Abstract
Endocardial fibroelastosis (EFE) refers to the thickening of the ventricular endocardium as a result of de novo deposition of subendocardial fibrous tissue layers during neonatal heart development. The origin of EFE fibroblasts is proposed to be postnatal endocardial cells that undergo an aberrant endothelial-to-mesenchymal transition (EndMT). Genetic lineage tracing of endocardial cells with the inducible endocardial Cre line Npr3-CreER and the endothelial cell tracing line Cdh5-CreER on an EFE-like model did not reveal any contribution of neonatal endocardial cells to fibroblasts in the EFE-like tissues. Instead, lineage tracing of embryonic epicardium by Wt1-CreER suggested that epicardium-derived mesenchymal cells (MCs) served as the major source of EFE fibroblasts. By labeling MCs using Sox9-CreER, we confirmed that MCs of the embryonic heart expand and contribute to the majority of neonatal EFE fibroblasts. During this pathological process, TGFβ signaling, the key mediator of fibroblasts activation, was highly upregulated in the EFE-like tissues. Targeting TGFβ signaling by administration of its antagonist bone morphogenetic protein 7 effectively reduced fibroblast accumulation and tissue fibrosis in the EFE-like model. Our study provides genetic evidence that excessive fibroblasts in the EFE-like tissues mainly originate from the epicardium-derived MCs through epicardial to mesenchymal transition (EpiMT). These EpiMT-derived fibroblasts within the EFE-like tissues could serve as a potential therapeutic target.
Similar content being viewed by others
Introduction
Endocardial fibroelastosis (EFE) refers to the pathological condition that often causes sudden death in infants and children1,2,3,4. EFE is characterized by diffuse profound thickening of the endocardium with abnormal deposition of collagen and elastin predominantly in the left ventricle2,3,5,6,7,8. The accumulated collagen and elastin form a de novo subendocardial fibrous tissue layer, which encapsulates the underlying myocardium. It is thought that EFE limits ventricular diastolic compliance and restricts ventricular growth9,10. EFE is correlated to a variety of diseases such as hypoplastic left heart syndrome (HLHS) and bacterial myocarditis. It has been reported that fetal intervention and postnatal surgical removal of EFE tissues could renew left ventricular growth and restore biventricular physiology in some cases of HLHS11,12. Nevertheless, effective measures are still needed to inhibit formation of EFE tissues. Therefore, understanding the pathophysiological progression of EFE would help provide new insights into the search for the potential therapeutic targets in treatment of EFE.
The main cellular population of EFE tissues is fibroblasts that generates extracellular matrix. Excessive deposition of fibroblasts and extracellular matrix leads to development of fibrosis, which is one of the main pathological features of EFE. To date, little is known about the cellular origins of EFE. It would therefore be helpful to unravel the cellular origins of EFE fibroblasts in order to delineate the pathogenesis of EFE and thereby design potential therapeutic strategies. As its name suggests, EFE is largely restricted to the endocardial tissue. It has been reported that the origin of EFE fibroblasts lies in the endocardial to mesenchymal transition13. Moreover, the ability to surgically remove the thickened fibroelastic layer without myocardial contamination suggests, but does not prove, that the fibrotic tissue might be derived from endocardium12.
During murine heart development, at approximately embryonic day (E)12.5, epicardial cells migrate into the myocardium and give rise to mesenchymal cells (MCs) via the epithelial-to-mesenchymal transition (EpiMT). These epicardial-derived mesenchymal cells (Epi-MCs) then contribute to several lineages of the heart, including fibroblasts, pericytes and smooth muscle cells (SMCs)14,15,16,17,18. In addition, at E9.5, endocardial cells undergo endothelial-to-mesenchymal transition (EndMT) and populate the endocardial cushions with MCs. Recent studies have suggested that these endocardial-derived mesenchymal cells (Endo-MCs) could migrate into the myocardium and differentiate into fibroblasts, pericytes and SMCs19,20,21. These studies suggest that both Epi-MCs and Endo-MCs could contribute to cardiac fibroblasts during heart development. In normal adult hearts, lineage tracing experiments with the Wt1-Cre line has shown that over 90% fibroblasts in the ventricular walls and 30% fibroblasts in the ventricular septum are derived from the embryonic epicardium19. Moreover, fate mapping of endothelium using the Tie2-Cre and Ve-cadherin (Cdh5)-CreER lines has demonstrated a contribution of embryonic endocardial-derived fibroblasts complementary to those of the epicardium. Embryonic endocardium contributes about 12% fibroblasts in the left ventricular walls and 64% fibroblasts in the ventricular septum19. Therefore, fibroblasts of normal adult hearts are mainly derived from the embryonic Epi-MCs with a minor contribution from the embryonic Endo-MCs19.
Following cardiac injury, it has been reported that the endothelium and hematopoietic cells are origins of cardiac fibroblasts in various fibrosis models including ascending aorta constriction, chronic allograft rejection, myocardial infarction, diabetes and the angiotensin-induced cardiac fibrosis model22,23,24,25,26. Notably, inhibition of EndMT during fibrosis reduces fibroblast deposition and facilitates heart repair22. Fibrotic EndMT and circulating fibroblast progenitors represent two potential therapeutic targets for fibrosis27. However, recent studies argued that new fibroblasts in injured heart are mainly derived from resident fibroblasts, which originate from embryonic Epi-MCs and Endo-MCs19,21. Lineage tracing studies for adult endothelium (Ve-cadherin-CreER), adult epicardium (Wt1-CreER and Tbx18-CreER) or hematopoietic cells (Vav-Cre) have not provided convincing evidence for their contribution to fibroblasts following myocardial stress19. Moreover, neither transplantation studies nor parabiosis experiments support the notion that new cardiac fibroblasts are derived from bone marrow or circulating cells21.
EFE is a specialized form of fibrosis that involves excessive fibroblast deposition in the thickened endocardial layer. Here, by virtue of multiple independent Cre knock-in mouse lines, we confirmed that postnatal endothelium, including both endocardium and coronary endothelium, did not contribute to fibrotic fibroblasts in mouse EFE-like models. Instead, embryonic MCs labeled by Sox9-CreER gave rise to the majority of EFE fibroblasts. Specifically, Epi-MCs but not Endo-MCs were the major sources of fibroblasts within the EFE-like tissues. We also detected high expression of TGFβ in the EFE-like tissues, and treatment of mice with bone morphogenetic protein 7 (BMP7), an antagonist for TGFβ signaling, effectively reduces fibroblast accumulation and tissue fibrosis. Our results suggest that therapeutic strategies in reducing pathogenic EFE fibroblasts should shift from the focus of targeting the postnatal EndMT-derived fibroblasts to pathways regulating development of the embryonic EpiMT-derived fibroblasts.
Results
Mouse EFE-like model of unloaded heterotopically transplanted hearts
The pathogenesis of human EFE remains obscure. It often occurs in fetal or neonatal hearts with hypoplastic left heart syndrome (HLHS), which may result from aortic stenosis and left ventricular dysfunction. From clinical observations, the presence of EFE has a strong relationship with the decreased left ventricular blood flow. To mimic the conditions of human fetal/neonatal hearts with EFE, an EFE-like model with a nonworking hemodynamically unloaded heart has been introduced to study EFE for years13,28. In this established model, reduced ventricular and intracavitary blood flow are found, recapitulating the conditions in human fetal hearts with HLHS when the aortic valve closes. Moreover, fibrosis in the EFE-like model may result from ischemia due to reduced blood flow in the hemodynamically unloaded heart.
To investigate the origin of fibroblasts within EFE-like tissues, we first developed the EFE-like mouse model, in which neonatal hearts were heterotopically transplanted into the abdomen of adult mice, as previously described28,29. Such surgery procedures on immature donor hearts could model the pathogenesis of human EFE, which is associated with reduced blood flow at the neonatal stage. Briefly, postnatal day (P)7 mouse heart was collected and implanted as a heterotopic infra-renal graft, where the ascending aorta was surgically linked to the infra-renal aorta and the pulmonary artery was linked to the inferior vena cava anastomoses28,29. The donor hearts were then analyzed at day 12 following transplantation (Figure 1A and 1B). Human EFE has the gross appearance of subendocardial fibrosis; thus we examined the morphology and degree of fibrosis of the heterotopic EFE-like heart transplants by H&E staining, sirius red staining and Collagen staining, respectively. Compared with the native hearts, excessive extracellular matrix was deposited in the subendocardial layers of left ventricles of the heterotopic heart transplants (Figure 1C and Supplementary information, Figure S1A-S1D). To confirm fibrous tissue deposition in the subendocardial layers of the heterotopic heart transplants, we stained heart sections for the fibroblast marker platelet-derived growth factor receptor alpha (PDGFRa); and found that deposition of PDGFRa+ fibroblasts was significantly increased in heterotopic heart transplants (Supplementary information, Figure S1E). We also stained heart sections for an additional, widely used yet less specific fibroblast marker fibroblast-specific protein-1 (FSP1)13, which also labels some endothelial cells and leukocytes30. Similarly, we observed a significant increase in FSP1+ cells in the heterotopic heart transplants compared with the native hearts (Supplementary information, Figure S1F). Taken together, our results showed a successful mouse model recapitulating pathogenesis of EFE via heterotopic heart transplantation.
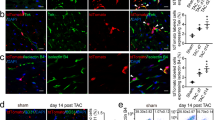
Postnatal endocardial and vascular endothelial cells do not contribute to fibroblasts in the EFE-like tissues. (A) Schematic showing experimental strategy. Donor heart from postnatal day (P)7 mouse was implanted into the peritoneal cavity of 8-week-old mouse. Donor hearts were analyzed at day 12 following transplantation. (B) Cartoon showing the heterotopic heart transplant model. The ascending aorta was anastomosed to the infrarenal aorta (Ao), and the pulmonary artery (PA) was anastomosed to the infrarenal vena cava (IVC). (C) Sirius red staining on heart sections shows EFE-like tissues within the inner layer of LV. (D) Schematic figure showing the experimental strategy for lineage tracing of endocardial cells using Npr3-CreER;R26-tdTomato mice. Tamoxifen was administered 2 days before heart transplantation. (E) Immunostaining for tdTomato, PDGFRa and CDH5 on donor heart sections. (F) Immunostaining for tdTomato, FSP1 and CDH5 on donor heart sections shows that endocardial cells do not contribute to FSP1+CDH5− fibroblasts in the EFE-like tissues. Boxed regions are magnified in the right panels. (G) Cartoon showing no lineage conversion from endocardial cells to fibroblasts. (H) Schematic showing the experimental strategy for lineage tracing of vascular endothelial cells using Cdh5-CreER;R26-tdTomato mice. (I) Immunostaining for tdTomato and PDGFRa on donor heart sections. (J) Immunostaining for tdTomato, FSP1 and CDH5 on donor heart sections shows that vascular endothelial cells (ECs) do not contribute to FSP1+CDH5− fibroblasts in the EFE-like tissues. Boxed regions are magnified in the right panels. (K) Cartoon showing no lineage conversion from vascular ECs to fibroblasts. Scale bars, 400 μm in C; 100 μm in E, F, I, J. LV, left ventricle; RV, right ventricle.
Lineage tracing of postnatal endothelium shows no evidence for EndMT in EFE-like tissues
Endothelial cells in the heart include both the endocardial and coronary endothelial cells. We generated the Npr3-CreER mouse line that specifically marked the endocardium at embryonic stages31,32. To test if Npr3-CreER labels postnatal endocardial endothelial cells, we crossed Npr3-CreER with the reporter line Rosa26-loxp-stop-loxp-tdTomato (R26-tdTomato)33 and treated Npr3-CreER;R26-tdTomato pups at postnatal day (P) 5 with tamoxifen. The hearts were then collected at 48 h post treatment. Immunostaining for the tracing reporter tdTomato and the coronary endothelial marker FABP4 on heart sections showed that Npr3-CreER labeled endocardial cells but not coronary endothelial cells (Supplementary information, Figure S2A). In addition to the endocardium, we found that Npr3-CreER also labeled a subset of epicardial cells (Supplementary information, Figure S2B).
A previous study suggests that fibroblasts in the EFE-like tissues are mainly derived from aberrant EndMT13. To verify if the endocardium contributes to fibroblasts within EFE-like tissues, we administered a neonatal Npr3-CreER;R26-tdTomato mouse with tamoxifen at P5, and implanted the P7 heart into the abdomen of an adult wild-type mouse. At day 12 following implantation, the donor heart was harvested and analyzed (Figure 1D). Immunostaining for tdTomato, fibroblast marker PDGFRa and endothelial cell-specific marker CDH5 showed that Npr3-CreER-labeled endocardial cells did not contribute to any fibroblast within the EFE-like tissues (Figure 1E). Immunostaining for tdTomato, CDH5 and an additional fibroblast marker, FSP1, confirmed that endocardial cells did not contribute to any FSP1+CDH5− fibroblast in the EFE-like tissues (Figure 1F). We also traced the neonatal endocardium in native Npr3-CreER;R26-tdTomato hearts following tamoxifen induction at P5 and heart collection at P19. In normal hearts, Npr3-CreER labeled both endocardial and epicardial cells at P19, but not vascular endothelial cells (Supplementary information, Figure S2C and S2D). In P19 mouse hearts, neither PDGFRa+ nor FSP1+ cells were derived from endocardial cells (Supplementary information, Figure S2E and S2F). Collectively, these data demonstrated that neonatal endocardial cells did not contribute to fibroblasts in either EFE-like or normal hearts (Figure 1G). These data also proved that Npr3-CreER-labeled neonatal epicardium did not contribute to fibroblasts in either EFE-like or normal hearts.
We next asked if vascular endothelial cells contribute to new fibroblasts through EndMT22. To this end, we employed Cdh5-CreER;R26-tdTomato to address vascular EndMT in EFE-like tissues, as it has been shown that Cdh5-CreER labels most coronary vascular endothelial cells34. Tamoxifen was administered 2 days before the Cdh5-CreER;R26-tdTomato heart was transplanted to the recipient mouse. The donor heart was harvested and analyzed at day 12 following transplantation (Figure 1H). Immunostaining for tdTomato and PDGFRa or FSP1 showed that Cdh5-CreER-labeled endothelial cells did not contribute to PDGFRa+ or FSP1+CDH5− cells (Figure 1I and 1J). Taken together, our lineage tracing data from Npr3-CreER and Cdh5-CreER lines demonstrated that endocardial and vascular endothelial cells did not contribute to fibroblasts within the EFE-like tissues (Figure 1G and 1K), suggesting that EndMT is unlikely to be the major mechanism for development of EFE fibroblasts.
Embryonic cardiac MCs are the major source of EFE fibroblasts
Given that some studies have demonstrated that fibroblasts deposited following aortic banding are derived solely from resident fibroblasts, which originate from embryonic Epi-MCs and embryonic Endo-MCs19,21, we hypothesized that the fibroblasts within EFE-like tissues were derived from preexisting cardiac MCs within the myocardium before birth. It has been reported that the transcription factor SRY-type box (Sox) 9, which plays critical roles in cardiac valve formation and epicardial migration at embryonic stages, is highly expressed in epicardial cells, Epi-MCs and Endo-MCs35,36,37. Thus we employed the Sox9-CreER mouse line to investigate if cardiac MCs contribute to EFE fibroblasts.
We first collected Sox9-CreER embryos from E9.5 to E11.5 and examined the expression of the estrogen receptor (ESR), which served as a surrogate for Sox9 expression. Immunostaining for ESR and CDH5 on E9.5 Sox9-CreER embryonic sections indicated that expression of ESR/Sox9 was mainly restricted to mesenchymal cells in the atrioventricular canal cushion, a subset of proepicardial cells and epicardial cells, but not in endocardial cells of the ventricular walls (Figure 2A and 2B). Co-staining for ESR and the (pro)epicardial marker TBX18 confirmed expression of ESR/Sox9 by proepicardial cells and epicardial cells (Figure 2A and 2C). At E10.5 and E11.5, immunostaining for ESR and CDH5 or TBX18 revealed similar ESR expression patterns compared to E9.5. Expression of ESR was detected in mesenchymal cells of the atrioventricular canal cushion and epicardial cells, but not in endocardial cells of the ventricular walls (Figure 2A and Supplementary information, Figure S3A-S3D). To further confirm that Sox9 was expressed in EndMT-derived cushion MCs, we collected embryos of Npr3-CreER;R26-tdTomato at E10.5, which had been treated with tamoxifen at E8.5, and stained for tdTomato, Sox9 and CDH5. The immunostaining data showed co-localization of Sox9 and tdTomato in the atrioventricular cushion (Supplementary information, Figure S3E), indicating that Sox9 was expressed in endocardial-derived MCs.
Sox9 labels mesenchymal cells at early embryonic stages. (A) Cartoon showing the expression map of Sox9 at E9.5 to E11.5. Blue coloring indicates Sox9+ cells including endocardial cushion mesenchymal cells, proepicardial and epicardial cells. (B) Immunostaining for CDH5 and ESR, where ESR is a surrogate of Sox9, on E9.5 Sox9-CreER embryonic sections shows that ESR/Sox9 is expressed in cushion mesenchymal cells and proepicardial cells, but not in ventricular endocardial cells at E9.5. B1 and B2 are magnified images of boxed regions in B. (C) Immunostaining for ESR and TBX18 shows that ESR is detected in proepicardial cells and epicardial cells at E9.5. White arrowheads indicate ESR+ proepicardial cell and yellow arrowheads indicate ESR+ epicardial cells. (D) Lineage tracing strategy using Sox9-CreER;R26-tdTomato embryos. Tamoxifen was administered at E9.5 and embryos were collected for analysis at E14.5. (E) Immunostaining for tdTomato and PDGFRb on heart sections of E14.5 Sox9-CreER;R26-tdTomato embryos. E1 and E2 are magnified images of boxed regions in E. (F) Immunostaining for tdTomato and PDGFRa on heart sections of Sox9-CreER;R26-tdTomato mice at E14.5 confirms that tdTomato labels epicardial-derived mesenchymal cells. a, atrium; AVC, atrioventricular canal; Epi, epicardium; PE, proepicardium; v, ventricle. Scale bar, 100 μm.
Having validated expression of CreER/Sox9 in the early embryonic heart, we utilized Sox9-CreER for genetic lineage tracing of MCs in the EFE-like model. We crossed the Sox9-CreER mouse strain with the reporter line R26-tdTomato to examine the labeling efficiency of MCs by Sox9-CreER. Tamoxifen was administered at E9.5 and hearts were harvested at E14.5 (Figure 2D). Immunostaining for tdTomato and the endothelial maker PECAM on E14.5 heart sections showed that tdTomato was robustly detected in the endocardial cushion, epicardium and subepicardial regions (Supplementary information, Figure S4B). Some of these tdTomato+ cells were located in close proximity to PECAM+ endothelial cells, but they were rarely endothelial cells (0.63% ± 0.23%, n = 4; Supplementary information, Figure S4B). Instead, immunostaining for the epicardial marker TBX18 showed that a high percentage of epicardial cells were tdTomato+ cells (95.42% ± 1.51%, n = 4; Supplementary information, Figure S4C). We also performed co-staining for tdTomato and the embryonic cardiac mesenchymal cell markers PDGFRa or platelet derived growth factor receptor beta (PDGFRb); and found that Sox9-CreER efficiently labeled cushion MCs and epicardial-derived MCs (Figure 2E and 2F). Taken together, Sox9-CreER labeled cardiac MCs that included both Endo-MCs and Epi-MCs at early-to-mid embryonic stages.
We next followed the cell fate of Sox9-CreER-labeled cardiac MCs to the postnatal stage (Figure 3A). We collected normal hearts at P19 and stained heart sections for PDGFRa, PDGFRb and alpha smooth muscle actin (aSMA), which are markers for fibroblasts, pericytes and SMCs, respectively, at this stage. We found that tdTomato+ cells constituted 91.18% ± 1.32% of PDGFRa+ fibroblasts, 93.60% ± 1.39% of PDGFRb+ pericytes, and 96.86% ± 1.74% of aSMA+ SMCs (n = 4; Figure 3B-3E). These data suggested that Sox9-CreER-labeled cardiac MCs contributed to the majority of fibroblasts, pericytes and SMCs in normal heart during postnatal homeostasis.
Sox9-CreER labeled embryonic mesenchymal cells contribute to fibroblasts, pericytes and smooth muscle cells in the native hearts. (A) Schematic showing the strategy for lineage tracing of Sox9+ cells using Sox9-CreER;R26-tdTomato mice. (B) Quantification of the percentage of labeled cells by Sox9-CreER at P19. n = 4. (C) Immunostaining for tdTomato and fibroblast marker PDGFRa shows that Sox9-CreER labels most PDGFRa+ fibroblasts at P19. Arrowheads indicate tdTomato+PDGFRa+ fibroblasts. Boxed region is magnified in the right panels. (D) Immunostaining for tdTomato and pericyte marker PDGFRb shows that Sox9-CreER labels most PDGFRb+ pericytes at P19. Arrowheads indicate tdTomato+PDGFRb+ pericytes. (E) Immunostaining for tdTomato and aSMA shows that Sox9-CreER labels most aSMA+ smooth muscle cells at P19. Arrowheads indicate tdTomato+aSMA+ smooth muscle cells. Scale bar, 100 μm. LV, left ventricle.
As hematopoietic cells have been reported as potential origins of fibroblasts23,24,26, we therefore examined whether Sox9-CreER labels resident myeloid cells or circulating hematopoietic cells. First we collected P7 hearts from Sox9-CreER;R26-tdTomato mice, which were administered with tamoxifen at E9.5. Immunostaining for tdTomato along with hematopoietic marker CD45 or macrophage marker F4/80 and CD11b suggested that Sox9-CreER did not label resident myeloid cells in hearts (Supplementary information, Figure S5A-S5C). Next, we collected the blood cells of Sox9-CreER;R26-tdTomato mice, and found that there were very few tdTomato+ hematopoietic cells (Supplementary information, Figure S5D-S5E). Collectively, Sox9-CreER rarely labeled hematopoietic cells.
To determine the contribution of cardiac MCs to EFE fibroblasts, we developed an EFE-like model using Sox9-CreER;R26-tdTomato donor hearts. Similar to the native heart study, tamoxifen was administered at E9.5 to label MCs in donor hearts. Donor hearts were collected and implanted in recipient mice at P7. At day 12 following transplantation, the donor hearts were harvested and analyzed (Figure 4A). Immunostaining for tdTomato and PDGFRa showed that the majority of PDGFRa+ fibroblasts within the EFE-like tissues were tdTomato+ (88.68% ± 0.98%; n = 5; Figure 4B-4D). We also carried out flow cytometry analysis to study the percentage of tdTomato+ fibroblasts in the EFE-like model, and found that 84.85% ± 0.98% of PDGFRa+ fibroblasts were labeled by tdTomato (Figure 4E-4G). To further confirm if MCs give rise to fibroblasts, we also co-stained for an additional fibroblast marker FSP1, and detected tdTomato+FSP1+CDH5− fibroblasts within the EFE-like tissues (Figure 4H). Our results suggested that most EFE fibroblasts were derived from Sox9-CreER-labeled embryonic MCs.
Embryonic cardiac mesenchymal cells (MCs) contribute to the majority of fibroblasts in the EFE-like tissues. (A) Schematic showing strategy for labeling embryonic cardiac MCs using Sox9-CreER;R26-tdTomato mice. Tamoxifen was administered at E9.5 and hearts were collected and transplanted into recipients at P7. The donor hearts were collected for analysis at day 12 following transplantation. (B, C) Immunostaining for tdTomato and PDGFRa shows that tdTomato labels PDGFRa+ fibroblasts (arrowheads) in the EFE-like tissues. Boxed region in B is magnified in C. (D) Quantification of the percentage of tdTomato+PDGFRa+ fibroblasts within the EFE-like tissues (n = 5). (E, F) Representative flow cytometric analysis of the percentage of tdTomato labeled PDGFRa+ cells within the EFE-like hearts. (G) Quantification results of tdTomato labeled PDGFRa+ cells within the EFE-like models by flow cytometric analysis. For each group, n = 4. (H) Immunostaining for tdTomato, CDH5 and FSP1 shows that tdTomato labels FSP1+CDH5− cells (arrowheads) within the EFE-like tissues. Scale bars, 100 μm. LV, left ventricle.
Embryonic endocardium minimally contributes to the EFE fibroblasts
As cardiac MCs include Endo-MCs and Epi-MCs, we asked to what extent these MCs from each respective compartment contribute to fibroblasts within EFE-like tissues. To address this question, we employed endocardial- and epicardial-specific inducible Cre lines for fate mapping analysis on their descendants in EFE-like models.
Cardiac endocardium represents a unique population of endothelial cells that expresses NFATC1 at early embryonic stages38,39,40. For tracing the descendants of embryonic endocardium including Endo-MCs, we generated a tamoxifen-inducible Nfatc1-2A-CreER knock-in line (Supplementary information, Figure S6A). First, we analyzed expression of ESR, which is a surrogate for NFATC1 expression. Whole-mount staining showed that ESR was expressed in the endocardium of Nfatc1-2A-CreER embryos at E8.5 (Supplementary information, Figure S6B). Immunostaining for ESR and CDH5 revealed that ESR expression was specific to the endocardial cells at E9.5 (Supplementary information, Figure S6C). Next, to determine the Endo-MCs labeling by Nfatc1-2A-CreER, we performed lineage tracing experiments by crossing Nfatc1-2A-CreER with the reporter R26-tdTomato line. Tamoxifen was administered at E8.5 and embryos were collected at E10.5 (Supplementary information, Figure S6D). Co-staining for tdTomato and the endothelial cell marker PECAM on E10.5 embryonic sections showed that Nfatc1-2A-CreER labeled endocardial cells and endocardial-derived cushion mesenchymal cells (Supplementary information, Figure S6E). We also followed the fate of Nfatc1-derived cells to the postnatal stage, and collected Nfatc1-2A-CreER;R26-tdTomato hearts at P19 (Supplementary information, Figure S6D). Immunostaining for tdTomato and the fibroblast marker PDGFRa on heart sections showed that a subset of tdTomato+ cells were PDGFRa+ and these were distributed within atrioventricular valves, the ventricular septum and the subendocardial myocardium (Supplementary information, Figure S6F-S6H). Our results are consistent with a recent study of the endocardial origin of cardiac fibroblasts19.
To examine the endocardial contribution to EFE fibroblasts, we developed EFE-like models using Nfatc1-2A-CreER;R26-tdTomato heart transplantation (Figure 5A). On the EFE heart sections, we co-stained tdTomato and PDGFRa, and identified tdTomato+PDGFRa+ fibroblasts in the subendocardial EFE-like tissues (Figure 5B). Quantification of the percentage of labeled fibroblasts within the EFE-like tissues showed that 5.96% ± 0.74% of fibroblasts were derived from the Nfatc1-2A-CreER-labeled embryonic endocardium (Figure 5C). Immunostaining for FSP1 and CDH5 also identified endocardial-derived tdTomato+FSP1+CDH5− cells (Figure 5D). We also carried out flow cytometric analysis to study the percentage of tdTomato+ fibroblasts within the EFE-like model, and found that about 8.20% ± 0.41% of PDGFRa+ fibroblasts were labeled by tdTomato (Figure 5E-5G). To test how endocardial-derived fibroblasts respond to injury, we collected donor hearts at day 3 post transplantation, and stained for tdTomato, PDGFRa and proliferation marker Ki67 (Figure 5H). Expression of Ki67 in tdTomato+ fibroblasts suggested that EndMT-derived fibroblasts proliferated resulting in excessive fibrosis in EFE.
Nfatc1-2A-CreER-labeled embryonic endocardial cells contribute to fibroblasts in the EFE-like tissues. (A) Schematic showing strategy for lineage tracing of embryonic endocardial cells in an EFE-like model using Nfatc1-2A-CreER;R26-tdTomato mice. (B) Immunostaining for tdTomato and PDGFRa on heart sections shows that a subset of PDGFRa+ fibroblasts was tdTomato+ (arrowheads) in the EFE-like tissues. (C) Quantification of the percentage of tdTomato+PDGFERa+ fibroblasts within the EFE-like tissues. n = 5. (D) Immunostaining for tdTomato and FSP1 on heart sections shows that a subset of FSP1+ cells were tdTomato+ in the EFE-like tissues. Arrowheads indicate tdTomato+FSP1+CDH5− cells. (E, F) Representative flow cytometric analysis of the percentage of tdTomato+ fibroblasts in the EFE-like model. (G) Quantification results of tdTomato-labeled PDGFRa+ cells within the EFE-like model by flow cytometric analysis. For each group, n = 4. (H) Immunostaining for tdTomato, PDGFRa and Ki67 on sections of the EFE-like hearts from Nfatc1-2A-CreER;R26-tdTomato mice. The mice were treated with tamoxifen at E8.5. The donor hearts were transplanted at P7 and collected at day 3 post transplantation. The arrowheads indicate tdTomato+PDGFRa+Ki67+ cells. Scale bars, white, 100 μm; yellow, 25 μm. LV, left ventricle.
We next employed another inducible Cre line Npr3-CreER that specifically labels the embryonic endocardium for analysis in the EFE-like model (Figure 6A). Co-staining of tdTomato and PDGFRa on the donor heart sections of Npr3-CreER;R26-tdTomato showed that tdTomato labeled some PDGFRa+ fibroblasts within the EFE-like tissues (Figure 6B). Quantification of the percentage of tdTomato+ cells in the PDGFRa+ population within the EFE-like tissues showed that 10.41% ± 2.41% of fibroblasts were derived from the Npr3-CreER-labeled embryonic endocardium (Figure 6C). Immunostaining for FSP1 and CDH5 on EFE heart sections of Npr3-CreER;R26-tdTomato also identified a small number of tdTomato+FSP1+CDH5− cells within the EFE-like tissues (Figure 6D). Flow cytometric analysis of the percentage of tdTomato-labeled PDGFRa+ cells in the EFE-like models showed that about 8.12% ± 0.28% fibroblasts were derived from the Npr3-CreER-labeled embryonic endocardium (Figure 6E-6G). Immunostaining for tdTomato, PDGFRa and Ki67 on the sections of Npr3-CreER;R26-tdTomato donor hearts, which were collected at day 3 post transplantation, showed proliferation of embryonic EndMT-derived fibroblasts after injury (Figure 6H). Taken together, our results demonstrated that the embryonic endocardium contributed to EFE fibroblasts, and their contribution (about 6-10%) did not represent a major source of EFE fibroblasts.
Npr3-CreER-labeled embryonic endocardial cells contribute to fibroblasts within the EFE-like tissues. (A) Schematic showing the strategy for lineage tracing of embryonic endocardial cells in the EFE-like model using Npr3-CreER;R26-tdTomato mice. (B) Immunostaining for tdTomato and PDGFRa on heart sections shows that a subset of PDGFRa+ fibroblasts were tdTomato+ (arrowheads) within the EFE-like tissues. (C) Quantification of the percentage of tdTomato+PDGFRa+ fibroblasts in the EFE-like tissues. n = 5. (D) Immunostaining for tdTomato and FSP1 on heart sections shows that a subset of FSP1+ cells were tdTomato+ within the EFE-like tissues. The arrowheads indicate tdTomato+FSP1+CDH5− cells. (E, F) Representative flow cytometric analysis of the percentage of tdTomato+ fibroblasts in the EFE-like model. (G) Quantification results of tdTomato+ fibroblasts within the EFE-like model by flow cytometric analysis. For each group, n = 4. (H) Immunostaining for tdTomato, PDGFRa and Ki67 on sections of the EFE-like hearts from Npr3-CreER;R26-tdTomato mice. The mice were treated with tamoxifen at E8.5. The donor hearts were transplanted at P7 and collected at day 3 post transplantation. The arrowheads indicate tdTomato+PDGFRa+Ki67+ cells. Scale bars, 100 μm. LV, left ventricle.
Embryonic epicardium contributes to the majority of EFE fibroblasts
As embryonic endocardium minimally contributed to EFE fibroblasts, we hypothesized that the embryonic epicardium (Epi-MCs) differentiated into the majority of fibroblasts within EFE-like tissues. To test this, we crossed the epicardial Cre line Wt1-CreER16 with the reporter line R26-tdTomato and examined the lineage conversion of epicardium in the EFE-like model. First, we performed fate mapping of the embryonic epicardium in native hearts. Tamoxifen was administered at E10.5 and the hearts were harvested and analyzed at P19 (Figure 7A). Immunostaining for tdTomato and PDGFRa was performed on heart sections of Wt1-CreER;R26-tdTomato. We found that the majority of PDGFRa+ fibroblasts in the ventricles were tdTomato+ (86.63% ± 2.87%, Figure 7B). These data suggested that the embryonic epicardium was the major source of fibroblasts in native hearts, which is in accordance with previous studies15,37,41. We then developed EFE-like models using Wt1-CreER;R26-tdTomato mice and examined the contribution of embryonic epicardium to EFE fibroblasts (Figure 7C). Co-staining of tdTomato and PDGFRa showed the co-localization of tdTomato and PDGFRa within the EFE-like tissues (Figure 7D). Immunostaining for tdTomato, FSP1 and CDH5 also identified tdTomato+FSP1+CDH5− cells within the subendocardial EFE-like tissues of the left ventricles (Figure 7E and 7F). Quantification of the percentage of tdTomato+PDGFRa+ cells in the EFE-like tissues showed that 65.10% ± 8.55% of fibroblasts were derived from Wt1-CreER-labeled embryonic epicardium and their derivatives (Figure 7G). Flow cytometric analysis showed that 62.58% ± 0.71% of PDGFRa+ cells were labeled by tdTomato (Figure 7H-7J). Immunostaining for tdTomato, PDGFRa and Ki67 on the sections of Wt1-CreER;R26-tdTomato donor hearts, which were collected at day 3 post transplantation, also confirmed proliferation of EpiMT-derived fibroblasts after injury (Figure 7K). Taken together, our results demonstrated that the majority of fibroblasts within EFE-like tissues originated from the epicardium. We also compared the proliferation rates of epicardial- and endocardial-derived fibroblasts at day 3 post transplantation, and did not find a significant difference (Figure 7L), which suggests that both EpiMT- and EndMT-derived fibroblasts responded to the injury and contributed to the fibrosis formation. Indeed, our results are consistent with previous work21.
Embryonic epicardial cells contribute to the fibroblasts within the EFE-like tissues. (A) Schematic showing strategy for lineage tracing of embryonic epicardial cells in native hearts using Wt1-CreER;R26-tdTomato mice. (B) Immunostaining for tdTomato and PDGFRa on heart section shows that the majority of PDGFRa+ fibroblasts are tdTomato+. (C) Schematic showing the strategy for lineage tracing of embryonic epicardial cells in the EFE-like model using Wt1-CreER;R26-tdTomato mice. (D) Immunostaining for tdTomato and PDGFRa on heart sections showed PDGFRa+ fibroblasts were tdTomato+ (arrowheads) in the EFE-like tissues. (E, F) Immunostaining for tdTomato and FSP1 on heart sections. Boxed region in E is magnified in F. Yellow arrowheads indicate tdTomato+FSP1+CDH5− cells. (G) Quantification of the percentage of tdTomato+PDGFRa+ fibroblasts. (H, I) Representative flow cytometric analysis of the percentage of tdTomato+PDGFRa+ fibroblasts in the EFE-like model. (J) Quantification results of tdTomato+PDGFRa+/PDGFRa+ within the EFE-like model by flow cytometric analysis. n = 5. (K) Immunostaining for tdTomato, PDGFRa and Ki67 on sections of the EFE-like hearts from Wt1-CreER;R26-tdTomato mice. The mice were administered with tamoxifen at E10.5. The donor hearts were transplanted at P7 and collected at day 3 post transplantation. The arrowheads indicate tdTomato+PDGFRa+Ki67+ cells. (L) Quantification of the percentage of proliferating fibroblasts from different origins. Scale bars, 100 μm. LV, left ventricle; RV, right ventricle.
Postnatal epicardium does not give rise to EFE fibroblasts
Previous lineage tracing data suggested that Npr3-CreER-labeled postnatal endocardium and epicardium did not contribute to EFE fibroblasts (Figure 1D-1F; Supplementary information, Figure S2A and S2B). To further examine whether postnatal epicardium generates EFE fibroblasts, we traced the fate of postnatal epicardium using Wt1-CreER;R26-tdTomato mice. To test the labeling of Wt1-CreER;R26-tdTomato in native postnatal hearts, tamoxifen was administered at P5 and hearts were harvested at P19 (Figure 8A). Immunostaining for tdTomato and PECAM on heart sections showed that Wt1-CreER labeled epicardial cells and some coronary endothelial cells (Figure 8B). Immunostaining for PDGFRa and tdTomato on heart sections showed that WT1-CreER did not label fibroblasts at the postnatal stages (Figure 8C). Next, we developed the EFE-like model using Wt1-CreER;R26-tdTomato mice. Tamoxifen was administered at P5, and hearts were collected and implanted at P7. At day 12 following transplantation, the donor Wt1-CreER;R26-tdTomato hearts were harvested and analyzed (Figure 8D). Immunostaining for tdTomato, CDH5 and PDGFRa on the EFE heart sections showed that tdTomato+ cells within the EFE-like tissues were CDH5+ but PDGFRa− (Figure 8E and 8F). Our results suggested that the postnatal epicardium did not contribute to EFE fibroblasts.
Postnatal epicardial cells do not contribute to the fibroblasts within the EFE-like tissues. (A) Schematic figure showing the strategy for lineage tracing of epicardial cells in native hearts at postnatal stage using Wt1-CreER;R26-tdTomato mice. (B) Immunostaining for tdTomato and PECAM on heart sections shows that postnatal Wt1-CreER labels coronary endothelial cells and epicardial cells. (C) Immunostaining for tdTomato and PDGFRa shows that Wt1-CreER does not label PDGFRa+ fibroblasts at postnatal stage. (D) Schematic figure showing the lineage tracing strategy of postnatal epicardial cells in the EFE-like model using Wt1-CreER;R26-tdTomato mice. (E,F) Immunostaining for tdTomato and PDGFRa on heart sections shows that Wt1-CreER labeled cells (including epicardial cells and some endothelial cells) do not contribute to PDGFRa+ fibroblasts within the EFE-like tissues. Boxed region in E is shown in F. Scale bar, 100 μm. epi, epicardium; LV, left ventricle.
Blocking the TGFβ signaling pathway effectively reduces fibroblast accumulation in EFE-like model
The TGFβ pathway has been reported to be a key mediator for cardiac fibroblast activation and shown to play important roles in cardiac fibrosis42,43. Previous gain- and loss-of-function studies have demonstrated the pivotal role of TGFβ in inducing fibrosis44,45,46,47. Similarly, inhibition of TGFβ has been shown to ameliorate fibrosis43,48. To test whether TGFβ signaling participates in fibroblast accumulation or fibrosis formation in the EFE-like model, we stained with anti-TGFβ1 and anti-TGFβ2 antibodies on the sections of native hearts and EFE-like hearts. In the native hearts at P19, neither TGFβ1 nor TGFβ2 was detected in the myocardium (Figure 9A). However, expression of TGFβ1 and TGFβ2 was significantly higher in the myocardium of left ventricles from the EFE-like hearts (Figure 9A). As an internal control, we have also checked the level of TGFβ in the right ventricle. Our results showed that the expression level of TGFβ1 and TGFβ2 was much lower in the right ventricles compared to that of the left ventricles (Figure 9A and 9B), which could partially explain why excessive fibrosis often occurs in the left ventricles rather than the right ventricles in EFE patients and EFE-like models.
Blocking TGFβ signaling pathway reduces fibroblast accumulation within the EFE-like model. (A) Immunostaining for TGFβ1 or TGFβ2 shows that both TGFβ1 and TGFβ2 are highly expressed in the left ventricles of EFE-like models. Scale bar, 100 μm. (B) Cartoon shows that TGFβ1 and TGFβ2 are highly expressed in the EFE-like tissue. (C) Representative sirius red staining on sections of the EFE-like hearts, which were treated with PBS or BMP7. Scale bars, 500 μm. (D) Quantification results of the relative fibrotic area of the EFE-like hearts, which are treated with PBS or BMP7. P < 0.05. For each group, n = 7. (E) Immunostaining for PDGFRa on sections of the EFE-like hearts, which were treated with PBS or BMP7. Scale bars, 100 μm. LV, left ventricle. RV, right ventricle.
Previous studies reported that bone morphogenetic protein 7 (BMP7) could antagonize the TGFβ signaling pathway through a direct Smad-dependent counteraction13,22,49. We therefore treated the recipient mice with BMP7 (30 μg/kg) on every alternate day following transplantation. At day 12 post transplantation, we collected the EFE-like hearts and performed sirius red staining, and found that the accumulation of fibroblasts within EFE-like hearts was significantly reduced after BMP7 administration compared to that of the control group (Figure 9C and 9D). PDGFRa staining also confirmed less fibroblast accumulation in the left ventricles after BMP7 treatment (Figure 9E). Taken together, our results show that TGFβ plays a pivotal role in fibroblast accumulation and inhibition of the TGFβ signaling could ameliorate fibrosis in the EFE-like model.
Discussion
EFE refers to a specialized type of fibrosis with excessive fibroblasts deposited in the thickened subendocardial layer. During EFE, proliferation of fibrous and elastic tissues may eventually lead to decreased compliance and impaired diastolic functions6. These excessive fibroblasts are mainly restricted to the subendocardial layer rather than throughout the myocardium. As indicated by its name endocardial fibroelastosis, EFE, is thought to be largely associated with endocardial cells and/or their derivatives, such as excessive fibroblast formation from endocardial to mesenchymal transition13. Here, we provide direct genetic lineage tracing evidence that, rather than the endocardium, the epicardium and its derived mesenchymal cells (Epi-MCs) contributed to the majority of fibroblasts within EFE-like tissues. These Epi-MCs populated the myocardium at the embryonic stage and generated excessive fibroblasts within EFE-like tissues at the postnatal stage. Unexpectedly, the endocardium contributed minimally to fibroblasts within EFE-like tissues. Rather than postnatal endocardial to mesenchymal transition, endocardial cells generated Endo-MCs at the embryonic stage and it was such MCs that contributed to fibroblast formation in postnatal EFE. Our study has discovered a new, major cellular origin of EFE and it suggests that the location of pathogenesis might not reflect its cellular origin. We also found that TGFβ1 and TGFβ2 were highly expressed in the myocardium of left ventricle and that inhibition of the TGFβ signaling with BMP7 ameliorated fibrosis in the EFE-like model. Our comprehensive lineage tracing studies from developmental to pathogenetic stages provide new insights into understanding the fundamental pathomechanisms underlying EFE.
In this study, we used an established EFE-like model13,28. This transplant model may create a cardiac pathology resembling human EFE, but it is still unknown if the underlying pathobiology of this model can accurately recapitulate human EFE. There are some differences between this model and human EFE, such as lack of cardio-respiratory coupling and negative intrathoracic pressures affecting the heart. Therefore, we term this transplant model as an “EFE-like” model. Further investigations are needed to extrapolate our findings to the clinical settings of human EFE. Even though this model has been developed and studied for years, there remains a paucity of investigation into the mechanism underlying fibrosis in the EFE-like model. Fibrosis can be a result of tissue ischemia with reduced blood flow into the hemodynamically unloaded heart. Moreover, inflammatory responses within the heart could also lead to proliferation and migration of mesenchymal cells that deposit extracellular matrix8,50,51. Nevertheless, we could not exclude the possible effects of the immune responses in the EFE-like model. Other causes such as nutrient deprivation may also be responsible for the fibrosis. Thus the precise cellular and molecular mechanisms underlying fibrosis in EFE merit further investigation with an improved model system.
As different origins of cells may determine distinct potential therapeutic targets, it is important to revisit the cellular origin of fibroblasts within the EFE-like tissues. Lineage tracing technology has been utilized to uncover the cellular origin and cell fate determination during development and diseases52. We took advantage of the bacteriophage Cre-Loxp system to permanently label Cre-expressing cells and all of their descendants53. By virtue of the constitutively active Cre lines (such as Tie2-Cre), any cells that expressed Cre from early development would be irreversibly labeled, thus constitutively active Cre lines do not allow us to distinguish the postnatal cell fate transition from early developmental lineage conversion54. A recent study using the constitutive Tie2-Cre reported that fibroblasts within EFE-like tissues originate from endothelial cells via aberrant EndMT13. It should be noted that Tie2-Cre labels not only postnatal endocardial cells, but also embryonic endocardial cells during early development and their derivatives such as Endo-MCs55. Indeed, Tie2-Cre labeled endocardial cells that undergo EndMT and convert into primitive mesenchymal progenitors expressing PDGFRa and PDGFRb20. These endocardial-derived MCs contribute to pericytes, vascular SMCs and a few fibroblasts in the developing heart at later embryonic stages19,20. Thus a previous study employing Tie2-Cre13 could not distinguish postnatal endocardial cells from early endocardium-derived MCs. In addition, Tie2-Cre labels both coronary vascular endothelial cells and endocardial cells, and it is hard to distinguish whether fibroblasts are derived from pre-existing endocardial cells or vascular endothelial cells13. Indeed, recent work reported that endocardium-derived fibroblasts labeled by Tie2-Cre and epicardium-derived fibroblasts proliferate and respond similarly to cardiac injury21. From temporal and spatial perspectives, it is critical to employ mouse lines that harbor inducible Cre recombinase under endocardium-specific promoter to address if postnatal endocardial cells give rise to excessive fibroblasts within EFE-like tissues.
To readdress the cellular origin of EFE fibroblasts, we generated new mouse lines for inducible lineage tracing with temporal and spatial control of Cre activity. By using the inducible endocardial Cre line Npr3-CreER31,32, we genetically labeled endocardial cells at P5 and found that most endocardial cells did not contribute to fibroblasts within EFE-like tissues. We further confirmed this finding by using the endothelial cell tracing line Cdh5-CreER, which labeled both endocardial cells and vascular endothelial cells with high efficiency. These data convincingly demonstrated that postnatal endocardial cells did not generate fibroblasts within EFE-like tissues. Furthermore, we identified Sox9-derived MCs at the embryonic stage and our fate mapping data showed that most fibroblasts within EFE-like tissues were derived from Sox9-derived MCs. By using Nfatc1-2A-CreER, Npr3-CreER or Wt1-CreER lines that respectively labeled endocardial or epicardial cells, respectively, we found that the majority of EFE fibroblasts were derived from the embryonic epicardium rather than embryonic endocardium. Collectively, these inducible lineage tracing lines with more restricted spatial and temporal Cre activity presented a new model that the resident epicardium-derived MCs, but not the endocardial cells, were the major source of EFE fibroblasts (Figure 10).
Proposed model for EFE fibroblast origins based on genetic lineage tracing in this study. Majority of subendocardial EFE fibroblasts are derived from embryonic MCs, which are derivatives of endocardium via EndMT and epicardium via EpiMT. Inducible lineage tracing studies showed that the embryonic epicardium is the major source for EFE fibroblasts (> 60%), while the embryonic endocardium contribute to minority of EFE fibroblasts (∼6%-10%). Neither postnatal epicardial cells nor endothelial cells (including endocardium and coronary endothelium) contribute to EFE fibroblasts.
Another issue that needs to be addressed for lineage tracing is the specificity of fibroblast markers. Recent studies suggested that several molecular markers used to define fibroblasts are either non-specific or only specific for certain subsets of fibroblast population rather than the whole-fibroblast pool. Such issues could account for results of the discordant origin mentioned in above studies56. For instance, the typical fibroblast marker vimentin has also been found to be expressed by other cell types, including endothelial cells57,58. Another marker discoidin domain receptor 2 is also expressed by both SMCs and leukocytes59,60. Collagens such as Collagen1A1 are more enriched in fibrosis tissues. However, collagens are extracellular matrix proteins and are not suitable for localizing fibroblasts with lineage reporter proteins. Collagen1A1-GFP fusion reporter driven by endogenous gene could be helpful to specifically label fibroblasts19. Furthermore, periostin, which is also supposed to label activated fibroblasts, is, in fact, an extracellular matrix protein61. aSMA is supposed to be highly expressed by activated fibroblasts termed “myofibroblasts”. However, aSMA is also highly expressed by SMCs, and labels only 15% of collagen-positive cells as fibrosis tissues19,62. FSP1 and thymus cell antigen-1 are putative markers for subgroups of fibroblasts. However, they are also expressed by a subset of endothelial cells and leukocytes19,30. Since previous work demonstrating endocardial contribution to fibroblasts mainly used FSP1 as the fibroblast cell marker13,22, we included FSP1 together with the endothelial cell marker CDH5 to distinguish FSP1+CDH5− and FSP1+CDH5+ cells within EFE-like tissues. We also found that FSP1 was expressed in leucocytes and co-localized with CD45, CD11b and F4/80 in native hearts and EFE-like hearts (Supplementary information, Figure S7). Previous evidence for EndMT-derived fibroblasts was based on results of lineage tracing via Tie2-Cre, which actually labels both endothelial cells and resident FSP1+ leucocytes13. Recent studies suggested that PDGFRa comprehensively and strongly marked fibroblasts19, despite that PDGFRa is also expressed in mesenchymal stem cell-like progenitor cells of the neonatal mouse heart63. Therefore, we used PDGFRa as a fibroblast marker to determine the lineage of fibroblasts within fibrous EFE-like tissues.
Although we provided new evidence suggesting that embryonic epicardium and their derived MCs were the major source of EFE fibroblasts, and the TGFβ signaling pathway plays important roles in EFE formation, additional studies are needed to delineate more signals regulating expansion of epicardium-derived fibroblasts and the relationship between EFE and pathogenesis of HLHS. The signaling pathways governing Epi-MCs expansion in EFE fibrosis may include both developmental program of EpiMT as well as signals regulatory fibroblast proliferation such as Shh, Notch, PDGF and FGF27,37,64,65,66,67,68. Therefore, detailed molecular mechanisms regulating Epi-MCs cell fate and function merit further investigations. Together, understanding the cellular origin of EFE fibroblasts and mechanisms underlying cell fate determination and expansion could potentially uncover new therapeutic candidates for treatment of EFE.
Materials and Methods
Mice
All animal experiments were carried out in strict accordance with the guidelines in the Institutional Animal Care and Use Committee (IACUC) of the Institute for Nutritional Sciences, Shanghai Institutes for Biological Sciences, Chinese Academy of Science. Tamoxifen was administered by oral gavage at the indicated time points (0.1-0.2 mg/g of body weight). Caesarean section was performed on pregnant mice receiving tamoxifen to obtain perinatal pups. R26-tdTomato, Npr3-CreER, Cdh5-CreER, Wt1-CreER and Sox9-CreER mice were described previously16,32,69,70,71. The Nfatc1-2A-CreER mouse line was generated by homologous recombination using CRISPR/Cas9 technology. The 2A peptide sequence derived from porcine teschovirus-1 was upstream of the cDNA encoding CreER and polyadenylation sequence. This 2A-CreER cassette was introduced into the 9th exon of Nfatc1, and corrected targeting was verified by long PCR genotyping. This mouse line was generated by Shanghai Biomodel Organism. Co, Ltd. All mice were maintained on the C57BL6/ICR background. Genomic DNA for genotyping was prepared from embryonic yolk sac or mouse tail. Tissues were lysed by incubation with Proteinase K overnight at 55 °C. DNA was precipitated by adding isopropanol, and washed in 70% ethanol. Then genomic DNA was diluted in 200 μl distilled water.
Animal model of unloaded heterotopic heart transplantation
The donor pups at postnatal day 7 were anesthetized by intraperitoneal injection of pentobarbital sodium, and then received 60 IU heparin. The hearts were collected through a midline thoracic incision. The vena cava, aorta and pulmonary artery were identified, isolated, ligated and transected. The donor hearts were then placed in cold high potassium Krebs-Henseleit solution as previously described until the time of implantation72. Seven-day-old donor hearts were implanted into the peritoneal cavity of 8-week-old ICR mice in an unloaded configuration as previously described28,29. Briefly, the ascending aorta was anastomosed to the infrarenal aorta, and the pulmonary artery was anastomosed to the infrarenal vena cava. At day 12 following implantation, the recipients were sacrificed and the donor hearts were collected and analyzed. To test the fibrosis inhibition of BMP7, the recipient mice were treated with BMP7 (Abcam, ab87055) or PBS by intraperitoneal injection every other day at a concentration of 30 μg/kg since transplantation. Then the donor hearts were collected at day 12 post transplantation for analysis.
Immunofluorescent staining
Immunofluorescent staining was performed as previously described31. Briefly, embryonic hearts were collected and washed with PBS to remove excessive blood. After fixed in 4% PFA at 4 °C for 20 min to 1 h, tissues were dehydrated in 30% sucrose for several hours or overnight until they sank to the bottom. The tissues were embedded in OCT (Sakura) and serial cryosections of 8-10 μm thickness were collected. After air drying for 30 min, slides were blocked with PBS supplemented with 0.1% triton X-100 and 5% normal donkey serum (Jackson Immuno Research) for 30 min at room temperature, followed by incubation with the first antibody at 4 °C overnight. The following first antibodies were used: tdTomato (Rockland, 600-401-379), tdTomato (ChromoTek, ABIN334653), PECAM (BD, 553370), CDH5 (R&D, AF1002), FABP4 (Abcam, ab13979), PDGFRa (eBioscience, 14-1401-81), PDGFRa (R&D, AF1062), PDGFRb (eBioscience, 14-1402), aSMA (Sigma, F3777), ESR (Abcam, ab27595), Tbx18 (Santa Cruz, sc-17869), FSP1 (Dako, A5114), Sox9 (Millipore, AB5535), CD11b (BD, 550282), CD45 (eBioscience, 47-0451), F4/80 (Abcam, ab6640), Collagen I (Abcam, ab34710), Collagen III (Southernbiotech, 1330-01), Ki67 (Thermo Scientific, RM-9106-S0), TGFβ1 (Santa Cruz, sc-146), TGFβ2 (Abcam, ab36495), Tnni3 (Abcam, ab56357). Signals were developed with Alexa fluorescence antibodies (Invitrogen). For weak signals, we used HRP-conjugated secondary antibodies and developed using tyramide signal amplification kit (PerkinElmer). Images were acquired by Olympus confocal microscope (FV1200) or Zeiss microscope (AXIO Zoom. V16).
Whole-mount immunostaining for estrogen receptor
We performed whole-mount immunostaining as described previously32. Embryos were collected in PBS and fixed in 4% PFA at 4 °C overnight. The embryos were then dehydrated through a methanol gradient (25%, 50%, 75% and 100% methanol in PBS) for 15 min in each condition at room temperature. After that, the embryos were rehydrated through 100%, 75%, 50% and 25% methanol in PBS for 15 min in each condition at room temperature. After pretreatment in blocking buffer (5% donkey serum and 0.1% Triton X-100 in PBS) for 1 h at 4 °C, the embryos were incubated with ESR antibody (Abcam, ab27595) at 4 °C overnight. The embryos were then washed in 0.1% Triton X-100/PBS for four times, which each lasted for 1 h. After incubation with the secondary antibody (Vector lab, MP-7401) for 2 h at room temperature, the embryos were washed in 0.1% Triton X-100/PBS for 4 h. The ImmPACT DAB kit (Vector, sk-4150) was used to develop the color to the desired condition. Images were acquired by a Leica stereomicroscope (M165FC).
Sirius red staining and H&E staining
Hearts were collected in PBS buffer and fixed in 4% PFA at 4 °C for 1 h. The hearts were then dehydrated in 30% sucrose overnight and embedded in OCT. Serial cryosections of 8-10 μm thickness were collected. For sirius red staining, sections were re-fixed with Bouin's solution at room temperature for 12 h. Next, sections were stained with 0.1% fast green for 3 min. After pretreatment with 1% acetic acid for 1 min, the sections were stained with 0.1% sirius red solution for 1 min followed by a serial of dehydration procedures in ethanol and xylene. The slides were then mounted in Permount Mounting Medium. For H&E staining, slides were incubated in Hematoxylin A solution for 3 min, and then rinsed in running tap water for 1 min. The slides were then treated with 1% concentrated hydrochloric acid in 70% ethanol for 1 min and rinsed in running tap water for 1 min. After soaking in 1% ammonia water for 1 min and washed by running tap water, the slides were stained with Eosin-Y solution for 10 s followed by dehydration in ethanol and xylene. The slides were lastly mounted in neural balsam. Images were acquired by Zeiss microscope (AXIO Zoom. V16).
Cell isolation and fluorescence-activated cell sorting
Cells were isolated from EFE-like hearts as described previously73. Briefly, EFE-like hearts were dissected and carefully cut into small pieces with fine scissors. And then the hearts were digested by trypsinase and collagenase type II at 37 °C. After digestion, the lysis buffer containing cells were passed through 70-mm cell strainers. After centrifuging at 500× g for 3 min at 4 °C, non-cardiomyocytes were obtained in the suspension liquid. For identification of live cells, the LIVE/DEAD Fixable Violet Dead Cell Stain Kit (Life Technology, L34955, 1:1 000) was used according to the manufacturer's instruction. Then the isolated cells were stained with PDGFRa-APC (eBioscience, 17-1401, 1:200) for 30 min. Flow cytometry was performed using a BD FACS Aria flow cytometry system (BD Biosciences, San Jose, CA, USA). Data were analyzed using FlowJo software (Tree Star, Ashland, Oregon, USA).
Statistical analysis
All data were collected from at least three independent experiments as indicated. Besides flow cytometry, we also counted the number of EFE fibroblasts via immunostaining on tissue sections. For analyzing the labeling percentage of EFE fibroblasts by tdTomato, we collected five heart samples for each experiment. At least eight tissue sections from each heart were collected for immunostaining; and images were taken with at least five fields in each section for analysis. The percentage of contribution to EFE fibroblasts was calculated as tdTomato+PDGFRa+ cells in PDGFRa+ cells within EFE-like tissues. All data were presented as mean values ± SEM. Statistical comparisons between data sets were done by a two-sided unpaired Student's t test for comparing differences between two groups. P < 0.05 was considered to be statistically significant.
Author Contributions
HZ and BZ conceived and designed the study, analyzed the data and wrote the manuscript. HZ, XH, KL, JT, LH, WP, QL, YL, XT, YW and Libo Z bred the mice and performed experiments. YY, HW, RH, FW, TC, QW, ZQ, Li Z and KL provided valuable comments, analyzed the data and edited the manuscript. BZ supervised the study.
Competing Financial Interests
The authors declare no competing financial interests.
References
Rosahn PD . Endocardial fibroelastosis: old and new concepts. Bull NY Acad Med 1955; 31:453–472.
De Letter EA, Piette MH . Endocardial fibroelastosis as a cause of sudden unexpected death. Am J Forensic Med Pathol 1999; 20:357–363.
Stehbens WE, Delahunt B, Zuccollo JM . The histopathology of endocardial sclerosis. Cardiovasc Pathol 2000; 9:161–173.
Weinberg T, Himelfarb AJ . Endocardial fibroelastosis (so-called fetal endocarditis). A report of two cases occurring in siblings. Bull Johns Hopkins Hosp 1943; 72:299.
Grellner W, Kaferstein H, Sticht G . Combination of fatal digoxin poisoning with endocardial fibroelastosis. Forensic Sci Int 1997; 89:211–216.
Moller JH, Lucas RV Jr, Adams P Jr, Anderson RC, Jorgens J, Edwards JE . Endocardial fibroelastosis: a clinical and anatomic study of 47 patients with emphasis on its relationship to mitral insufficiency. Circulation 1964; 30:759–782.
Gilbert-Barness E, Barness LA . Nonmalformative cardiovascular pathology in infants and children. Pediatr Dev Pathol 1999; 2:499–530.
Lurie PR . Changing concepts of endocardial fibroelastosis. Cardiol Young 2010; 20:115–123.
McElhinney DB, Vogel M, Benson CB, et al. Assessment of left ventricular endocardial fibroelastosis in fetuses with aortic stenosis and evolving hypoplastic left heart syndrome. Am J Cardiol 2010; 106:1792–1797.
McElhinney DB, Marshall AC, Wilkins-Haug LE, et al. Predictors of technical success and postnatal biventricular outcome after in utero aortic valvuloplasty for aortic stenosis with evolving hypoplastic left heart syndrome. Circulation 2009; 120:1482–1490.
Emani SM, Bacha EA, McElhinney DB, et al. Primary left ventricular rehabilitation is effective in maintaining two-ventricle physiology in the borderline left heart. J Thorac Cardiovasc Surg 2009; 138:1276–1282.
Emani SM, McElhinney DB, Tworetzky W, et al. Staged left ventricular recruitment after single-ventricle palliation in patients with borderline left heart hypoplasia. J Am Coll Cardiol 2012; 60:1966–1974.
Xu X, Friehs I, Zhong Hu T, et al. Endocardial fibroelastosis is caused by aberrant endothelial to mesenchymal transition. Circ Res 2015; 116:857–866.
Merki E, Zamora M, Raya A, et al. Epicardial retinoid X receptor alpha is required for myocardial growth and coronary artery formation. Proc Natl Acad Sci USA 2005; 102:18455–18460.
Cai CL, Martin JC, Sun Y, et al. A myocardial lineage derives from Tbx18 epicardial cells. Nature 2008; 454:104–108.
Zhou B, Ma Q, Rajagopal S, et al. Epicardial progenitors contribute to the cardiomyocyte lineage in the developing heart. Nature 2008; 454:109–113.
Wilm B, Ipenberg A, Hastie ND, Burch JB, Bader DM . The serosal mesothelium is a major source of smooth muscle cells of the gut vasculature. Development 2005; 132:5317–5328.
Volz KS, Jacobs AH, Chen HI, et al. Pericytes are progenitors for coronary artery smooth muscle. eLife 2015; 4:e10036.
Moore-Morris T, Guimaraes-Camboa N, Banerjee I, et al. Resident fibroblast lineages mediate pressure overload-induced cardiac fibrosis. J Clin Invest 2014; 124:2921–2934.
Chen Q, Zhang H, Liu Y, et al. Endothelial cells are progenitors of cardiac pericytes and vascular smooth muscle cells. Nat Commun 2016; 7:12422.
Ali SR, Ranjbarvaziri S, Talkhabi M, et al. Developmental heterogeneity of cardiac fibroblasts does not predict pathological proliferation and activation. Circ Res 2014; 115:625–635.
Zeisberg EM, Tarnavski O, Zeisberg M, et al. Endothelial-to-mesenchymal transition contributes to cardiac fibrosis. Nat Med 2007; 13:952–961.
Mollmann H, Nef HM, Kostin S, et al. Bone marrow-derived cells contribute to infarct remodelling. Cardiovasc Res 2006; 71:661–671.
van Amerongen MJ, Bou-Gharios G, Popa E, et al. Bone marrow-derived myofibroblasts contribute functionally to scar formation after myocardial infarction. J Pathol 2008; 214:377–386.
Widyantoro B, Emoto N, Nakayama K, et al. Endothelial cell-derived endothelin-1 promotes cardiac fibrosis in diabetic hearts through stimulation of endothelial-to-mesenchymal transition. Circulation 2010; 121:2407–2418.
Xu J, Lin SC, Chen J, et al. CCR2 mediates the uptake of bone marrow-derived fibroblast precursors in angiotensin II-induced cardiac fibrosis. Am J Physiol Heart Circ Physiol 2011; 301:H538–547.
von Gise A, Pu WT . Endocardial and epicardial epithelial to mesenchymal transitions in heart development and disease. Circ Res 2012; 110:1628–1645.
Friehs I, Illigens B, Melnychenko I, Zhong-Hu T, Zeisberg E, Del Nido PJ . An animal model of endocardial fibroelastosis. J Surg Res 2013; 182:94–100.
Asfour B, Hare JM, Kohl T, et al. A simple new model of physiologically working heterotopic rat heart transplantation provides hemodynamic performance equivalent to that of an orthotopic heart. J Heart Lung Transplant 1999; 18:927–936.
Kong P, Christia P, Saxena A, Su Y, Frangogiannis NG . Lack of specificity of fibroblast-specific protein 1 in cardiac remodeling and fibrosis. Am J Physiol Heart Circ Physiol 2013; 305:H1363–H1372.
Zhang H, Pu W, Tian X, et al. Genetic lineage tracing identifies endocardial origin of liver vasculature. Nat Genet 2016; 48:537–543.
Zhang H, Pu W, Li G, et al. Endocardium minimally contributes to coronary endothelium in the embryonic ventricular free walls. Circ Res 2016; 118:1880–1893.
Madisen L, Zwingman TA, Sunkin SM, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci 2010; 13:133–140.
Zhang H, Pu W, Liu Q, et al. Endocardium contributes to cardiac fat. Circ Res 2016; 118:254–265.
Akiyama H, Chaboissier MC, Behringer RR, et al. Essential role of Sox9 in the pathway that controls formation of cardiac valves and septa. Proc Natl Acad Sci USA 2004; 101:6502–6507.
Garside VC, Cullum R, Alder O, et al. SOX9 modulates the expression of key transcription factors required for heart valve development. Development 2015; 142:4340–4350.
Smith CL, Baek ST, Sung CY, Tallquist MD . Epicardial-derived cell epithelial-to-mesenchymal transition and fate specification require PDGF receptor signaling. Circ Res 2011; 108:e15–26.
Ranger AM, Grusby MJ, Hodge MR, et al. The transcription factor NF-ATc is essential for cardiac valve formation. Nature 1998; 392:186–190.
de la Pompa JL, Timmerman LA, Takimoto H, et al. Role of the NF-ATc transcription factor in morphogenesis of cardiac valves and septum. Nature 1998; 392:182–186.
Chang CP, Neilson JR, Bayle JH, et al. A field of myocardial-endocardial NFAT signaling underlies heart valve morphogenesis. Cell 2004; 118:649–663.
Acharya A, Baek ST, Huang G, et al. The bHLH transcription factor Tcf21 is required for lineage-specific EMT of cardiac fibroblast progenitors. Development 2012; 139:2139–2149.
Lijnen PJ, Petrov VV, Fagard RH . Induction of cardiac fibrosis by transforming growth factor-beta(1). Mol Genet Metab 2000; 71:418–435.
Bujak M, Frangogiannis NG . The role of TGF-beta signaling in myocardial infarction and cardiac remodeling. Cardiovasc Res 2007; 74:184–195.
Tomita H, Egashira K, Ohara Y, et al. Early induction of transforming growth factor-beta via angiotensin II type 1 receptors contributes to cardiac fibrosis induced by long-term blockade of nitric oxide synthesis in rats. Hypertension 1998; 32:273–279.
Nakajima H, Nakajima HO, Salcher O, et al. Atrial but not ventricular fibrosis in mice expressing a mutant transforming growth factor-beta(1) transgene in the heart. Circ Res 2000; 86:571–579.
Rosenkranz S, Flesch M, Amann K, et al. Alterations of beta-adrenergic signaling and cardiac hypertrophy in transgenic mice overexpressing TGF-beta(1). Am J Physiol Heart Circ Physiol 2002; 283:H1253–1262.
Schultz Jel J, Witt SA, Glascock BJ, et al. TGF-beta1 mediates the hypertrophic cardiomyocyte growth induced by angiotensin II. J Clin Invest 2002; 109:787–796.
Kuwahara F, Kai H, Tokuda K, et al. Transforming growth factor-beta function blocking prevents myocardial fibrosis and diastolic dysfunction in pressure-overloaded rats. Circulation 2002; 106:130–135.
Zeisberg M, Yang C, Martino M, et al. Fibroblasts derive from hepatocytes in liver fibrosis via epithelial to mesenchymal transition. J Biol Chem 2007; 282:23337–23347.
Ueha S, Shand FH, Matsushima K . Cellular and molecular mechanisms of chronic inflammation-associated organ fibrosis. Front Immunol 2012; 3:71.
Neustein HB, Lurie PR, Fugita M . Endocardial fibroelastosis found on transvascular endomyocardial biospsy in children. Arch Pathol Lab Med 1979; 103:214–219.
Kretzschmar K, Watt FM . Lineage tracing. Cell 2012; 148:33–45.
Nagy A . Cre recombinase: the universal reagent for genome tailoring. Genesis 2000; 26:99–109.
Tian X, Pu WT, Zhou B . Cellular origin and developmental program of coronary angiogenesis. Circ Res 2015; 116:515–530.
Zhang H, von Gise A, Liu Q, et al. Yap1 is required for endothelial to mesenchymal transition of the atrioventricular cushion. J Biol Chem 2014; 289:18681–18692.
Moore-Morris T, Cattaneo P, Puceat M, Evans SM . Origins of cardiac fibroblasts. J Mol Cell Cardiol 2016; 91:1–5.
Zeisberg EM, Kalluri R . Origins of cardiac fibroblasts. Circ Res 2010; 107:1304–1312.
Lane EB, Hogan BL, Kurkinen M, Garrels JI . Co-expression of vimentin and cytokeratins in parietal endoderm cells of early mouse embryo. Nature 1983; 303:701–704.
Afonso PV, McCann CP, Kapnick SM, Parent CA . Discoidin domain receptor 2 regulates neutrophil chemotaxis in 3D collagen matrices. Blood 2013; 121:1644–1650.
Hou G, Wang D, Bendeck MP . Deletion of discoidin domain receptor 2 does not affect smooth muscle cell adhesion, migration, or proliferation in response to type I collagen. Cardiovasc Pathol 2012; 21:214–218.
Kaur H, Takefuji M, Ngai CY, et al. Targeted ablation of periostin-expressing activated fibroblasts prevents adverse cardiac remodeling in mice. Circ Res 2016; 118:1906–1917.
Weber KT . Monitoring tissue repair and fibrosis from a distance. Circulation 1997; 96:2488–2492.
Chong JJ, Chandrakanthan V, Xaymardan M, et al. Adult cardiac-resident MSC-like stem cells with a proepicardial origin. Cell Stem Cell 2011; 9:527–540.
Lavine KJ, Yu K, White AC, et al. Endocardial and epicardial derived FGF signals regulate myocardial proliferation and differentiation in vivo. Dev Cell 2005; 8:85–95.
Lavine KJ, White AC, Park C, et al. Fibroblast growth factor signals regulate a wave of Hedgehog activation that is essential for coronary vascular development. Genes Dev 2006; 20:1651–1666.
Lavine KJ, Kovacs A, Ornitz DM . Hedgehog signaling is critical for maintenance of the adult coronary vasculature in mice. J Clin Invest 2008; 118:2404–2414.
de la Pompa JL, Epstein JA . Coordinating tissue interactions: Notch signaling in cardiac development and disease. Dev Cell 2012; 22:244–254.
Luna-Zurita L, Prados B, Grego-Bessa J, et al. Integration of a Notch-dependent mesenchymal gene program and Bmp2-driven cell invasiveness regulates murine cardiac valve formation. J Clin Invest 2010; 120:3493–3507.
Zhang H, Pu W, Tian X, et al. Genetic lineage tracing identifies endocardial origin of liver vasculature. Nat Genet 2016; 48:537–543.
Wang Y, Nakayama M, Pitulescu ME, et al. Ephrin-B2 controls VEGF-induced angiogenesis and lymphangiogenesis. Nature 2010; 465:483–486.
Xu Z, Wang W, Jiang K, et al. Embryonic attenuated Wnt/beta-catenin signaling defines niche location and long-term stem cell fate in hair follicle. eLife 2015; 4:e10567.
Friehs I, del Nido PJ . Increased susceptibility of hypertrophied hearts to ischemic injury. Ann Thorac Surgery 2003; 75:S678–684.
Yu W, Huang X, Tian X, et al. GATA4 regulates Fgf16 to promote heart repair after injury. Development 2016; 143:936–949.
Acknowledgements
We thank Baojin Wu, Guoyuan Chen, Zhonghui Weng and Aimin Huang for the animal husbandry; and Wei Bian for his technical help. We thank Shanghai Biomodel Organism Science & Technology Development Co., Ltd for mouse generation. We thank Ralf Adams at Max Plank Institute for providing the Cdh5-CreER mouse line and Hongkui Zeng for reporter lines. We also thank other members of our laboratory for insightful discussion and technical help throughout this study. This work was supported by Strategic Priority Research Program of the Chinese Academy of Sciences (CAS, XDB19000000), The National key Research & Development Program of China (2017YFC1001300 and 2016YFC1300600), National Natural Science Foundation of China (91639302, 31625019, 31571503, 31501172, 31601168), Youth Innovation Promotion Association of CAS (2015218), Key Project of Frontier Sciences of CAS (QYZDB-SSW-SMC003), International Cooperation Fund of CAS, National Program for Support of Top-notch Young Professionals, Shanghai Science and Technology Commission (14JC1407300, 17ZR1449600, 17ZR1449800), Shanghai Yangfan Project (15YF1414000, 16YF1413400) and Rising-Star Program (15QA1404300), China Postdoctoral Science Foundation (2015M581669, 2016T90387, 2016LH0042), President Fund of Shanghai Institutes for Biological Sciences (SIBS), Astrazeneca, Boehringer Ingelheim, Sanofi-SIBS Fellowship and Research Grants Council of Hong Kong (24110515, 14111916).
Author information
Authors and Affiliations
Corresponding authors
Additional information
( Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary information
Supplementary information, Figure S1
Endocardial fibroelastosis (EFE) tissue lining the left ventricle after heterotopic heart transplantation. (PDF 1240 kb)
Supplementary information, Figure S2
Npr3-CreER labeled endocardial or epicardial cells do not contribute to fibroblasts in native hearts. (PDF 1700 kb)
Supplementary information, Figure S3
Sox9 is expressed in cushion mesenchymal cells and epicardiai cells at E10.5 - E11.5. (PDF 1205 kb)
Supplementary information, Figure S4
Sox9-CreER labels embryonic cardiac mesenchymal cells and epicardial cells, but not endothelial cells. (PDF 1263 kb)
Supplementary information, Figure S5
Sox9-CreER does not label hematopoietic cells. (PDF 747 kb)
Supplementary information, Figure S6
Embryonic endocardial cells contribute to fibroblasts in the native hearts. (PDF 1181 kb)
Supplementary information, Figure S7
FSP1 is expressed in leucocytes. (PDF 1018 kb)
Rights and permissions
About this article
Cite this article
Zhang, H., Huang, X., Liu, K. et al. Fibroblasts in an endocardial fibroelastosis disease model mainly originate from mesenchymal derivatives of epicardium. Cell Res 27, 1157–1177 (2017). https://doi.org/10.1038/cr.2017.103
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cr.2017.103
Keywords
This article is cited by
-
Tongxinluo Activates PI3K/AKT Signaling Pathway to Inhibit Endothelial Mesenchymal Transition and Attenuate Myocardial Fibrosis after Ischemia-Reperfusion in Mice
Chinese Journal of Integrative Medicine (2024)
-
Endocardial fibroelastosis in infants and young children: a state-of-the-art review
Heart Failure Reviews (2023)
-
Genetic lineage tracing identifies cardiac mesenchymal-to-adipose transition in an arrhythmogenic cardiomyopathy model
Science China Life Sciences (2023)
-
Recessive ciliopathy mutations in primary endocardial fibroelastosis: a rare neonatal cardiomyopathy in a case of Alstrom syndrome
Journal of Molecular Medicine (2021)
-
Reassessing endothelial-to-mesenchymal transition in cardiovascular diseases
Nature Reviews Cardiology (2018)