clinical Epigenetics (2017)
-
RGS10 deficiency ameliorates the severity of disease in experimental autoimmune encephalomyelitis
Journal of Neuroinflammation (2016)