Abstract
Pseudohypoparathyroidism is a rare endocrine disorder that can be caused by genetic (mainly maternally inherited inactivating point mutations, although intragenic and gross deletions have rarely been reported) or epigenetic alterations at GNAS locus. Clinical and molecular characterization of this disease is not that easy because of phenotypic, biochemical and molecular overlapping features between both subtypes of the disease. The European Consortium for the study of PHP (EuroPHP) designed the present work with the intention of generating the standards of diagnostic clinical molecular (epi)genetic testing in PHP patients. With this aim, DNA samples of eight independent PHP patients carrying GNAS genetic and/or epigenetic defects (three patients with GNAS deletions, two with 20q uniparental disomy and three with a methylation defect of unknown origin) without GNAS point mutations were anonymized and sent to the five participant laboratories for their routine genetic analysis (methylation-specific (MS)-MLPA, pyrosequencing and EpiTYPER) and interpretations. All laboratories were able to detect methylation defects and, after the data analysis, the Consortium compared the results to define technical advantages and disadvantages of different techniques. To conclude, we propose as first-level investigation in PHP patients copy number and methylation analysis by MS-MLPA. Then, in patients with partial methylation defect, the result should be confirmed by single CpG bisulphite-based methods (ie pyrosequencing), whereas in case of a complete methylation defect without detectable deletion, microsatellites or SNP genotyping should be performed to exclude uniparental disomy 20.
Similar content being viewed by others
Introduction
Pseudohypoparathyroidism (PHP) is a rare disorder characterized by hypocalcaemia, hyperphosphataemia and elevated parathyroid hormone (PTH) levels as a result of end-organ resistance to this hormone.1, 2, 3 PHP in association with obesity and clinical features of Albright hereditary osteodystrophy (AHO), which includes short stature, variable degree of mental retardation, brachydactyly and heterotopic ossifications, is classified as PHP1A (OMIM 103580).3, 4, 5, 6 In most cases (70–80%), PHP1A is caused by maternally inherited heterozygous inactivating mutations in the coding sequence of Gsα (exons 1–13 of GNAS), whereas paternally inherited mutations are associated with AHO alone, in the absence of hormone resistance – a variant termed ‘pseudopseudohypoparathyroidism’ (PPHP) (OMIM 612463).3, 4 Both diseases are characterized by a decreased responsiveness of Gsα to the in vitro stimulation of the GPCR-cAMP-PKA pathway. Beside point mutations, in very few patients, large deletions including part or the whole gene have been reported7, 8, 9 (Figure 1).
Schematic representation of GNAS locus including the genetic deletions described. Maternal STX16 deletions cause isolated A/B loss of methylation;11, 13, 20, 21 maternal deletion of NESP55 leads to isolated A/B loss of methylation with hemizygosity in NESP55.22 However, maternal deletions affecting AS exons 3 and 4 result in a loss of methylation at all maternal GNAS imprints.15, 23 Gross deletions affecting some or all the DMRs of GNAS locus and leading to an apparent methylation defect has also been published.7, 8, 9
The other main form of PHP is PHP type 1B (PHP1B; OMIM 603233), characterized by PTH resistance and sometimes TSH resistance, but usually patients have neither additional endocrine abnormalities nor further clinical features. PHP1B subjects display paternal-specific patterns of cytosine methylation within differentially methylated regions (DMRs) of their maternally inherited GNAS alleles,10, 11 suggesting a loss of imprinting (LoI) as the basis of the disorder.
Most PHP1B cases, showing LoI at all GNAS-DMRs, are the uniquely affected individuals of their family and thereafter considered as sporadic (sporPHP1B).10, 11, 12, 13 The postzygotic occurrence of this epigenetic defect is supported by the observation of a partial LoI in some patients.14, 15 In a small subset of these patients (10–25% according to various reports), uniparental disomy of chromosome 20q has been identified as the cause of the GNAS methylation anomaly.16, 17, 18, 19 On the other hand, some cases are familial with an autosomal dominant mode of inheritance (AD-PHP1B).10 AD-PHP1B is typically associated with a loss of methylation restricted to the exon A/B DMR owing to maternally inherited microdeletions within STX1611, 13, 20, 21 or NESP55,22 which likely harbour a cis-acting control element crucial for the establishment of the methylation imprint at exon A/B. In addition, deletions removing the entire NESP55 DMR as well as part of GNAS-AS transcript have also been identified in some AD-PHP1B kindreds in whom affected individuals show loss of all maternal GNAS imprints.15, 23
However, clinical and molecular analysis for PHP is not easy to achieve because of different reasons. First, beyond the classic PHP type 1 classification, our groups and others demonstrated a genetic overlap between PHP1A and PHP1B, reporting patients with mild AHO features and methylation defects.24, 25, 26, 27 Second, Gsα activity has also been reported to be decreased not only in patients with GNAS mutations (PHP1A) but also in patients with methylation defects at the GNAS locus.28 Third, as mentioned above, methylation defects at the GNAS locus may be partial and undetected by non-quantitative methods of methylation analysis.
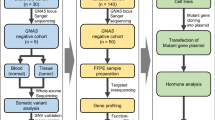
The aim of this work was to provide an external quality assessment (EQA) among five different European laboratories with the purpose of raising and maintaining the standards of diagnostic clinical molecular (epi)genetic testing in PHP patients. The idea was formed in 2012 after the first meeting of the EuroPHP Consortium supported by the European Society of Pediatric Endocrinology (ESPE). The main issue reflected by participating clinicians and scientists was the classification of different degrees of methylation defects, that is, partial versus complete, at GNAS-DMRs (see Supplementary Figure 1). Therefore, eight patients with known GNAS molecular defects were selected, anonymized and sent to participant laboratories. Each laboratory was asked to characterize the (epi)genetic variation, explain the causative defect (if possible) and propose further analyses (if needed).
Materials and methods
A group of clinician and scientist members of the EuroPHP Consortium designed an external quality assessment (EQA) for the (epi)genetic analysis of patients with PHP not caused by point mutations at the GNAS gene.
Scheme design
The five participant laboratories received aliquots of the same eight samples of PHP patients without point mutations at GNAS gene. They were asked to perform molecular analysis using their in-house methods and to send back the raw data and the genetic interpretation.
Results were compared and discussed at a workshop in Lübeck, Germany in 2013, focused on diagnostic flow, methodologies, interpretation of results and reporting, with the final aim to formulate Best Practice Guidelines for the molecular diagnosis of PHP supported by the ESPE.
Bisulphite modification
Approximately 0.5–1 μg DNA was subjected to sodium bisulphite treatment and purified using the EZ GOLD methylation kit (Zymo, Irvine, CA, USA), EpiTect bisulphite kit (Qiagen, Hilden, Germany) or MethylDetector bisulphite modification kit (Active Motif, Carlsbad, CA, USA), according to the manufacturer’s instructions. Experiments were run in duplicate or triplicate, depending on the laboratory.
Methylation detection
Methylation-specific-MLPA
Allelic dosage and methylation analyses of GNAS were carried out by methylation-specific (MS)-MLPA using SALSA ME031 kit (MRC-Holland, Amsterdam, The Netherlands). The analysis of MS-MLPA PCR products was performed on an ABI3500 (Lab no. 5) or ABI3130xl (Lab no. 3) genetic analysers, and analysed using the GeneMapper v.4.1, PeakScanner v.1.0 (Applied Biosystems, Foster City, CA, USA) and Coffalyser v.9.4 (MRC-Holland) softwares, as described previously.29
Pyrosequencing
Standard bisulphite PCR using commercial or home-made primers targeting DMRs at NESP55, AS, XLαs and exon A/B were used for amplification (see Supplementary Table 1). The entire biotinylated PCR product was mixed with 40 μl of binding buffer and 2 μl (10 mg/ml) of streptavidin-coated polystyrene beads. Bead–amplicon complexes were captured on a vacuum prep tool (Qiagen) and PCR products were denatured using 0.2 M NaOH. The denatured DNA was resuspended in 0.3 μ M of sequencing primer dissolved in annealing buffer and primer annealing was achieved by heating the sample at 80 °C for 2 min before cooling to room temperature. The pyrosequencing reaction was carried out on a PyroMark Q96 or Q24 instrument (Qiagen). Peak heights were determined using the PyroMark Q24 v.2.0.6.20 software.14, 26 The percentage of CpG methylation for each DMR was calculated as the mean of the percentage of methylation at each CpG.
Sequenom EpiTYPER system
NESP55, XLαs and exon A/B methylation was studied via the Sequenom EpiTYPER using primers and conditions reported previously.30 Briefly, PCR amplification reactions for each amplicon consisted of 20 ng of bisulphite-treated DNA, 1 × of Megamix Gold (Microzone Ltd, Haywards Heath, UK) and 0.2 μ M of primers (Life Technologies, Carlsbad, CA, USA), and 2 μl PCR products were used as template for in vitro transcription and RNAaseA cleavage for the T-reverse reaction (Sequenom hMC, San Diego, CA, USA). Samples were spotted on a 384-SpectroCHIP (Sequenom) and analysed by the MassARRAY Analyser Compact MALDI-TOF MS and the EpiTYPER software (Sequenom). The average of CpG methylation (as percentage) for each amplicon, which was analysed on MassARRAY in triplicate, was calculated for both control and patient samples.
The position of CpGs analysed is shown in Supplementary Table 2.
Statistical analysis
Methylation was reported as a percentage. Outliers (by each laboratory and for each sample) were defined and identified as any data point more than 1.5 interquartile ranges below the first quartile or above the third quartile and calculated by Dixon’s Q test for outliers.31, 32 These data were removed for the calculi.
An exploratory data analysis of the sample studied (mean and standard deviations and median and interquartile ranges for values of biomarkers measured by each instrument assessed) was performed. Based on the distribution of the data, median and interquartile ranges were used for comparisons. In addition, boxplots of each biomarker were performed.
For healthy controls (50% of methylation), the coefficient of variation for each DMR at each laboratory was computed. The coefficient of variation is a measure of dispersion around the mean. It represents the ratio of the standard deviation to the mean.
The nonparametric Kruskall–Wallis test for independent samples was used to evaluate the interlaboratory and intertechnique variability for each sample.
Furthermore, reference values were calculated for each DMR and stratified by methylation percentage (0, 50 and 100%) using nonparametric methods, computing the 2.5th and 97.5th percentile and their 95% confidence intervals (CIs).
All statistical analyses were performed using the SAS System statistical package (version 9.2) and figures were developed by R statistical software (R Foundation for Statistical Computing, Vienna, Austria; http://www.R-project.org/). After Bonferroni correction, a P-value of <0.05 was deemed to indicate statistical significance.
Results
Two laboratories (Lab nos. 2 and 4) performed pyrosequencing analysis, Lab no. 1 used the Sequenom EpiTYPER technology, whereas Lab nos. 3 and 5 used both MS-MLPA and pyrosequencing analysis. Lab no. 3’s methylation data are for MS-MLPA and ones from Lab no. 5 are for pyrosequencing; these techniques were the ones used for routine diagnosis.
Bisulphite modification was performed by EZ GOLD methylation kit (Lab nos. 4 and 5), Epitec bisulphite kit (Lab no. 2) and MethylDetector bisulphite modification kit (Lab no. 1), and posterior experiments were run on PyromarkQ96 (Lab nos. 3 and 4), PyromarkQ24 (Lab nos. 2 and 5) and EpiTYPER (Lab no. 1) (see Supplementary Table 3).
Genomic regions analysed
The five laboratories studied different or overlapping regions of the four GNAS-DMRs (Supplementary Figure 2). In brief, for GNAS-AS, Lab nos. 2, 3 and 5 analysed the same region, whereas Lab no. 4 analysed an independent and non-overlapping one. For NESP55, Lab nos. 2 and 3 analysed the same region and Lab nos. 4 and 5 studied the same CpGs in one of their assays. Different regions were analysed for the other two DMRs. The number of the CpG islands analysed by the different groups is around 8–12 for each DMR. In one laboratory (Lab no. 1), the GNAS analysis comprised three DMRs (NESP55, XLαs and A/B).
Controls’ results
All laboratories developed their assays after checking the normal values for control people (expected 50% of methylation), with similar results independent of the technique or bisulphite treatment used. Lab no. 1 showed the lowest methylation values for all the DMRs (Supplementary Figure 3A). Variation coefficients varied from 8.1 (Lab no. 4) to 59.9 (Lab no. 1) for NESP55, from 6.8 (Lab no. 4) to 11.4 (Lab no. 2) for AS, from 7.9 (Lab no. 4) to 39 (Lab no. 1) for XLαs and from 4.1 (Lab no. 4) to 40.8 (Lab no. 1) for A/B. Variation coefficient for the same CpG positions analysed by the different techniques were compared, when possible, revealing the highest variation value for EpiTYPER (data not shown).
Three labs used specific samples as standards for 0% methylation controls, being either pat20UPD DNA sample (Lab nos. 3–5) or commercial 0% (Cells-to-CpG Unmethylated gDNA Control; Applied Biosystems) (Lab no. 5). Based on the results, commercial control was thereafter excluded for subsequent analysis because it was not adjusted to 0% (Supplementary Figure 3B).
As 100% methylation control, Lab no. 4 used a patient-derived DNA sample with paternal deletion of the GNAS locus, whereas Lab no. 5 used both paternal 20q deletion and commercial 100% (Cells-to-CpG Methylated gDNA Control; Applied Biosystems), both with similar results, except for NESP55 where the commercial DNA maintains the fully methylated result (100%) and the pat20q deletion presents nearly 0% of methylation as expected (Supplementary Figure 3C).
Samples’ results
Anonymized samples were sent to the five laboratories for methylation analysis of GNAS-DMRs and interpretation of the data was performed regardless of clinical information.
Selected samples included:
-
a proven genetic defect at GNAS (genetic variants described at http: //www.LOVD.nl/GNAS): paternal (OMIM no. 612463) and maternal deletion at the GNAS locus (EQA no. 1, GNAS_00212 and EQA no. 2, GNAS_00213, respectively), and intragenic maternal deletion at the GNAS gene (EQA no. 4, GNAS_00211; Fernandez-Rebollo et al8);
-
a cytogenetic defect involving GNAS: pat20qUPD (EQA no. 5);
-
partial methylation defects at the whole GNAS locus either due to a mosaic of pat20qUPD (EQA no. 8; Maupetit-Mehouas et al19) or from unknown origin (EQA nos. 3, 6 and 7).
Table 1 summarizes the results of our GNAS analysis and the proposed genetic interpretation, whereas Figure 2 shows the data sustaining the proposal. Even if analysed regions were not exactly the same for most DMRs (see Supplementary Figure 2), all labs were able to define accurately the methylation defect of analysed samples, independently of the method used.
Results of methylation quantification for different analysed samples. Median is indicated by filled black dot, and the 25th and 75th quantiles (IQR: interquartile range) are determined by the range of the box. The ends of the whiskers represent the lowest datum still within 1.5 IQR of the lower quartile, and the highest datum still within 1.5 IQR of the upper quartile. Points beyond the whiskers are potential outliers. For Lab no. 5, M represents the results obtained by MS-MLPA and P those of pyrosequencing. (a) All labs suggested complete LOM at NESP55 and GOM for the rest of DMRs for EQA no. 1 methylation results. (b) and (c) (EQA nos. 5 and 2, respectively) presented equal methylation patterns: complete GOM at NESP55 and LOM for the rest of DMRs, being analogous among different labs. (d) All labs proposed isolated LOM at A/B for EQA no. 4. (e) Methylation results for EQA nos. 3, 6–8 were comparable; therefore, only figure for EQA no. 8 is shown. Methylation quantification indicates a partial GOM at NESP55 and partial LOM at the rest of DMRs.
In particular, all labs identified the methylation alteration at the GNAS locus, but only those using MLPA were able to attribute the defect to genomic deletions within the locus (see samples in EQA nos. 1, 2 and 4), owing to the additional information provided by the MS-MLPA kit on methylation-independent probes against most GNAS exons, as well the STX16 gene.
All labs gave a similar interpretation for EQA nos. 3, 6–8 and some of them (Lab nos. 4 and 5) pointed out the need for further studies to conclude the exact underlying molecular determinant of the observed methylation defect, as it could be a primary epigenetic defect or secondary to genetic defects as UPD or deletions.
It was noted that uniparental disomy was unanimously suggested by laboratories when methylation defects affected all GNAS-DMRs. Alternatively, laboratories not performing MS-MLPA also proposed to exclude deletions as the primary genetic defect by further investigations.
Although the MS-MLPA reaction is robust and reproducible, the use of different DNA extraction methods may determine a different DNA degradation pattern and the carryover of different contaminants (ie heparin, melanin, etc), thus possibly influencing the peak pattern. To minimize non-biologic differences between EQA samples and in-lab healthy references, we purified EQA samples by ethanol precipitation. Nevertheless, we could estimate the acceptable quality of experiments, but we were not able to fully eliminate variability and the copy analysis could not be properly ascertained for some of the samples (EQA nos. 1 and 2).
Statistical analyses of methylation percentages among labs showed significant differences for most samples, although they could not be associated with either the lab or the analysed region (Supplementary Table 4A). When techniques were compared, statistical differences were also observed (Supplementary Table 4B).
One important limiting step of the study was the definition of boundaries for a normal, partial and complete methylation defect. According to results obtained by different techniques, we tried to define cutoffs for partial and complete methylation defects, and data from this analysis for each DMR are summarized in Table 2. Pyrosequencing reference values for methylation defects were a methylation percentage higher than 70% for complete GoM at NESP55 (50% methylation range: 32–68%), and lower than 20% at GNAS-AS (50%: 35–55%), lower than 16% at XLαs (50%: 30–57) and lower than 14% at exon A/B (50%: 35–63%) for complete LoM. Regarding MS-MLPA, reference values were established as higher than 90% for GoM at NESP55 (50%: 38–61%), and lower than 16% at GNAS-AS (50%: 42–62%), lower than 17% at XLαs (50%: 36–62%) and lower than 8% at exon A/B (50%: 38–65%) for complete LoM. For EpiTYPER, cutoff values were as follows: higher than 92% for hypermethylation at NESP55 (50%: 5–61%), and lower than 28% for hypomethylation at XLαs (50%: 13-61%) and lower than 26% for hypomethylation at exon A/B (50%: 11–62%).
Discussion
More than 70 years ago, Albright et al33 described PHP as an entity characterized by hypocalcaemia and hyperphosphataemia with a reduced calcaemic and phosphaturic response to injected bovine parathyroid extract, leading to the hypothesis of a resistance to PTH action. Now, PHP encompasses a heterogeneous group of rare metabolic disorders characterized by end-organ resistance to the action of PTH and whose (epi)genetic defect(s) are been actively investigated (for a review, see Mantovani34).
Clinical diagnosis of PHP1A and PHP1B is hampered by (epi)genetic and clinical overlaps.24, 25, 26, 27 It is therefore of utmost importance to establish a reliable identification and quantification of the methylation at GNAS-DMRs to (i) provide an accurate diagnosis of PHP to patients and physicians, (ii) orientate genetic and cytogenetic investigations depending on the methylation pattern and (iii) identify patients without any known primary defect causing the methylation anomalies to nourish the research.
In the present work, a group of experts in clinical and molecular aspects on PHP analyzed the (epi)genetic status of eight independent PHP patients without point mutations at GNAS gene. Methylation status was studied by pyrosequencing, Sequenom EpiTYPER and MS-MLPA.
First of all, technique validation was analyzed by measuring the coefficient of variation for 50% methylation control and differences seem to be due to the used technique. For the experiments, all labs used DNA from healthy people as 50% methylation control and some of them do also include in-house or commercial 0 and 100% DNA controls. Based on our experience, in-house 0 and 100% controls show more robust data, so we suggest to use them.
Previous studies suggested that pyrosequencing with specific primers after bisulphite treatment of the genomic DNA is more sensitive in detecting methylation defects rather than MS-MLPA, as it is a precise quantitative method.35, 36 Despite statistical differences in methylation quantification, our data show that the (epi)genetic interpretation for each patient was essentially the same, independently of the technique used, even for cases presenting a partial methylation defect. Differences among techniques, reported previously, are more marked in case of partial defects with very subtle methylation defects, which are mainly present in imprinting disorders with a postzygotic methylation establishment.36 Additional patients with partial methylation defects (infrequent in PHP) should be analyzed to confirm differences among techniques.
On the other hand, MS-MLPA experiments are useful not only for methylation quantification but also for the identification of putative deletions or duplications causative of some methylation defects, an essential information for proper genetic counselling.37 In the present work, only Lab nos. 3 and 5 could identify deletions carried by EQA nos. 1, 2 and 4. However, MS-MLPA presents the limitation that reference and index samples should be extracted by the same method and have a similar concentration to minimize non-biologic differences and structural variation (see hhtp: //www.mlpa.com). This could be the cause for the difficulty found for copy analysis by MS-MLPA in EQA nos. 1 and 2.
We also highlight that an identical interpretation was obtained independently of the analysed region, indicating that methylation defects in PHP are not CpG specific, in contrast to other diseases.38, 39 The validation of such data in PHP patients with methylation defects not caused by genetic alterations is important, as biased results have been shown in other imprinting disorders owing to the variation of methylation at neighbouring CpGs within the DMRs.40, 41
Based on our results, partial methylation defects are difficult to relate with threefold standard deviation, as in previous reports.42 Thus, we propose to define partial methylation defects at GNAS for each technique and at each DMR, and we confirm that it is essential to use in-house 0, 50 and 100% controls for methylation analysis. Partial methylation defects may arise from a postzygotic occurrence of the epigenetic defect.19 The distribution of mosaic genetic defects can vary widely among different tissues of a patient, and similar discrepancies can also be found for methylation defects.43 This implies that partial ‘epimutations’ detected in lymphocytes of PHP patients might be explained by mosaicism; thus, strict methylation cutoff values for partial GoM or LoM are not useful in this condition. Moreover, low or undetectable (epi)mutations in lymphocytes do not exclude a high percentage of mutated cells in target tissues like the proximal renal tubule.
In conclusion, this EQA study showed that, in spite of small differences in methylation percentages and the use of different methods, the overall results as well as the consequent (epi)genetic interpretation were very similar in all involved labs. MS-MLPA demonstrated the advantage, besides the estimation of methylation frequencies at all four GNAS-DMRs, to detect large deletions within the coding sequence of Gsα, alternative GNAS exons or STX16, allowing to pick out patients affected with sporPHP1B or AD-PHP1B in one experiment. Therefore, this method proved to be suitable to identify concomitantly both genetic and epigenetic GNAS defects in PHP patients. Nevertheless, this method is sensitive to DNA impurities and DNA isolation methods, and thus it is strongly recommended to perform DNA isolation in the lab performing the assay.
According to the long-standing experience of involved groups in performing the clinical and molecular diagnosis of PHP and to the results of this EQA study, we propose, as a further workflow for PHP patients without GNAS inactivating mutations, to perform copy and methylation analysis by MS-MLPA. Subsequently, in patients with a partial methylation defect, confirm the result by pyrosequencing or any other quantitative CpG-specific bisulphite-based method. If complete methylation defect is found, in the absence of a detectable deletion, microsatellite or SNP genotyping should be finally performed to exclude uniparental disomy.
Change history
12 March 2015
This article has been amended since online publication. A corrigendum also appears in this issue.
References
Mantovani G, Spada A : Mutations in the Gs alpha gene causing hormone resistance. Best Pract Res Clin Endocrinol Metab 2006; 20: 501–513.
Kelsey G : Imprinting on chromosome 20: tissue-specific imprinting and imprinting mutations in the GNAS locus. Am J Med Genet C 2010; 154C: 377–386.
Weinstein LS, Yu S, Warner DR, Liu J : Endocrine manifestations of stimulatory G protein alpha-subunit mutations and the role of genomic imprinting. Endocr Rev 2001; 22: 675–705.
Levine MA : Pseudohypoparathyroidism: from bedside to bench and back. J Bone Miner Res 1999; 14: 1255–1260.
Bastepe M, Juppner H : Editorial: Pseudohypoparathyroidism and mechanisms of resistance toward multiple hormones: molecular evidence to clinical presentation. J Clin Endocrinol Metab 2003; 88: 4055–4058.
Miric A, Vechio JD, Levine MA : Heterogeneous mutations in the gene encoding the alpha-subunit of the stimulatory G protein of adenylyl cyclase in Albright hereditary osteodystrophy. J Clin Endocrinol Metab 1993; 76: 1560–1568.
Genevieve D, Sanlaville D, Faivre L et al: Paternal deletion of the GNAS imprinted locus (including Gnasxl) in two girls presenting with severe pre- and post-natal growth retardation and intractable feeding difficulties. Eur J Hum Genet 2005; 13: 1033–1039.
Fernandez-Rebollo E, Garcia-Cuartero B, Garin I et al: Intragenic GNAS Deletion involving exon A/B in pseudohypoparathyroidism type 1A resulting in an apparent loss of exon A/B methylation: potential for misdiagnosis of pseudohypoparathyroidism type 1B. J Clin Endocrinol Metab 2010; 95: 765–771.
Mitsui T, Nagasaki K, Takagi M, Narumi S, Ishii T, Hasegawa T : A family of pseudohypoparathyroidism type Ia with an 850-kb submicroscopic deletion encompassing the whole GNAS locus. Am J Med Genet A 2011; 158A: 261–264.
Liu J, Litman D, Rosenberg MJ, Yu S, Biesecker LG, Weinstein LS : A GNAS1 imprinting defect in pseudohypoparathyroidism type IB. J Clin Invest 2000; 106: 1167–1174.
Bastepe M, Pincus JE, Sugimoto T et al: Positional dissociation between the genetic mutation responsible for pseudohypoparathyroidism type Ib and the associated methylation defect at exon A/B: evidence for a long-range regulatory element within the imprinted GNAS1 locus. Hum Mol Genet 2001; 10: 1231–1241.
Liu J, Nealon JG, Weinstein LS : Distinct patterns of abnormal GNAS imprinting in familial and sporadic pseudohypoparathyroidism type IB. Hum Mol Genet 2005; 14: 95–102.
Bastepe M, Frohlich LF, Hendy GN et al: Autosomal dominant pseudohypoparathyroidism type Ib is associated with a heterozygous microdeletion that likely disrupts a putative imprinting control element of GNAS. J Clin Invest 2003; 112: 1255–1263.
Maupetit-Mehouas S, Mariot V, Reynes C et al: Quantification of the methylation at the GNAS locus identifies subtypes of sporadic pseudohypoparathyroidism type Ib. J Med Genet 2011; 48: 55–63.
Chillambhi S, Turan S, Hwang DY, Chen HC, Juppner H, Bastepe M : Deletion of the noncoding GNAS antisense transcript causes pseudohypoparathyroidism type Ib and biparental defects of GNAS methylation in cis. J Clin Endocrinol Metab 2010; 95: 3993–4002.
Bastepe M, Lane AH, Juppner H : Paternal uniparental isodisomy of chromosome 20q – and the resulting changes in GNAS1 methylation – as a plausible cause of pseudohypoparathyroidism. Am J Hum Genet 2001; 68: 1283–1289.
Fernandez-Rebollo E, Lecumberri B, Garin I et al: New mechanisms involved in paternal 20q disomy associated with pseudohypoparathyroidism. Eur J Endocrinol 2010; 163: 953–962.
Bastepe M, Altug-Teber O, Agarwal C, Oberfield SE, Bonin M, Juppner H : Paternal uniparental isodisomy of the entire chromosome 20 as a molecular cause of pseudohypoparathyroidism type Ib (PHP-Ib). Bone 2010; 48: 659–662.
Maupetit-Mehouas S, Azzi S, Steunou V et al: Simultaneous hyper- and hypomethylation at imprinted loci in a subset of patients with GNAS epimutations underlies a complex and different mechanism of multilocus methylation defect in pseudohypoparathyroidism type 1b. Hum Mutat 2013; 34: 1172–1180.
Linglart A, Gensure RC, Olney RC, Juppner H, Bastepe M : A novel STX16 deletion in autosomal dominant pseudohypoparathyroidism type Ib redefines the boundaries of a cis-acting imprinting control element of GNAS. Am J Hum Genet 2005; 76: 804–814.
Elli FM, de Sanctis L, Peverelli E et al: Autosomal dominant pseudohypoparathyroidism type Ib: a novel inherited deletion ablating STX16 causes loss of Imprinting at the A/B DMR. J Clin Endocrinol Metab 2014; 99: E724–E728.
Richard N, Abeguile G, Coudray N et al: A new deletion ablating NESP55 causes loss of maternal imprint of A/B GNAS and autosomal dominant pseudohypoparathyroidism type Ib. J Clin Endocrinol Metab 2012; 97: E863–E867.
Bastepe M, Frohlich LF, Linglart A et al: Deletion of the NESP55 differentially methylated region causes loss of maternal GNAS imprints and pseudohypoparathyroidism type Ib. Nat Genet 2005; 37: 25–27.
Perez de Nanclares G, Fernandez-Rebollo E, Santin I et al: Epigenetic defects of GNAS in patients with pseudohypoparathyroidism and mild features of Albright's hereditary osteodystrophy. J Clin Endocrinol Metab 2007; 92: 2370–2373.
Unluturk U, Harmanci A, Babaoglu M et al: Molecular diagnosis and clinical characterization of pseudohypoparathyroidism type-IB in a patient with mild Albright's hereditary osteodystrophy-like features, epileptic seizures, and defective renal handling of uric acid. Am J Med Sci 2008; 336: 84–90.
Mantovani G, de Sanctis L, Barbieri AM et al: Pseudohypoparathyroidism and GNAS epigenetic defects: clinical evaluation of albright hereditary osteodystrophy and molecular analysis in 40 patients. J Clin Endocrinol Metab 2010; 95: 651–658.
Mariot V, Maupetit-Mehouas S, Sinding C, Kottler ML, Linglart A : A maternal epimutation of GNAS leads to Albright osteodystrophy and PTH resistance. J Clin Endocrinol Metab 2008; 93: 661–665.
Zazo C, Thiele S, Martin C et al: Gsalpha activity is reduced in erythrocyte membranes of patients with psedohypoparathyroidism due to epigenetic alterations at the GNAS locus. J Bone Miner Res 2011; 26: 1864–1870.
Perez-Nanclares G, Romanelli V, Mayo S et al: Detection of hypomethylation syndrome among patients with epigenetic alterations at the GNAS locus. J Clin Endocrinol Metab 2012; 97: E1060–E1067.
Izzi B, Decallonne B, Devriendt K et al: A new approach to imprinting mutation detection in GNAS by Sequenom EpiTYPER system. Clin Chim Acta 2010; 411: 2033–2039.
Dixon WJ : Analysis of extreme values. Ann Math Statist 1950; 21: 488–506.
Dixon WJ : Processing data for outliers. Biometrics 1953; 9: 74–89.
Albright F, Burnett CH, Smith PH, Parson W : Pseudohypoparathyroidsm – an example of ‘Seabright syndrome’. Endocrinology 1942; 30: 922–932.
Mantovani G : Clinical review: pseudohypoparathyroidism: diagnosis and treatment. J Clin Endocrinol Metab 2011; 96: 3020–3030.
Lee BH, Kim GH, Oh TJ et al: Quantitative analysis of methylation status at 11p15 and 7q21 for the genetic diagnosis of Beckwith–Wiedemann syndrome and Silver–Russell syndrome. J Hum Genet 2013; 58: 604–610.
Christians A, Hartmann C, Benner A et al: Prognostic value of three different methods of MGMT promoter methylation analysis in a prospective trial on newly diagnosed glioblastoma. PLoS One 2012; 7: e33449.
Mantovani G, Linglart A, Garin I, Silve C, Elli FM, de Nanclares GP : Clinical utility gene card for: pseudohypoparathyroidism. Eur J Hum Genet 2013; 21: doi:10.1038/ejhg.2012.211.
Hubertus J, Zitzmann F, Trippel F et al: Selective methylation of CpGs at regulatory binding sites controls NNAT expression in Wilms tumors. PLoS One 2013; 8: e67605.
De Smet C, Loriot A, Boon T : Promoter-dependent mechanism leading to selective hypomethylation within the 5' region of gene MAGE-A1 in tumor cells. Mol Cell Biol 2004; 24: 4781–4790.
Kannenberg K, Weber K, Binder C, Urban C, Kirschner HJ, Binder G : IGF2/H19 hypomethylation is tissue, cell, and CpG site dependent and not correlated with body asymmetry in adolescents with Silver–Russell syndrome. Clin Epigenet 2012; 4: 15.
Romanelli V, Magano L, Fernandez L et al: Hypomethylation gradient at the 11p15 different methylated region 2 (DmR2) in BWs patients. Eur J Hum Genet 2008; 16: 132–133.
Court F, Martin-Trujillo A, Romanelli V et al: Genome-wide allelic methylation analysis reveals disease-specific susceptibility to multiple methylation defects in imprinting syndromes. Hum Mutat 2013; 34: 595–602.
Begemann M, Spengler S, Kanber D et al: Silver–Russell patients showing a broad range of ICR1 and ICR2 hypomethylation in different tissues. Clin Genet 2011; 80: 83–88.
Acknowledgements
This work was supported by the Euro-Pseudohypoparathyroidism network (EuroPHP) grant from the European Society for Pediatric Endocrinology Research Unit (to AL), grants from the University of Luebeck (awarded to ST), Attractivité Grant from Paris-Sud University 2013 (to AL), ANR EPIFEGROW (Agence nationale de la recherche) (to AL), French Society of Pediatric Endocrinology and Diabetology (to VG), the Italian Ministry of Health (to GM: GR-2009-1608394), recurrent funding from INSERM U986 (to AL, VG and CS) and Instituto de Salud Carlos III (PI10/0148 and PI13/00467 to GPdN). KF and BI are supported by research Grants G.0490.10N and G.0A23.14N from the FWO-Vlaanderen (Belgium); IG by FIS-Program (I3SNS-CA10/01056) and GPdN by I3SNS Program of the Spanish Ministry of Health (CP03/0064; SIVI 1395/09). All members of the EuroPHP are members of the EUCID.net (COST action BM1208 on imprinting disorders; http: //www.imprinting-disorders.eu).
Author information
Authors and Affiliations
Consortia
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Members of the EuroPHP Consortium
Belgium (Kathleen Freson and Benedetta Izzi, Department of Cardiovascular Sciences, Center for Molecular and Vascular Biology, University of Leuven), France (Agnes Linglart, Caroline Silve, Stephanie Maupetit-Mehouas and Virginie Grybek, INSERM U986, Hôpital Bicêtre, Le Kremlin Bicêtre, Paris; and Anne Barlier, Laboratory of Biochemistry and Molecular Biology, Conception Hospital, Aix-Marseille University, Marseille), Germany (Olaf Hiort, Susanne Thiele, Bettina Brix and Ralf Werner, Division of Experimental Paediatric Endocrinology and Diabetes, Department of Paediatrics, University of Luebeck), Italy (Giovanna Mantovani, Francesca M Elli, Paolo Bordogna and Valentina Boldrin, Department of Clinical Sciences and Community Health, University of Milan, Endocrinology and Diabetology Unit, Fondazione IRCCS Ca’ Granda Ospedale Maggiore Policlinico, Milan; and Luisa de Sanctis, Department of Public Health and Pediatrics, University of Turin, Reggina Margherita Children’s Hospital, Turin), Spain (Guiomar Perez de Nanclares, Intza Garin and Arrate Pereda, Molecular (Epi)Genetics Laboratory, BioAraba National Health Institute, Hospital Universitario Araba-Txagorritxu, Vitoria-Gasteiz; and Beatriz Lecumberri, Endocrinology Service, Hospital Universitario La Paz, Madrid), Turkey (Serap Turan, Marmara University School of Medicine Hospital, Istanbul) and United Kingdom (Deborah JG Mackay, Faculty of Medicine, University of Southampton, Southampton).
Supplementary Information accompanies this paper on European Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Garin, I., Mantovani, G., Aguirre, U. et al. European guidance for the molecular diagnosis of pseudohypoparathyroidism not caused by point genetic variants at GNAS: an EQA study. Eur J Hum Genet 23, 438–444 (2015). https://doi.org/10.1038/ejhg.2014.127
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2014.127
This article is cited by
-
Diagnosis and management of pseudohypoparathyroidism and related disorders: first international Consensus Statement
Nature Reviews Endocrinology (2018)
-
Genome-wide DNA methylation analysis of pseudohypoparathyroidism patients with GNAS imprinting defects
Clinical Epigenetics (2016)
-
Prenatal molecular testing for Beckwith–Wiedemann and Silver–Russell syndromes: a challenge for molecular analysis and genetic counseling
European Journal of Human Genetics (2016)
-
A multiplex oligonucleotide ligation-PCR as a complementary tool for subtyping of Salmonella Typhimurium
Applied Microbiology and Biotechnology (2015)