Abstract
The 185delAG* BRCA1 mutation is encountered primarily in Jewish Ashkenazi and Iraqi individuals, and sporadically in non-Jews. Previous studies estimated that this is a founder mutation in Jewish mutation carriers that arose before the dispersion of Jews in the Diaspora ∼2500 years ago. The aim of this study was to assess the haplotype in ethnically diverse 185delAG* BRCA1 mutation carriers, and to estimate the age at which the mutation arose. Ethnically diverse Jewish and non-Jewish 185delAG*BRCA1 mutation carriers and their relatives were genotyped using 15 microsatellite markers and three SNPs spanning 12.5 MB, encompassing the BRCA1 gene locus. Estimation of mutation age was based on a subset of 11 markers spanning a region of ∼5 MB, using a previously developed algorithm applying the maximum likelihood method. Overall, 188 participants (154 carriers and 34 noncarriers) from 115 families were included: Ashkenazi, Iraq, Kuchin-Indians, Syria, Turkey, Iran, Tunisia, Bulgaria, non-Jewish English, non-Jewish Malaysian, and Hispanics. Haplotype analysis indicated that the 185delAG mutation arose 750–1500 years ago. In Ashkenazim, it is a founder mutation that arose 61 generations ago, and with a small group of founder mutations was introduced into the Hispanic population (conversos) ∼650 years ago, and into the Iraqi–Jewish community ∼450 years ago. The 185delAG mutation in the non-Jewish populations in Malaysia and the UK arose at least twice independently. We conclude that the 185delAG* BRCA1 mutation resides on a common haplotype among Ashkenazi Jews, and arose about 61 generations ago and arose independently at least twice in non-Jews.
Similar content being viewed by others
Introduction
More than 3000 pathogenic mutations and sequence alterations have been reported within the BRCA1 (MIM# 113705) and BRCA2 (MIM# 600185) since they were identified in the mid 1990s (http://research.nhgri.nih.gov/bic/). In several populations, the spectrum of mutations is rather limited, reflecting a single or a limited number of ‘founder mutation(s)’: the Dutch,1 Icelandic,2 the Polish,3 Russian,4 and the Norwegian5 populations. Notably, among Ashkenazi Jews (ie, Jews of east European ancestry), three mutations in BRCA1 (185delAG, 5382InsC) and BRCA2 (6174delT) occur frequently.6 One of these mutations (185delAG* BRCA1) is also reported among non-Ashkenazi (primarily Iraqi origin) Jews,7 and sporadically among non-Jewish populations (http://research.nhgri.nih.gov/bic/).8, 9, 10, 11, 12, 13, 14, 15, 16 The occurrence of an identical mutation in ethnically diverse populations may stem from either a founder effect or may reflect a so-called mutational hotspot. Earlier studies conducted in a Jewish 185delAG *BRCA1 mutation carrier showed that in Ashkenazim this is a seemingly founder mutation that shares a common haplotype,7, 16, 17, 18 but in Indian12 and Pakistani13 non-Jewish mutation carriers the haplotype was reportedly different, suggesting that this mutation arose independently in these populations. As the majority of these earlier studies were based on data generated from a few intragenic markers, obtained from a single-genotyped individual from a carrier family, phasing and estimation of the age when the mutation arose were suboptimal. The aim of the present study was to overcome these caveats by using multiple polymorphic markers on multiple family members to allow defining the shared haplotype, phasing, and comparing the haplotype in ethnically diverse 185delAG*BRCA1 mutation carriers.
Materials and methods
Participant identification and recruitment
Israel
The study population was recruited from among individuals counseled and tested at one of three oncogenetics services located at the Sheba Medical Center, Tel-Hashomer, the Rambam Medical Center, Haifa, or the Rivkah Ziv Medical center in Zefat, since 1 January 2000. Participants recruited were either diagnosed with breast cancer or ovarian cancer, or in the minority of cases were asymptomatic individuals from ‘high-risk breast/ovarian cancer families based on well-accepted criteria.19 The study was approved by the local IRBs, and each patient gave informed consent.
USA
Hispanics in New Mexico (all of Colonial New Mexico Hispanic ancestry)20, 21 and California (primarily of Mexican ancestry) with a personal or family history of breast and/or ovarian cancer were enrolled in an IRB-approved registry after informed consent and underwent genetic counseling and BRCA testing within the City of Hope Clinical Cancer Genetics Community Research Network (CCGCRN). The CCGCRN is a consortium of 14 US cancer center and community-based clinics that provide GCRA to individuals with a personal or a family history of cancer.22 A blood sample, demographic data, and four- to five-generation pedigrees were obtained, including reported ethnicity and country/state of origin, for each grandparental lineage. Clinical details (eg, age at diagnosis, pathology report, and/or death certificate when possible) were obtained for relatives affected with breast and/or ovarian cancer. A bilingual cancer risk counselor conducted GCRA sessions for Spanish-speaking patients, or translation services were provided, with adapted counseling aides and Spanish consent forms.22, 23
Malaysia
Patients were recruited as part of a study into the genetic factors of breast cancer in Malaysia's multiethnic population, using previously described identification and recruitment schemes that were ethically approved.24
England
Breast and ovarian cancer families have been tested for BRCA1/2 mutations since 1996 in the Manchester region of North-West England. The region covers a population of ∼4.5 million people. Women who attend the specialist genetic clinics in the region with a family history of breast or ovarian cancer have a detailed family tree elicited with all first-, second-, and, if possible, third-degree relatives recorded. If a BRCA1 or BRCA2 mutation is identified, further attempts are made to ensure that all individuals at risk of inheriting the family mutation are represented on the pedigree.25 Once a family-specific pathogenic BRCA1/2 mutation is identified, predictive testing is offered to all blood relatives. Where possible, all affected women with breast or ovarian cancer are tested to establish the true extent of BRCA1/2 involvement in the family.
DNA extraction-Peripheral blood leukocyte DNA was extracted using the PUREgene kit (Gentra Inc., Minneapolis, MN, USA) using the manufacturer’s recommended protocol.
Analysis for the 185delAG BRCA1 mutation
Israel
Analysis for the BRCA1 185delAG mutation was carried out using a PCR-directed mutagenesis assay to introduce a restriction site that distinguishes between the wild-type and the mutant allele, as previously described and used by us.6, 26 Confirmation of any suspect sample was done using sequencing of the same amplicon.
USA
BRCA testing was performed at Myriad Genetic Laboratories (MGL), Inc. (Salt Lake City, UT, USA), including full sequencing of exons and flanking intronic segments.
Malaysia
Full sequencing of exons and intron–exon boundaries, and MLPA analysis were performed as previously described.24
England
Mutation screening involves a whole-gene sequencing of exons and intron–exon boundaries, and MLPA analysis for large deletions.27 Among 335 non-Jewish families with pathogenic BRCA1 mutations, 5 (1.5%) harbored the 185delAG*BRCA1 mutation.
Allelotyping for the BRCA1 locus
To determine the haplotype structure of the BRCA1*185delAG mutation, the following markers were used: three intragenic short-tandem repeats, D17S855, D17S1322, and D17S1323, and 12 flanking perigenic markers, D17S1147, D2171801, D17S1299, D17S1814, D17S1818, and D17S1867 upstream to BRCA1. D17S951, D17S1789, D17S1861, D17S931, D17S1827, and D17S1795 were downstream to BRCA1. All markers were dinucleotide STRs, except for D17S1299 (tetranucleotide) and D17S1322 (tri-nucleotide). It is noteworthy that the three intragenic markers were also used by Neuhausen et al.18 The primer sequences for all markers were retrieved from the Genome DataBase online database (www.gdb.org). All markers span approximately 12.5 Mbp around the BRCA1 locus (Figure 1). Genomic DNA from each subject was amplified by PCR for each marker. The forward primers of each pair of primers were labeled with FAM for the analysis of the Amplicons. A volume of 2 μl of each PCR product was mixed with 0.5 μl of the TAMRA 500 internal size standard (Applied Biosystems Inc., Foster City, CA, USA) and 12 μl of formamide. Samples were read on the ABI Prism 3100 using the GeneScan Software (Applied Biosystems). The GeneScan raw data were analyzed using the Genotyper software (Foster City, CA, USA) to obtain the allele repeat in base pairs.
Haplotype construction and age of the mutation
Age Estimates using the maximum likelihood method
To estimate the age of the mutation (or, more precisely, the number of generations as the most recent common ancestor (MRCA) of the 185delAG carriers), we used the method that was first used to estimate the age of several BRCA1 mutations, including the mutation that is the focus of the present study, 185delAG* BRCA1,18 and then extended and applied to BRCA2 mutations,28 and which has been used in several other similar studies, most recently in an analysis of the mutation BRCA1*c.5266dupC (5382insC).29 This method uses maximum likelihood and allows for both recombination and mutational events at the marker loci as means of altering a presumed ancestral haplotype. Phased haplotypes were used if these could be inferred from available family data; otherwise, all possible haplotypes were constructed from multi locus genotype data and weighted according to their probability. For each value of G (the number of generations since the MRCA), the relative likelihood that each haplotype is descended from the ancestral haplotype via mutation and recombination is calculated compared with the likelihood that it is a totally independent haplotype (ie, an independent recurrent 185delAG mutation on a different haplotype background). The value of G that maximizes this likelihood is obtained through iterative search. Ninty-five percent support intervals were constructed by identifying those points, GL and GU, where the likelihood differed from the maximum by 0.86 (corresponding to a chi-squared likelihood ratio statistic of 3.84, eg, P=0.05). To examine the likely genetic history of the 185delAG mutation, we analyzed separately each of several defined subgroups in which a sufficient number of samples were available for analysis. These subgroups were Ashkenazi Jewish, Iraqi, and Hispanic. In the case of the Ashkenazi and Iraqi subgroups, there were a few samples of mixed origins (eg, Iraqi–Turkish). We ran the analyses by both including them and excluding them from the relevant group. For these, the reference haplotype was determined by choosing the two most frequent alleles at each marker, and found the reference haplotype and the number of generations that provided the best fit to the data. Haplotypes/multilocus genotypes for all 115 families analyzed in this study are given in Supplementary Table 1.
Assumed genetic map
The recombination rates between markers were assumed to be those estimated in Kong et al.30 Physical positions of the STRs and SNPs were those from the Human Reference sequence, build 3.7. For markers present on the deCODE map, we used the genetic positions in centimorgans as reported there, whereas for those not on the deCODE map we estimated the genetic position from the proportion of physical distance between the known markers and then translated this to the genetic scale. This has the effect of using locally defined relationships between physical and genetic distance, and thus can accommodate the reported recombination suppression in this region.31 As our method uses marker allele frequencies in the calculation of the likelihood, we estimated these frequencies from the unlinked allele (not on the assumed haplotype bearing the 185delAG mutation) of the chromosomes in the sample. Marker positions and assumed allele frequencies are given in Supplementary Table 2.
Marker mutation rates
As a baseline, we used the rates for the nine dinucleotide and two tri/tetranucleotide microsatellite markers as estimated from CEPH data by Weber and Wong32 of 0.0006 and 0.002, respectively, for a mutation of a single-repeat unit. We assumed that the probability of changes of n repeat units in a given meiosis was (0.0006 or 0.002)n for n=2, 3, 4 and that for more than four repeats was taken to be equal to that for four repeat units. Because of the imprecision of these rates (and model), we introduced another parameter into the likelihood and jointly estimated the number of generations and a multiplier of the assumed marker mutation rates described above. Thus, to a certain extent, we let the data inform the proper marker mutation rates. In addition to the true underlying marker mutation rates, this also allows for potential genotyping errors to be accounted for in the model. We found that the best fit to our data was when the recombination rate was 1.25x that of Weber and Wong.32
Cluster analysis of 185delAG haplotpyes
To graphically present all 115 haplotypes, we performed hierarchical clustering analysis. To measure (in some sense) the similarity of haplotypes, we used a variation of the likelihood method described above, and calculated the likelihood of each haplotype paired with every other haplotype, assuming that the first was the ‘reference haplotype’ and assuming G=1. From these pairwise likelihoods, we calculated Dij=−(Lij+Lji – Lii−Ljj), where Lij is the log likelihood derived from the comparison of haplotype i with haplotype j, as a measure of the distance between haplotypes i and j. Note that because of the assumption that one of the two haplotypes is the reference and the other is derived from that reference, Lij≠Lji. The distance matrix composed of the (Dij) has the properties of being symmetric, positive, and Dii=0. This distance matrix was then used as an input to a hierarchical cluster analysis using the Ward measure of inter-cluster similarity, as implemented in STATA v.11.0 (StataCorp., Austin, TX, USA).
Results
Participants’ characteristics
Overall, there were 188 participants in the study (Table 1): 54 Ashkenazim (from 38 families- of whom 46 were carriers and 8 noncarriers from 5 families). Ninety-seven non-Ashkenazi Jews were from the following origins: Iraq, Kuchin India, Syria, Turkey, Iran, Lebanon, and Bulgaria. In 18 non-Ashkenazi families, 24 noncarriers were also genotyped with the BRCA1-associated markers. In addition, 24 Hispanics, from 17 families of self-declared Mexican origin all 185delAG* BRCA1 mutation carriers from the San Luis Valley, CO, USA, Arizona or California, were genotyped, as were three Malaysian non-Jewish individuals from three independent families, and 10 individuals from five families recruited in the UK. Of the 188 participants, 16 were men. There were 64 women diagnosed with breast cancer (mean age at diagnosis being (± SD) 42.7 ± 10.15 years), and an additional four women had bilateral breast cancer. Twenty women were diagnosed with ovarian or peritoneal cancer (mean age at diagnosis was 49.9 ± 7.5 years) and the remainder (n=88) were asymptomatic carriers, with a mean age at counseling (data available for the Israeli patients only, n=78) being 42.3 ± 8.6 years.
Estimation of the age of 185delAG
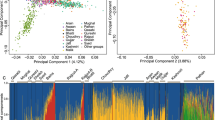
Of the markers genotyped, analysis was restricted to 11 markers, as the four most distant ones (D17S1867, D17S18181, D17S1827, D17S795) were too distant and only added noise to the analysis. For the whole sample consisting of 115 haplotypes/unphased multilocus genotypes, the maximum likelihood estimate of the time to the MRCA of the haplotypes was 59 generations (95% confidence interval (CI), 51–69 generations). It is more informative, however, to look at the results for specific subpopulations rather than the whole data set. In this case, the Ashkenazi Jewish set showed the greatest degree of haplotype diversity with an estimated time to MRCA of 61 generations (95% CI, 47–77 generations), compared with 31 (95% CI, 19–47 genrations) for the Hispanic 185delAG carriers and 23 generations (95% CI, 17–33 generations) in the Iraqi population. Figure 2 shows the results of the cluster analysis as a dendrogram. The heights of the vertical lines are proportional to the distance at which the clusters joined. One can see immediately that, for example, the three Malaysian haplotypes are similar to each other but quite different from the rest of the haplotypes in the data set, indicating that in this group the 185delAG mutation arose independently on a different haplotypic background. The English haplotypes tended to fall into two groups, one a separate haplotype, which has previously been indicated18 and appears to be relatively common in the north of England (Yorkshire haplotype). However, two of the English haplotypes fit very well within the Ashkenazi Jewish haplotype, and most likely represent members of the Jewish community in Manchester that is of Ashkenazi origin. The Hispanic haplotypes seem to fall into two major groups, although both seem to cluster with a mix of Ashkenazi and Iraqi haplotypes.
Cluster analysis of the 185delAG* BRCA1 mutation carriers. The Letter beneath each node describes the origin of the sample: J: Ashkenazi Jews; I: Iraqi Jews; H: US Hispanics; M: Malaysians: E: English. 1–8 refer to other Jewish singleton samples as follows: 1: Sephardic; 2: Ashkenazi/Iran; 3: Kuchin, India; 4: Iran; 5: Syria/Turkey; 6: Turkey/Iran; 7: Lebanese/Bulgarian; 8: non-Ashkenazi. Some similar haplotypes of the same group have already been combined at the first step and could not be individually displayed on the diagram.
Discussion
The results of the present study suggest that the 185delAG*BRCA1 mutation is indeed a founder mutation in Jewish mutation carriers that arose about 1200 years ago in Ashkenazi Jews, and that through the migration of a small subset of founder mutation carriers was introduced into the Hispanic population about 650 years ago, and to the Jewish–Iraqi community about 450 years ago. These numbers are based on calculating 20 years/generation.33 Using a 30 year/generation and using the upper limits of the estimated the MRCA the estimated ages are 2300, 1400, and 1000 years ago for the Ashkenazi, Hispanics, and the Iraqis, respectively.
These results are somewhat unexpected, as the prevailing notion that was based both on historical events of the Jewish people and the finding of a similar haplotype in Ashkenazi and non-Ashkenazi (primarily Iraqi) mutation carriers prompted the tentative conclusion that the 185delAG*BRCA1 mutation is an ancient Jewish mutation that arose before the dispersion of the Jews in the Diaspora about 2500 years ago.34, 35 Thus, the finding of a later age at which the mutation arose in Ashkenazim and that the date it was introduced into the Iraqi–Jewish community was significantly later in history is somewhat puzzling, and unaccounted for by historical events. Specifically, there are no records of any major influx of Ashkenazi populations from Europe back to the region of Modern-day Iraq. However, given the central role that Iraq and the area played in trade, the traditional involvement of Jewish Ashkenazim in trade, it is plausible that a few Jewish Ashkenazi individuals (including one or few founders) immigrated to Iraq as individuals and not as part of a big migration wave. Such a small and genealogically insignificant migration would not be captured by most historians.
Yet the results pertaining to the existence of the 185delAG*BRCA1 mutation and the date when it arose in Hispanics are in line with Jewish historical events. In 1492, the Jews in Spain were given the option by the Inquisition of either becoming Christians or be expelled from Spain. A subset of Jews elected to stay in Spain, and while openly converting to Christianity secretly maintained their Jewish way of life.36 Some of these individuals, called Conversos, later immigrated to the New World. The estimated contribution of Jewish linage in the current-day inhabitants of the Iberian Peninsula is estimated at 20%,37 and the contribution and existence of Sephardic Jews is also noted in South American communities.38 The age at which the mutation arose in the Hispanics, and the shared haplotype with Ashkenazim, is in line with these historic events, as well as with the specific history of the establishment of the New Mexico Colony in 1598 and descendants to the San Luis Valley in 1850.20, 35
The Jewish Ashkenazi 185delAG*BRCA1 mutation carriers all share a common haplotype, supporting the founder mutation theory. These data are consistent with our own previously published data,7 and those of others reporting on multi locus allelotyping of Jewish Ashkenazi mutation carriers.16, 18 The use of families and saturating the region for polymorphic markers facilitated an accurate evaluation of the age at which the mutation arose. Thus, the age at which the 185delAG*BRCA1 mutation arose in the Ashkenazi population reported herein (61 generations ago ∼1200 years ago) is consistent with the age estimated by Neuhausen et al,18 who estimated that the mutation arose around 1235 CE (90% CI ranging from 396 to 1536 CE) based on the 500-kb conserved region centromeric to the mutation. It is noteworthy that Im et al39 reported that 50% of 316 Ashkenazi 185delAG*BRCA1 mutation carriers share a common ancient 2.1-Mb haplotype at the same locus, making this a more recent mutation. On the basis of a much smaller number of cases, Pereira et al40 reported that there is a single common Ashkenazi 185delAG-bearing haplotype present in 15% of all Ashkenazi individuals.
In the present study, three Malaysian non-Jewish 185delAG*BRCA1 mutation carriers were also genotyped. Their haplotype seemed to cluster in a small genomic segment, suggesting that it arose independently from that of the Ashkenazi and other Jewish individuals. Yet, the small number of analyzed individuals precludes the ability to assess the precise age of the mutation. The so-called Yorkshire haplotype18 also seems to have arisen independently of the Jewish mutation carriers. These data may be interpreted to indicate that this is a mutational hotspot, but this notion remains speculative and should be stated cautiously. Yet some British mutation carriers have a haplotype indistinguishable from that of the Ashkenazi one. Indeed, Jewish presence in the British Isles is well documented since 1070, with earlier presence presumably dating even to Roman times. Jewish migration from Europe into current-day England, was ongoing until the expulsion of Jews from England in 1290 by King Edward I.41 Thus, it seems plausible that similar to the occurrence of the 185delAG* BRCA1 mutation in the Jewish–Iraqi population a few founders also migrated into the UK. Earlier studies reported the existence of the 185delAG *BRCA1 mutation in non-Jewish individuals of diverse ethnicities: Chilean,8 Spanish,9 Spanish Gypsies,10 Indian,11, 12 Pakistani,13 Egyptians,14 East Europeans,15 and other populations in Europe.1 In all cases where the haplotype of non-Jewish mutation carrying individuals was determined,8, 9, 10, 15 the haplotype was reportedly identical with that of Ashkenazim except for a few British families18 the Indian,12 and Pakistani13 mutation carriers. These latter families shared an identical haplotype, distinct from that of Ashkenazim. It is possible that these individuals share the same haplotype with the Malaysian non-Jews genotyped in the current study. However, without actual haplotyping of more families from that area, this remains speculative.
In conclusion, the 185delAG*BRCA1 mutation arose in the Ashkenazi population about 61 generations ago and was later introduced into the Sephardic and Iraqi–Jewish populations, and in non-Jewish individuals it has a different origin and may have arisen at least twice independent of the Jewish origin mutation.
References
Petrij-Bosch A, Peelen T, van Vliet M et al: BRCA1 genomic deletions are major founder mutations in Dutch breast cancer patients. Nat Genet 1997; 17: 341–345.
Thorlacius S, Struewing JP, Hartge P et al: Population-based study of risk of breast cancer in carriers of BRCA2 mutation. Lancet 1998; 352: 1337–1339.
Górski B, Cybulski C, Huzarski T et al: Breast cancer predisposing alleles in Poland. Breast Cancer Res Treat 2005; 92: 19–24.
Sokolenko AP, Mitiushkina NV, Buslov KG et al: High frequency of BRCA1 5382insC mutation in Russian breast cancer patients. Eur J Cancer 2006; 42: 1380–1384.
Heimdal K, Maehle L, Apold J, Pedersen JC, Møller P : The Norwegian founder mutations in BRCA1: high penetrance confirmed in an incident cancer series and differences observed in the risk of ovarian cancer. Eur J Cancer 2003; 39: 2205–2213.
Abeliovich D, Kaduri L, Lerer I et al: The founder mutations 185delAG and 5382insC in BRCA1 and 6174delT in BRCA2 appear in 60% of ovarian cancer and 30% of early-onset breast cancer patients among Ashkenazi women. Am J Hum Genet 1997; 60: 505–514.
Bar-Sade RB, Kruglikova A, Modan B et al: The 185delAG BRCA1 mutation originated before the dispersion of Jews in the diaspora and is not limited to Ashkenazim. Hum Mol Genet 1998; 7: 801–805.
Ah Mew N, Hamel N, Galvez M, Al-Saffar M, Foulkes WD : Haplotype analysis of a BRCA1: 185delAG mutation in a Chilean family supports its Ashkenazi origins. Clin Genet 2002; 62: 151–156.
Díez O, Osorio A, Robledo M et al: Prevalence of BRCA1 and BRCA2 Jewish mutations in Spanish breast cancer patients. Br J Cancer 1999; 79: 1302–1303.
Díez O, Doménech M, Alonso MC et al: Identification of the 185delAG BRCA1 mutation in a Spanish Gypsy population. Hum Genet 1998; 103: 707–708.
Kumar BV, Lakhotia S, Ankathil R, Madhavan J et al: Germline BRCA1 mutation analysis in Indian breast/ovarian cancer families. Cancer Biol Ther 2002; 1: 18–21.
Kadalmani K, Deepa S, Bagavathi S, Anishetty S, Thangaraj K, Gajalakshmi P : Independent origin of 185delAG BRCA1 mutation in an Indian family. Neoplasma 2007; 54: 51–56.
Rashid MU, Zaidi A, Torres D et al: Prevalence of BRCA1 and BRCA2 mutations in Pakistani breast and ovarian cancer patients. Int J Cancer 2006; 119: 2832–2839.
Ibrahim SS, Hafez EE, Hashishe MM : Presymptomatic breast cancer in Egypt: role of BRCA1 and BRCA2 tumor suppressor genes mutations detection. J Exp Clin Cancer Res 2010; 29: 82–91.
Cierniková S, Tomka M, Kovác M, Stevurková V, Zajac V : Ashkenazi founder BRCA1/BRCA2 mutations in Slovak hereditary breast and/or ovarian cancer families. Neoplasma 2006; 53: 97–102.
Mullineaux LG, Castellano TM, Shaw J et al: Identification of germline 185delAG BRCA1 mutations in non-Jewish Americans of Spanish ancestry from the San Luis Valley, Colorado. Cancer 2003; 98: 597–602.
Berman DB, Wagner-Costalas J, Schultz DC, Lynch HT, Daly M, Godwin AK : Two distinct origins of a common BRCA1 mutation in breast-ovarian cancer families: a genetic study of 15 185delAG-mutation kindreds. Am J Hum Genet 1996; 58: 1166–1176.
Neuhausen SL, Mazoyer S, Friedman L et al: Haplotype and phenotype analysis of six recurrent BRCA1 mutations in 61families: results of an international study. Am J Hum Genet 1996; 58: 271–280.
Lynch HT, Watson P, Tinley S et al: An update on DNA-based BRCA1/BRCA2 genetic counseling in hereditary breast cancer. Cancer Genet Cytogenet 1999; 109: 91–98.
Hordes SM : To the End of The Earth: A History of the Crypto-Jews of New Mexico. Columbia University Press: New York, 2005.
Duncan PR, Lin JT : Ingredients for success: a familial cancer clinic in an oncology practice setting. J Oncol Practice 2011; 7: 39–42.
Makriyianni I, Hamel N, Ward S, Foulkes WD, Graw S : BRCA1:185delAG found in the San Luis Valley probably originated in a Jewish founder. J Med Genet 2005; 42: e27–e29.
Weitzel JN, Lagos V, Blazer KR et al: Prevalence of BRCA mutations and founder effect in high-risk Hispanic families. Cancer Epidemiol Biomarkers Prev 2005; 14: 1666–1671.
Thirthagiri E, Lee SY, Kang P et al: Evaluation of BRCA1 and BRCA2 mutations and risk-prediction models in a typical Asian country (Malaysia) with a relatively low incidence of breast cancer. Breast Cancer Res 2008; 10: R59–R71.
Evans DG, Shenton A, Woodward E, Lalloo F, Howell A, Maher ER : Penetrance estimates for BRCA1 and BRCA2 based on genetic testing in a Clinical Cancer Genetics service setting. BMC Cancer 2008; 8: 155–163.
Shiri-Sverdlov R, Oefner P, Green L et al: Mutational analyses of BRCA1 and BRCA2 in Ashkenazi and non-Ashkenazi Jewish women with familial breast and ovarian cancer. Hum Mutat 2000; 16: 491–501.
Evans DG, Lalloo F, Cramer A et al: Addition of pathology and biomarker information significantly improves the performance of the Manchester scoring system for BRCA1 and BRCA2 testing. J Med Genet 2009; 46: 811–817.
Neuhausen SL, Godwin AK, Gershoni-Baruch R et al: Haplotype and phenotype analysis of nine recurrent BRCA2 mutations in 111 families: results of an international study. Am J Hum Genet 1998; 62: 1381–1388.
Hamel N, Feng BJ, Foretova L et al: On the origin and diffusion of BRCA1 c.5266dupC (5382insC) in European populations. Eur J Hum Genet 2011; 19: 300–306.
Kong A, Gudbjartsson DF, Sainz J et al: A high-resolution recombination map of the human genome. Nat Genet 2002; 31: 241–247.
Liu X, Barker DF : Evidence for effective suppression of recombination in the chromosome 17q21 segment spanning RNU2-BRCA1. Am J Hum Genet 1999; 64: 1427–1439.
Weber JL, Wong C : Mutation of human short tandem repeats. Hum Mol Genet 1993; 2: 1123–1128.
Nachman MW, Crowell SL : Estimate of the mutation rate per nucleotide in humans. Genetics 2000; 156: 297–304.
Atzmon G, Hao L, Pe'er I et al: Abraham's children in the genome era: major Jewish diaspora populations comprise distinct genetic clusters with shared Middle Eastern Ancestry. Am J Hum Genet 2010; 86: 850–859.
Ostrer H : A genetic profile of contemporary Jewish populations. Nat Rev Genet 2001; 2: 891–898.
Prinz J : The Secret Jews. Random House: New York, 1973.
Adams SM, Bosch E, Balaresque PL et al: The genetic legacy of religious diversity and intolerance: paternal lineages of Christians, Jews, and Muslims in the Iberian Peninsula. Am J Hum Genet 2008; 83: 725–736.
Velez C, Palamara PF, Guevara-Aguirre J et al: The impact of converso jews on the genomes of modern Latin Americans. Hum Genet 2012; 131: 251–263.
Im KM, Kirchhoff T, Wang X et al: Haplotype structure in Ashkenazi Jewish BRCA1 and BRCA2 mutation carriers. Hum Genet 2011; 130: 685–699.
Pereira LHM, Pineda MA, Rowe WH et al: The BRCA1 Ashkenazi founder mutations occur on common haplotypes and are not highly correlated with anonymous single nucleotide polymorphisms likely to be used in genome-wide case–control association studies. BMC Genet 2007; 8: 68–82.
Skinner P (ed): Jews in Medieval Britain. The Boydell Press: Woodbridge, 2003.
Acknowledgements
The work at City of Hope was supported in part by American Cancer Society Award #RSGT-00–263–01, and awards #1R03CA139588 and #1RC4CA153828 from the United States National Cancer Institute (PI: JNW). AL was supported by an award from Genesis UK. DGE and WGN are supported by the Manchester NIHR BRC. The Malaysian Breast cancer Genetic Study (MyBrCa) is supported by grants from the Malaysian Ministry of Science, Technology and Innovation, the Ministry of Higher Education (HIR Grant UM.C/HlR/MOHE/06) and charitable funding through Cancer Research Initiatives Foundation (CARIF).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest
Additional information
Supplementary Information accompanies the paper on European Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Laitman, Y., Feng, BJ., Zamir, I. et al. Haplotype analysis of the 185delAG BRCA1 mutation in ethnically diverse populations. Eur J Hum Genet 21, 212–216 (2013). https://doi.org/10.1038/ejhg.2012.124
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2012.124
Keywords
This article is cited by
-
Increased prevalence of the founder BRCA1 c.5309G>T and recurrent BRCA2 c.1310_1313delAAGA mutations in breast cancer families from Northerstern region of Morocco: evidence of geographical specificity and high relevance for genetic counseling
BMC Cancer (2023)
-
Founder vs. non-founder BRCA1/2 pathogenic alleles: the analysis of Belarusian breast and ovarian cancer patients and review of other studies on ethnically homogenous populations
Familial Cancer (2023)
-
Analysis of pathogenic variants in BRCA1 and BRCA2 genes using next-generation sequencing in women with triple negative breast cancer from South India
Molecular Biology Reports (2022)
-
DNA damage repair system in C57BL/6 J mice is evolutionarily stable
BMC Genomics (2021)
-
Age at diagnosis of cancer in 185delAG BRCA1 mutation carriers of diverse ethnicities: tentative evidence for modifier factors
Familial Cancer (2021)