Abstract
Reciprocal relationship usually exists between osteoblastogenesis and adipogenesis, with factors stimulating one of these processes at the same time inhibiting the other. In the present study, miR-30e was found to be involved in the reciprocal regulation of osteoblast and adipocyte differentiation. Our data indicated that miR-30e was induced in primarily cultured mouse bone marrow stromal cell, mesenchymal cell line C3H10T1/2 and preadipocyte 3T3-L1 after adipogenic treatment. Conversely, it was reduced in mouse stromal line ST2 and preosteoblast MC3T3-E1 after osteogenic treatment. Enforced expression of miR-30e in 3T3-L1 significantly suppressed the growth of the cells and induced the preadipocytes to differentiate into mature adipocytes, along with increased expression of adipocyte-specific transcription factors peroxisome proliferator-activated receptor-γ (PPARγ), CCAAT/enhancer binding protein-α (C/EBPα) and C/EBPβ, and the marker gene aP2. In contrast, inhibition of the endogenous miR-30e enhanced the cell growth and repressed preadipocytes to differentiate. Conversely, supplementing miR-30e activity blocked, whereas knocking down miR-30e enforced the preosteoblast MC3T3-E1 to fully differentiate. Furthermore, miR-30e overexpression stimulated adipocyte formation and inhibited osteoblast differentiation from marrow stromal cells. Low-density lipoprotein receptor-related protein 6 (LRP6), one of the critical coreceptor for Wnts, was shown to be a direct target of miR-30e by using the luciferase assay. Knockdown of LRP6 in 3T3-L1 cells downregulated β-catenin/T-cell factor (TCF) transcriptional activity and dramatically potentiated the differentiation of the cells into mature adipocytes. Taken together, the present work suggests that the expression of miR-30e is indispensable for maintaining the balance of adipocytes and osteoblasts by targeting the canonical Wnt/β-catenin signaling.
Similar content being viewed by others
Main
Recent emerging evidences confirmed that adipocytes and osteoblasts originate from a common mesenchymal precursor cell, thus inverse reciprocal relationship exists between osteoblastogenesis and adipogenesis, with factors stimulating one of these processes at the same time inhibiting the other.1, 2 Osteoblast development is governed by the activation of Wnt/β-catenin signaling and the expression of several master transcription factors, and among them Runt-related transcription factor (Runx2), osterix (Osx) and Dlx5 are of great importance.3, 4, 5, 6 In contrast, formation of terminally differentiated adipocytes in marrow requires the activity of peroxisome proliferator-activated receptor-γ (PPARγ) and members of the CCAAT/enhancer binding protein (C/EBP) family.7, 8, 9, 10 PPARγ is regarded as an essential transcriptional regulator of both adipocyte differentiation and lipid storage in mature adipocytes based on the fact that homozygous PPARγ-deficient embryonic stem cells were unable to differentiate into adipocytes and underwent osteoblast formation instead in vivo and in vitro.11, 12 Conversely, enhanced expression of PPARγ in bone marrow stromal progenitors led to increased marrow adiposity and the block of osteoblast differentiation, probably because of the suppression of the osteoblast-specific transcription factor Runx2.7, 13
Three members of C/EBP family, those are, C/EBPα, C/EBPβ and C/EBPγ, are believed to have key metabolic and differentiation missions in adipose tissue.14, 15, 16, 17 C/EBPα was induced late during adipogenesis,14 whereas C/EBPβ and C/EBPδ were induced early and transiently during adipocyte development.18
Although it is well documented, as mentioned above, that transcription factors and diffusible signals influence adipocyte/osteoblast development and differentiation, emerging evidences suggest that the posttranscriptional regulation of gene expression also has a crucial role in the same events.19, 20, 21 MicroRNAs (miRNAs) have been found to exert essential regulatory functions in adipocyte development.
miRNAs are short (19–25 nt) noncoding RNAs that mainly regulate gene expression in a posttranscriptional manner. Encoded by eukaryotic nuclear DNA, miRNAs function via base pairing with complementary sequences within mRNA molecules, usually resulting in gene silencing via translational repression or target degradation.22 Although core components of the miRNA pathway are conserved between plants and animals, miRNA repertoires in the two kingdoms function in quite different manner. Plant miRNAs usually perfectly or near-perfectly pair with their messenger RNA targets at either coding regions or untranslated regions, and thus induce gene repression through degrading their target transcripts. In contrast, animal miRNAs typically are only partially complementary to their mRNA targets at their 3′-UTR regions. However, the imperfect pairing may result in dramatic block of protein translation.
To date, a regulated expression pattern of particular miRNAs was reported throughout adipogenesis, suggesting a role for miRNAs in adipocyte commitment and differentiation. Reduced expression of miR-155, miR-221 and miR-222 were reported during adipogenic programming of human bone marrow-derived stromal cells, suggesting that they negatively regulate adipocyte differentiation. Conversely, ectopic expression of these miRNAs blocked adipogenesis, which was paralleled by the suppression of the master regulators PPARγ and C/EBPα.23 In contrast, expression of miR-637 significantly increased during adipocyte differentiation and decreased during osteoblast differentiation. miR-637 repressed osteogenesis and enhanced adipocyte differentiation by targeting the transcription factor osterix.24 miR-210 was found to be able to promote adipogenesis by targeting the transcription factor TCF7l2 in the Wnt/β-catenin signaling pathway.25 miR-204 and its homolog miR-211 were demonstrated to be induced during adipocyte differentiation. miR-204/miR-211 were able to target Runx2 and changed Runx2 protein levels, thus significantly altering differentiation fate of mesenchymal progenitor cells.26
The miR-30 family members include miR-30a, miR-30b, miR-30c, miR-30d and miR-30e, and all of them share the same seed sequence in their 5′ terminuses. The family is associated with cell differentiation, cellular senescence and apoptosis, and involved in the pathogenesis of tumors and other disorders of the nervous, genital, circulatory, alimentary and respiratory systems.27, 28, 29 Previous studies reported the downregulation of miR-30 family members during osteoblast differentiation from mouse preosteoblast cell lines.30, 31 miR-30a, miR-30b, miR-30c and miR-30d were demonstrated to be able to negatively regulate BMP-2-induced osteoblast differentiation by targeting Smad1.30 In contrast, miR-30 family members were upregulated during adipogenic differentiation of adipose tissue-derived stem cells, and miR-30a and miR-30d contributed to adipocyte formation.32 Our present work shows that miR-30e is upregulated following adipogenic treatment and downregulated following osteogenic treatment in mouse marrow stromal cells and adipocyte/osteoblast precursor lines. Furthermore, our present work provides novel evidences demonstrating that miR-30e is capable of promoting adipogenesis and repressing osteogenesis via direct regulation of canonical Wnt pathway.
Results
miR-30e family members were differentially regulated during adipocyte formation and osteoblast differentiation
To identify the miRNA expression signature in the marrow stromal cells in response to the osteogenic or adipogenic culture medium for 72 h, miRNA profiles of the cells were analyzed using TaqMan Rodent MicroRNA Array (Applied Biosystems, Foster City, CA, USA). The fold change of Ct value >1.8 was regarded as significant difference. In total, ∼59 miRNAs were found to be differentially expressed in osteogenic medium-treated cells compared with control, among which 18 were increased and 41 were decreased. In contrast, 84 miRNAs were found to be differentially expressed following adipogenic treatment compared with control, among which 76 were increased and 8 were decreased. Upon further analysis, only 14 miRNAs were found to be increased during adipocyte differentiation and at the same time decreased during osteoblast differentiation (Table 1). These miRNAs might conversely regulate osteoblast formation and adipocyte differentiation from the bone marrow stromal progenitors.
The miR-30 family members were all detected during both osteoblast and adipocyte differentiation. In brief, miR-30a and miR-30b were increased during adipogenesis but remained unchanged during osteoblastogenesis. miR-30c, miR-30d and miR-30e, however, were increased during adipocyte differentiation and at the same time decreased during osteoblast differentiation. The expression profiles of the miR-30 family members observed in the microarray analysis were validated using qPCR (Figures 1a–e). miR-30e was the one with greatest changes in either adipocytes or osteoblasts. The qPCR results showed that osteogenic and adipogenic treatments led to a 50% reduction and a 4.6-fold induction of miR-30e expression, respectively (Figures 1a–e).
miR-30e was induced during adipocyte formation and reduced during osteoblast differentiation. Real-time PCR showed that AIM treatment of marrow stromal cells for 72 h induced all miR-30 family members, whereas OIM reduced miR-30c, miR-30d and miR-30e levels and did not affect miR-30a and miR-30b (a–e). miR-30e was also induced in 3T3-L1 (f) and C3H10T1/2 (g) at 48 h after treatment with AIM. In contrast, miR-30e was reduced in a time-dependent manner in ST2 (h) and MC3T3-E1 (i) after treatment with OIM. Values represent the mean±S.D. of three experiments. *Significant versus Vehicle, P<0.05
To further demonstrate that change of miR-30e during adipocyte and osteoblast differentiation is not an accidental finding, we tested the expression of miR-30e in the mesenchymal cell line C3H10T1/2 and the preadipocyte 3T3-L1 at 48 h after treatment with adipogenic medium or in stromal ST2 cells after treatment with osteogenic medium. Both C3H10T1/2 and 3T3-L1 lines expressed increased levels of miR-30e when induced with adipocyte-inducing medium (AIM) to differentiate, with a 1.9-fold increase in 3T3-L1 and a 5.1-fold increase in C3H10T1/2, respectively (Figures 1f and g). In contrast, miR-30e expression was significantly reduced in ST2 cells after treatment with osteogenic medium at all time points examined, with the greatest decrease of 64% at 72 h (Figure 1h). Similarly, miR-30e expression was reduced in MC3T3-E1 cells at 24 and 48 h following osteogenic treatment as well (Figure 1i). We postulated that miR-30e might play a role in the processes of adipocyte and osteoblast differentiation.
Supplementing miR-30e activity suppressed cell proliferation in adipocyte progenitors and favored adipocyte differentiation
To study the effect of miR-30e on the differentiation of adipocytes, we first examined its effect on cell proliferation. Synthetic miR-30e mimics, or miR-30e inhibitor, along with their respective negative control (NC) were transiently transfected into 3T3-L1, and cell growth was assayed. As shown in Figure 2a, miR-30e mimics inhibited cell proliferation in 3T3-L1 by 13% and, conversely, miR-30e inhibitor promoted cell growth by 14% as compared with NC (Figure 2b).
Supplementing miR-30e activity suppressed 3T3-L1 proliferation and favored adipocyte differentiation. miR-30e mimic transfection inhibited, whereas miR-30e inhibitor promoted cell growth (a and b). miR-30e mimics induced adipocyte formation in the presence of AIM. Oil Red O extracted with isopropanol was measured at OD510 (c and d). The mRNA levels of PPARγ, C/EBPα, C/EBPβ and aP2 (e–h) and protein levels of PPARγ, C/EBPα and aP2 (i) were induced after transfection of mi-30e mimics. Values represent the mean±S.D. of three experiments. *Significant versus control, P<0.05
In the presence of adipogenic medium, the exposure to the miR-30e mimics resulted in a significant increase of differentiated adipocyte numbers (68% increase in Oil Red O staining) (Figures 2c and d). This was further confirmed by the induction of the adipocyte-specific transcription factors and marker genes. The mRNA levels of PPARγ, C/EBPα, C/EBPβ and aP2 were all elevated after transfection of miRNA mimics in a time-dependent manner (Figures 2e–h). Consistent with this, the western blotting showed that the protein levels of PPARγ, C/EBPα and aP2 were increased after adipogenic treatment in the cells transfected with miR-30e mimics (Figure 2i).
Furthermore, we made the lentivirus that overexpresses miR-30e precursor and infected the 3T3-L1 cells with the virus. The examination of the mature miR-30e by real-time PCR demonstrated the efficacy of the expression (10-fold increase) (Figure 3a). At 48 h after infection, the 3T3-L1 cells were subjected to adipogenic treatment for 4 days. As shown in Figures 3b–d, the miR-30e expressing virus promoted adipocyte formation and increased the mRNA levels of PPARγ, C/EBPα, C/EBPβ and aP2 at 48 h after adipogenic treatment as compared with the NC virus. Moreover, miR-30e expressing lentivirus also increased the protein levels of PPARγ, C/EBPα and aP2 at 72 h after adipogenic treatment (Figure 3e).
miR-30e expressing lentivirus promoted 3T3-L1 to fully differentiate. The miR-30e-expressing lentivirus increased mature miR-30e in 3T3-L1 (a). In the presence of AIM, miR-30e-expressing lentivirus induced adipocyte formation (b and c). The mRNA levels of PPARγ, C/EBPα, C/EBPβ and aP2 (d) and protein levels of PPARγ, C/EBPα and aP2 (e) were induced after infection of miR-30e virus. Values represent the mean±S.D. of three experiments. *Significant versus control, P<0.05
Inhibition of miR-30e reduced adipocyte differentiation
We also examined the effects of synthetic inhibitor of miR-30e on adipogenesis from preadipocyte 3T3-L1. The adipocyte formation was significantly blocked by the miR-30e inhibitor, evidenced by the reduced numbers of Oil Red O-positive adipocytes (Figures 4a and b). Consistent with this, PPARγ, C/EBPα, C/EBPβ and aP2 were all downregulated after inactivation of miR-30e by the inhibitor in a time-dependent manner (Figures 4c–f). Furthermore, western blotting showed that the miR-30e inhibitor decreased the protein levels of PPARγ, C/EBPα and aP2 after adipogenic treatment (Figure 4g).
Inactivation of miR-30e reduced 3T3-L1 differentiation. The miR-30e inhibitor transfection blocked the adipocyte formation (a and b). Transfection of miR-30e inhibitor decreased the mRNA expression levels of PPARγ, C/EBPα, C/EBPβ and aP2 at all the indicated time points (c–f), and reduced protein levels of PPARγ, C/EBPα and aP2 at 72 h (g). Values represent the mean±S.D. of three experiments. *Significant versus control, P<0.05
Perturbation of miR-30e in preosteoblasts altered osteoblast differentiation
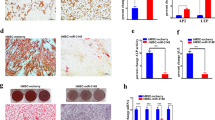
To further verify whether miR-30e could regulate osteoblast differentiation, we altered the expression of miR-30e in preosteoblast MC3T3-E1 by transfecting the miR-30e mimics or inhibitor into the cells. In the presence of osteogenic medium, miR-30e mimic transfection blocked osteoblast differentiation in MC3T3-E1 cells, as evidenced by the attenuated alkaline phosphatase (ALP) staining in comparison with the NC transfection (Figure 5a). Consistently, the mRNA levels of Runx2, osterix, osteocalcin, ALP and bone sialoprotein (BSP) were all significantly reduced by the miR-30e mimics at 48 h after osteogenic treatment (Figure 5b). Furthermore, the protein levels of Runx2, osterix and ALP were decreased in the cells transfected with miR-30e mimics at 72 h after osteogenic treatment (Figure 5c).
Perturbation of miR-30e in MC3T3-E1 altered osteoblast differentiation. The miR-30e mimic transfection blunted ALP staining (a), decreased mRNA levels of Runx2, osterix, osteocalcin, ALP and BSP (b) and reduced protein levels of Runx2, osterix and ALP (c). In contrast, miR-30e inhibitor transfection enhanced ALP staining (d), induced mRNA levels of Runx2, osterix, osteocalcin, ALP and BSP (e) and increased protein levels of Runx2, osterix and ALP (f). Values represent the mean±S.D. of three experiments. *Significant versus control, P<0.05
In contrast, the osteoblast differentiation of MC3T3-E1 was potentiated following transfection of the miR-30e inhibitor, as evidenced by the enhanced ALP staining (Figure 5d), the increased mRNA levels of Runx2, osterix, osteocalcin, ALP and BSP (at 48 h; Figure 5e) and elevated protein levels of Runx2, osterix and ALP (at 72 h) in the cells transfected with the miR-30e inhibitor (Figure 5f).
miR-30e overexpression stimulated adipocyte formation and inhibited osteoblast differentiation from mouse marrow stromal cells
To further demonstrate the adipogenic and osteogenic effects of miR-30e, we infected the primarily cultured mouse marrow stromal cells with the miR-30e-expressing virus. Real-time PCR revealed 7.6-fold increase of miR-30e level following infection with the miR-30e virus (Figure 6a). At 48 h after infection, the cells were subjected to adipogenic or osteogenic treatment. A 4-day adipogenic treatment led to significant increase of adipocyte formation (Figures 6b and c). The mRNA levels of PPARγ, C/EBPα, C/EBPβ and aP2 were significantly induced 48 h after adipogenic treatment in the cells overexpressing miR-30e (Figure 6d). Accordingly, the protein levels of PPARγ, C/EBPα and aP2 were increased 72 h after adipogenic treatment (Figure 6e).
miR-30e overexpression stimulated adipocyte formation and blocked osteoblast differentiation from marrow stromal cells. The miR-30e-expressing lentivirus increased mature miR-30e in primarily cultured mouse marrow stromal cells (a). In the presence of AIM, miR-30e-expressing lentivirus promoted adipocyte formation (b and c), increased mRNA levels of PPARγ, C/EBPα, C/EBPβ and aP2 (d) and induced protein levels of PPARγ, C/EBPα and aP2 (e). In the presence of OIM, miR-30e overexpression blunted ALP staining (f), decreased mRNA levels of Runx2, osterix, osteocalcin, ALP and BSP (g) and reduced protein levels of Runx2, osterix and ALP (h). Values represent the mean±S.D. of three experiments. *Significant versus control, P<0.05
In contrast, miR-30e overexpression negatively affected osteoblast differentiation, evidenced by the blunted ALP staining (Figure 6f). Furthermore, mRNA levels of Runx2, osterix, osteocalcin, ALP and BSP were reduced 48 h after osteogenic treatment (Figure 6g) in the cells infected with the miR-30e virus versus NC virus. Accordingly, the protein levels of Runx2, osterix and ALP were decreased 72 h after osteogenic treatment (Figure 6h).
LRP6 was a direct target of miR-30e
As is well known, miRNAs exert function through suppressing the expression of their target genes. To clarify the molecular mechanisms underlying the regulation of adipogenesis and osteogenesis by miR-30e, bioinformatic prediction of miRNA targets was performed with TargetScan (http://www.targetscan.org/) and PicTar (http://pictar.mdc-berlin.de/). Although hundreds of different targets were predicted by the program, only a few targets are potentially associated with adipocyte and osteoblast differentiation. These include Runx2, Wnt7b and low-density lipoprotein receptor-related protein 6 (LRP6), the latter two of which are components of canonical Wnt signaling. The expressions of these three genes were detected at the undifferentiated state (data not shown).
In order to test whether the three genes are really targeted by miR-30e, the 3′-UTR fragments of the mRNAs containing the target sequences were PCR-amplified and cloned into the luciferase reporter vector, respectively (Figure 7a). The 3′-UTR constructs were then cotransfected into 293-AD cells along with miR-30e miRNA expression plasmid or a NC plasmid. Luciferase assay revealed that the miR-30e miRNA slightly decreased the luciferase activity of Runx2 3′-UTR construct by 26% (Figure 7b), whereas it had no impact on the 3′-UTR construct of Wnt7b (Figure 7c). In contrast, the luciferase activity of LRP6 3′-UTR construct was the one that decreased most significantly (by 50%; Figure 7d). These results suggested that Runx2 and LRP6, especially the latter, are likely the targets of miR-30e. Deletion mutation of the target sequence was thus made within the LRP6 3′-UTR construct. The luciferase activity of the mutant construct was significantly higher than that of the WT construct after cotransfection with the miR-30e expression plasmid (Figure 7e). Furthermore, enhanced expression of miR-30e in 3T3-L1 cells led to a decrease in LRP6 and β-catenin protein in 3T3-L1 cells (Figure 7f). In contrast, suppression of endogenous miR-30e increased LRP6 and β-catenin protein levels (Figure 7g).
miR-30e directly targeted LRP6. The 3′-UTR fragments of Runx2, Wnt7b and LRP6 were PCR-amplified and cloned (a). The miR-30e miRNA overexpression in AD-293 slightly decreased the luciferase activity of Runx2 construct (b), whereas it had no impact on Wnt7b construct (c). The luciferase activity of LRP6 construct was dramatically decreased following miR-30e overexpression (d). Deletion of the miR-30e-binding sequence from the LRP6 3′-UTR construct increased the luciferase activity (e). Furthermore, miR-30e mimic transfection induced, and miR-30e inhibitor transfection reduced, LRP6 and β-catenin protein in 3T3-L1 cells (f and g). Values represent the mean±S.D. of three experiments. *Significant versus control, P<0.05
LRP6 negatively regulated adipogenesis
The data we have shown above suggested that LRP6 is a direct target of miR-30e. To further demonstrate that LRP6 controls adipogenesis, we carried out LRP6 loss-of-function studies using 3T3-L1 cells. The infection of the adenovirus that expresses the LRP6 RNAi sequences efficiently downregulated the endogenous LRP6 mRNA by 78% (Figure 8a). Western blotting showed that knockdown of LRP6 led to a striking decrease in β-catenin protein (Figure 8b). Moreover, Topflash assay revealed a dramatic decrease of luciferase activity following knockdown of LRP6 in 3T3-L1 cells (Figure 8c). These demonstrated that altered LRP6 expression may change bioactivity of the β-catenin/T-cell factor (TCF) signaling. Furthermore, in the presence of AIM, the knockdown lentivirus potentiated adipocyte differentiation, evidenced by the increased number of Oil Red O-positive adipocytes (Figures 8d and e). The mRNA expression levels of PPARγ, C/EBPα, C/EBPβ and aP2 and protein levels of PPARγ, C/EBPα and aP2 were enhanced (Figures 8f and g). These results demonstrated that LRP6 restrains adipogenesis and suggested that the appropriate level of LRP6 expression is critical for maintaining the balance of adipocyte and osteoblast formation from their common progenitor cells.
LRP6 negatively regulated adipogenesis. The infection of the LRP6 RNAi adenovirus downregulated the endogenous LRP6 mRNA (a) and decreased β-catenin protein in 3T3-L1 cells (b). The luciferase activity of the β-catenin/TCF reporter was reduced following knockdown of LRP6 in 3T3-L1 cells (c). In the presence of AIM, the knockdown of LRP6 increased adipocyte formation (d and e), induced the mRNA levels of PPARγ, C/EBPα, C/EBPβ and aP2 at 48 h (f) and the protein levels of PPARγ, C/EBPα and aP2 at 72 h (g). The Values represent the mean±S.D. of three experiments. *Significant versus control, P<0.05
Discussion
In the case of microarray screening, previous studies usually examined miRNA profilings after osteogenic treatment alone or adipogenic stimulation alone. However, it is well documented that adipogenic factors at the same time usually block osteoblast differentiation, and vice versa. The miRNAs that play a role in adipogenesis or osteogenesis might follow this regular pattern as well. Thus, we carried out adipogenic treatment and osteogenic stimulation together in our microarray analysis. The result showed that 14 miRNAs were upregulated during adipogenesis and at the same time downregulated during osteogenesis, and miR-30e was among the most significantly changed. These data suggest that the expression of miR-30e might act as a switch to balance the differentiation between osteoblasts and adipocytes.
Although the mature miRNA sequences of all the members of miR-30 family share a common 8-mer conserved seed sequence, the flanking sequences of the members are substantially different from each other. They are expressed by genes localized in different chromosomes, and thus are often differentially expressed and regulated during biological processes. Li et al.33 reported that the expression levels of miR-30b and miR-30c were significantly induced whereas the levels of miR-30a, miR-30d and miR-30e were not altered after γ-radiation in CD34+ cells. In the present study, the microarray and qPCR validation data also revealed a differentially regulated expression pattern with the miR-30 family members. miR-30c, miR-30d and miR-30e were increased during adipocyte differentiation and decreased during osteoblast differentiation. However, miR-30a and miR-30b were increased during adipogenesis but remained unchanged during osteoblastogenesis.
Our study for the first time demonstrated that miR-30e could alleviate cell growth of 3T3-L1 cells. In addition, we also found that the expression of miR-30e was increased during adipogenesis, whereas it was decreased during osteoblast differentiation. To delineate the precise role of miR-30e in differentiation, we tested the effect of miR-30e on adipocyte differentiation and osteoblast formation. Our results showed that both synthetic miR-30e mimics and lentivirally expressed miR-30e enhanced the formation of adipocytes. Conversely, knockdown of miR-30e blocked differentiation of adipocytes. Concomitantly, supplementing miR-30e mimics to preosteoblasts blocked osteoblast differentiation, whereas knockdown of miR-30e expression by synthetic inhibitor promoted osteoblast differentiation. Furthermore, miR-30e-expressing lentivirus enforced adipocyte formation and mitigated osteoblast differentiation from marrow stromal cells. These findings provided evidences that miR-30e maintains the differentiation balance between adipocytes and osteoblasts.
The miRNAs orchestrate osteoblast/adipocyte differentiation through their regulation of diverse signaling molecules and pathways that include bone morphogenetic protein 2 (BMP2)/Smad, Wnt/β-catenin and Runx2 signaling pathways.26, 30, 34, 35, 36, 37, 38, 39, 40 To date, only a small number of predicted targets have been experimentally validated, and hence further investigation is needed to clarify how these miRNAs function. In an attempt to elucidate the regulatory machinery underlying the modulation of adipocyte/osteoblast differentiation by miR-30e, we analyzed the potential targets under the assistance of TargetScan and PicTar. Three targets were predicted that include Runx2, Wnt7b and LRP6. Runx2 was previously confirmed to be posttranscriptionally regulated by miR-204/211 during osteoblast/adipocyte formation.26 In our study Runx2 3′-UTR construct showed a slightly reduced luciferase activity upon overexpression of miR-30e, suggesting that there might be other genes mediating the effects of adipogenesis promoted by miR-30e. In paralleled experiments using 3′-UTR constructs we demonstrated that LRP6 rather than Wnt7b was more potently inhibited by miR-30e, and therefore is a direct target of miR-30e. We assumed that LRP6 may probably govern adipogenesis and osteoblast formation.
LRP6, as well as LRP5, another coreceptor for Wnts, is a single-pass transmembrane protein belonging to the low-density lipoprotein receptor (LDLR)-related protein family. Along with Frizzled, another serpentine transmembrane protein, LRP5/6 binds to the Wnt signal and transmits the Wnt signal. When the Wnt signal is off, the APC–GSK3β–Axin complex degrades β-catenin by proteosome-mediated degradation. In the presence of Wnt ligands, a trimeric complex comprising Wnt, Frizzled and LRP5/6 initiates the Wnt signaling and prevents the degradation of cytoplasmic β-catenin. β-Catenin then translocates to nucleus and interacts with TCF/LEF to form a functional transcription factor and regulates the expression of Wnt target genes.
The human Wnt coreceptors LRP5 and LRP6 share 70% amino acid identity and exhibit a highly conserved domain structure. Both LRP5 and LRP6 are critical for bone formation. Humans and mice lacking LRP5 have low BMD. One allele deletion of LRP6 further decreased BMD in mice lacking LRP5.41 In humans, LRP6 has been shown to be involved in features of metabolic syndrome and osteoporosis, although the underlying mechanisms remain to be clarified.42 Given the fact that osteoblasts and adipocytes share common mesenchymal progenitor cells and canonical Wnt signaling balances the reciprocal relationship between osteoblastogenesis and adipogenesis, we assume that LRP6 is likely to be implicated in adipogenesis.
It is noteworthy that LRP6 mRNA remains relatively stable during adipocyte differentiation upon stimulation with the AIM (data not shown). However, the LRP6 3′-UTR construct harboring a conserved binding sequence for miR-30e was the most significantly altered among the three potential target genes upon ectopic expression of miR-30e in 293-AD cells. Removal of the putative binding sequence from the LRP6 3′-UTR construct abolished the inhibition of the luciferase activity by miR-30e overexpression. In contrast, miR-30e only slightly decreased the luciferase activity of Runx2 3′-UTR construct and had no effect on Wnt7b. Further functional investigations demonstrated that loss of function enforced the preadipocyte differentiation to mature adipocyte. Moreover, LRP6 knockdown attenuated β-catenin level and β-catenin/TCF reporter activity in 3T3-L1 cells, suggesting that regulation of adipogenesis is through β-catenin/TCF signaling. These data provide evidences that LRP6 may be required for cell-fate decision of osteoblast/adipocyte progenitor cells.
Taken together, our study demonstrated that miR-30e reciprocally regulates adipocyte and osteoblast differentiation by directly targeting the canonical Wnt/LRP6/β-catenin/TCF signaling. It also suggests that miR-30e could be a therapeutic target for osteoporosis because of its capacity to maintain the balance between adipogenesis and osteogenesis.
Materials and Methods
MicroRNA array analysis
Bone marrow cells were isolated from femurs and tibias of 4-week-old C57 mice as described previously43 and cultured with α-MEM supplemented with 10% FBS in a 10-cm dish. When cells reached ∼80% confluence, they were trypsinized and plated into 25 cm2 flasks at the density of 3 × 105 cells/ml in α-MEM containing 10% FBS. When the cells reached 80% confluence, osteoblast-inducing medium (OIM, α-MEM containing 10% FBS, 50 μg/ml ascorbic acid and 5 mmol/l β-glycerophosphate) was supplemented for 3 days to induce osteoblast differentiation. For adipocyte differentiation, the cells were cultured in AIM (α-MEM containing 10% FBS, 0.5 μM dexamethasone, 0.25 mM methylisobutylxanthine, 5 μg/ml of insulin, and 50 μM indomethacin) for 3 days. Total RNA was extracted from the cells by using the Ambion mirVana miRNA Isolation Kit (Ambion, Austin, TX, USA). The expression profiles of miRNAs were analyzed using TaqMan Array Rodent MicroRNA A+B Cards Set v3.0. The miRNAs were amplified after specific reverse transcription and preamplification using Megaplex Assay Performance (Megaplex RT Primer Pools and Megaplex PreAmp pools, both from Applied Biosystems, Foster City, CA, USA), and normalized against RNU6B (U6 snRNA, an endogenous control assay designed in both cards). Relative expression was determined using the ΔΔCt method and a ≥32 Ct value was interpreted as amplification too low to quantify.
Quantitative RT-PCR
To confirm the reproducibility of miRNA expression profiles examined by miRNA array analysis, we measured their expression levels by RT-PCR using a miRNA reverse transcription kit and qPCR detection kit (Genecopoeia, Germantown, MD, USA). Briefly, RNA was extracted using miRNeasy columns (Qiagen, Valencia, CA, USA). RNA (1 μg) was first polyadenylated by poly(A) polymerase and then reverse-transcribed into cDNA using oligo-dT primer with a universal tag. Subsequently, miRNAs were PCR-amplified on a real-time PCR system using specific mature miRNA sequences (ordered from Genecopoeia) as forward primers and a universal reverse primer provided in the kit. The expression levels were normalized to U6, and measured by the comparative Ct (ΔΔCt) method. The qRT-PCR consisted of 40 cycles (95°C for 10 s, 60°C for 10 s and 72°C for 10 s) after an initial denaturation step (95°C for 2 min).
For mRNA expression analysis, total cellular RNA was prepared by a RNA isolation kit (Omega Bio-Tek, Norcross, GA, USA) according to the manufacturer’s protocol. Total RNA (1 μg) was used to synthesize cDNA using a first-strand cDNA Synthesis Kit (Thermo Scientific, Rockford, IL, USA). Real-time PCR amplifications were performed using gene specific primers and SYBR green real-time PCR kit (Thermo Scientific). The primers used were: PPARγ, Fw: 5′-CTTGACA GGAAAGACAACGG-3′; Rev: 5′-GCTTCTACGGATCGAAACTG-3′; C/EBPα, Fw: 5′-CTGATTCTTGCCAAACTGAG-3′; Rev: 5′-GAGGAAGCTAAGACCCACTAC-3′; C/EBPβ, Fw: 5′-TCACTTAAAGATGTTCCTGC-3′; Rev: 5′-TGTCTTCACTTTAATGCTCG-3′; aP2, Fw: 5′-AAATCACCGCAGACGACAGG-3′; Rev: 5′-GGCTCATGCCCTTTCATAAAC-3′; Osx, Fw: 5′-TCTCTCCATCTGCCTGACTC-3′; Rev: 5′-GTCAGCGTATGGCTTCTTTG-3′; ALP, Fw: 5′-GTTGGGGGT GCCCACGGT-3′; Rev: 5′-CCTTGGACAGAGCCATGTATG-3′; Runx2, Fw: 5′-TCCTGTAGATCCGAGCACCA-3′; Rev, 5′-CTGCTGCTGTTGTTGCTGTT-3′; Osteocalcin, Fw: 5′-TCTGACAAACCTTCATGTCC-3′, Rev: 5′-AAATAGTGATACCGTAGATGCG-3′; BSP, Fw: 5′-GAGCCAGGACTGCCGAAAGGA A-3′, Rev: 5′-CCGTTGTCTCCTCCGCTGCTGC-3′; and β-actin, Fw: 5′-TCAGGTTACTGG TTCGGTCTG-3′, Rev: 5′-ACCAGAGGCATACAGGGACAG-3′. The thermal cycling conditions were: an initial denaturation at 95°C for 7 min, followed by 40 cycles of 20 s of denaturing at 95°C, 20 s of annealing at 57°C and 20 s of extension at 72°C. The levels of the target gene expression were normalized to that of β-actin.
miRNA target site prediction
A search for predicted target mRNAs was performed using the data bases TargetScan (http://www.targetscan.org/) and PicTar (http://pictar.mdc-berlin.de/). Sequence conservation was examined using University of California Santa Cruz genome browser (http://genome.ucsc.edu/).
Cell growth analysis
Cells were plated in 96-well plates at 104 per well and were transfected with miRNAs. After transfection, the cells were cultured for 24 h. The effect of miR-30e on cell growth and viability was determined by the methylthiazolyldiphenyl-tetrazolium bromide (MTT) assay as described previously.44
Transfections
The synthetic mimics or inhibitors of miR-30e (Ribobio, Guangzhou, China) were transfected into 3T3-L1 or MC3T3-E1 cultures using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) to promote or inhibit miR-30e activity, respectively. NCs were used for both reactions. The final concentration of mimics and inhibitors was 75 and 100 nM, respectively. At 5 h after transfection, the transfection solution was replaced with complete culture medium. The cultures were supplemented with OIM when the cells reached 70% confluence to induce osteoblast differentiation, or with AIM when the cells reached 100% confluence to induce adipocyte differentiation.
Constructs and luciferase reporter assay
The luciferase-3′-UTR reporter constructs were generated by introducing the 3′-UTRs of the genes carrying the putative miR-30e-binding site into a modified pGL3-control vector (Promega, San Luis Obispo, CA, USA). We first amplified the 3′-UTR sequences by PCR using specific primers and mouse DNA as a template. The primers used were: LRP6, Fw: 5′-ACATATGTTTTACCTTAACAGGTGTCG-3′; Rev: 5′-GATATCTTAAATACTGCCAACCAACTGATAC-3′; Wnt7b, Fw: 5′-AGAATTCCGCCATTATTTTGCACATCC-3′; Rev: 5′-AGATATCCCCAGTGTAGGTCACAATAAC-3′; and Runx2, Fw: 5′-ACATATGCACAAGTGATTGGTTGAAC-3′; Rev: 5′- GATATCAGAGGAAGAAGAAGAAGACGAC-3′. The PCR products were first cloned into a PCR cloning vector pEasy T1-simple (Transgen, Beijing, China) and then digested using NdeI/EcoRV (for LRP6, Runx2) or EcoRI/EcoRV (for Wnt7b) restriction enzymes. The resultant fragments were subcloned, respectively, into a modified pGL3-control vector where NdeI, EcoR1 and EcoRV sites were introduced into the original XbaI site. Deletion of the putative miR-30e-binding site in the LRP6 3′-UTR was made by using the Quikchange II XL kit (Stratagene, La Jolla, CA, USA). All the inserts were verified by DNA sequencing. The 293-AD cells were cotransfected with the miR-30e lentiviral expression plasmid (described below) or the empty plasmid along with the 3′-UTR construct or its mutant in 24-well plates. The Renilla luciferase reporter plasmid pRL-SV40 (10 ng) was also included in the cotransfection mixture to monitor the transfection efficiency. Then, the cells were harvested and lysed for luciferase assay 36 h after transfection using a dual-luciferase reporter assay kit (Promega). The relative firefly luciferase activity was calculated by normalization to the Renilla luciferase activity.
In some experiments, 3T3-L1 cells were infected with LRP6 RNAi virus or a NC virus, followed by cotransfection with pRL-SV40, along with either Wnt signaling luciferase reporter construct Topflash or its NC, Fopflash (Addgene, Cambridge, MA, USA) using Lipofectamine 2000. Luciferase assays were performed 36 h after transfection.
Lentiviral, adenoviral construct packaging and infection
The miR-30e precursor sequence was PCR-amplified using the mouse DNA as the template. The primers were: Fw: 5′-CGAATTCAGAAGTGGGCATCCTAATTG-3′; Rev: 5′-AGGATCCAAACGAAGAGAGACAGTC-3′. The PCR fragment was restricted with EcoRI/BamHI enzymes and then cloned into the lentiviral vector pCDH-CMV-MCS-EF1-copGFP (System Biosciences, Mountain View, CA, USA).
The lentivirus was packaged by cotransfecting 293T cells with the above-mentioned lentiviral transfer plasmids, packaging (GAG/Pol and REV) plasmids and envelope (VSV-G) plasmid using the lentiviral packaging system (Jiman Biotech, Shanghai, China). The medium was changed 18 h after transfection and supernatant containing the virus was harvested, concentrated and titered (multiplicity of infection (MOI)) by analyzing the frequency of GFP-positive cells in a known number of 293T cells infected with serially diluted viral supernatant. The virus was used at an MOI of 20 to infect 3T3-L1 or mouse marrow stromal cells. The efficiency of infection was determined at 48 h by the number of GFP-positive cells that exceeded 90%. The cells were then treated with AIM to induce adipocyte differentiation or with OIM to induce osteoblast differentiation. The lentivirus packaged with the empty pCDH vector served as a control.
For the knockdown of LRP6 we used adenovirus system for shRNA expression. The shRNA sequence designed to target mouse LRP6 mRNA was: 5′-CGCACTACATTAGTTCCAAAT-3′. The coding sequence of the shRNA was cloned into plasmid pGensil-1, and then the LRP6 shRNA expression cassette driven by the hU6 promoter was excised and ligated into pGStrack-EGFP. The resultant shuttle plasmid was recombined with the pGSadeno vector, resulting in the adenoviral construct, which was further packaged and amplified in 293-AD cells. After several rounds of enrichment, high-titer adenovirus was collected and used at an MOI of 20 to infect 3T3-L1 cells.
ALP staining
The cells were cultured for 7 days (MC3T3-E1) or 14 days (marrow stromal stem cells) followed by ALP staining as described in previous studies.45 In brief, for ALP staining, cells were fixed in 10% formalin for 10 min and stained using 1-Step NBT/BCIP staining kit (Pierce, Thermo Scientific, Rockford, IL, USA) for 15 min. The cultures were then rinsed several times with water.
Oil red O staining
The 3T3-L1 cells transfected with the synthetic miRNA mimics or inhibitors were cultured in six-well plates for 3 days in the presence or absence of AIM (2/3 concentration for stimulatory studies or full concentration for inhibitory studies) as mentioned above followed by treatment of insulin alone for one more day. After incubation, the cells were gently washed twice with phosphate-buffered saline (PBS), and then fixed in 4% paraformaldehyde for 10 min. The samples were then washed twice with deionized water, and 60% saturated Oil Red O staining was carried out for 5 min. For Oil Red O quantification, 4% IGEPAL CA 630 (Sigma, St. Louis, MO, USA) in isopropanol was added to each well. Light absorbance by the extracted dye was measured at 510 nm.
Western blot analysis
Cells were lysed on ice for 30 min in a buffer containing 50 mM Tris-HCL pH 7.4, 150 mM NaCl, 1% Nonidet P-40 and 0.1% SDS supplemented with protease inhibitors (10 μg/ml leupeptin, 10 μg/ml pepstatin A and 10 μg/ml asprotinin) and phosphatase inhibitors (1 mM NaF and 1 mM Na3VO4). Proteins were fractionated by SDS-PAGE and transferred to nitrocellulose membranes. The membranes were blocked with 5% skim milk for 1 h and incubated with rabbit monoclonal anti-C/EBPα, anti-LRP6, anti-β-catenin (Abcam, Cambridge, MA, USA), mouse monoclonal anti-Runx2 (MBL, Woburn, MA, USA), rabbit polyclonal anti-osterix (Abcam), anti-ALP (Abgent, San Diego, CA, USA), anti-PPARγ, anti-aP2 or anti-β-actin antibodies (Proteintech, Wuhan, China). This was then followed by incubation with the corresponding horseradish peroxide-labeled IgG (1 : 5000) for 1 h. Finally, chemiluminescence reagent (Advansta, Menlo Park, CA, USA) was used to visualize the results, and β-actin was used as an internal control.
Statistical analysis
Data are expressed as mean±S.D. Statistical analysis was performed using the independent t-test. A P-value of <0.05 was considered to be statistically significant.
Abbreviations
- ALP:
-
alkaline phosphatase
- BMP2:
-
bone morphogenetic protein 2
- BSP:
-
bone sialoprotein
- C/EBP:
-
CCAAT/enhancer binding protein
- LRP6:
-
low-density lipoprotein receptor-related protein 6
- Osx:
-
osterix
- PPARγ:
-
peroxisome proliferator-activated receptor γ
- PBS:
-
phosphate-buffered saline
- Runx2:
-
runt-related transcription factor 2
- TCF:
-
T-cell factor
- MTT:
-
methylthiazolyldiphenyl-tetrazolium bromide
- miRNA:
-
microRNA
- MOI:
-
multiplicity of infection
References
Nuttall ME, Gimble JM . Controlling the balance between osteoblastogenesis and adipogenesis and the consequent therapeutic implications. Curr Opin Pharmacol 2004; 4: 290–294.
Gimble JM, Zvonic S, Floyd ZE, Kassem M, Nuttall ME . Playing with bone and fat. J Cell Biochem 2006; 98: 251–266.
Monroe DG, McGee-Lawrence ME, Oursler MJ, Westendorf JJ . Update on Wnt signaling in bone cell biology and bone disease. Gene 2012; 492: 1–18.
Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, Rosewell IR et al. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell 1997; 89: 765–771.
Nakashima K, Zhou X, Kunkel G, Zhang Z, Deng JM, Behringer RR et al. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell 2002; 108: 17–29.
Ryoo HM, Hoffmann HM, Beumer T, Frenkel B, Towler DA, Stein GS et al. Stage-specific expression of Dlx-5 during osteoblast differentiation: involvement in regulation of osteocalcin gene expression. Mol Endocrinol 1997; 11: 1681–1694.
Tontonoz P, Hu E, Spiegelman BM . Stimulation of adipogenesis in fibroblasts by PPAR gamma 2, a lipid-activated transcription factor. Cell 1994; 79: 1147–1156.
Siersbaek R, Nielsen R, Mandrup S . Transcriptional networks and chromatin remodeling controlling adipogenesis. Trends Endocrinol Metab 2012; 23: 56–64.
Lin FT, Lane MD . CCAAT/enhancer binding protein alpha is sufficient to initiate the 3T3-L1 adipocyte differentiation program. Proc Natl Acad Sci USA 1994; 91: 8757–8761.
Zanotti S, Stadmeyer L, Smerdel-Ramoya A, Durant D, Canalis E . Misexpression of CCAAT/enhancer binding protein beta causes osteopenia. J Endocrinol 2009; 201: 263–274.
Akune T, Ohba S, Kamekura S, Yamaguchi M, Chung UI, Kubota N et al. PPARgamma insufficiency enhances osteogenesis through osteoblast formation from bone marrow progenitors. J Clin Invest 2004; 113: 846–855.
Zhang J, Fu M, Cui T, Xiong C, Xu K, Zhong W et al. Selective disruption of PPARgamma 2 impairs the development of adipose tissue and insulin sensitivity. Proc Natl Acad Sci USA 2004; 101: 10703–10708.
Liu LF, Shen WJ, Zhang ZH, Wang LJ, Kraemer FB . Adipocytes decrease Runx2 expression in osteoblastic cells: roles of PPARgamma and adiponectin. J Cell Physiol 2010; 225: 837–845.
Freytag SO, Paielli DL, Gilbert JD . Ectopic expression of the CCAAT/enhancer-binding protein alpha promotes the adipogenic program in a variety of mouse fibroblastic cells. Genes Dev 1994; 8: 1654–1663.
Linhart HG, Ishimura-Oka K, DeMayo F, Kibe T, Repka D, Poindexter B et al. C/EBPalpha is required for differentiation of white, but not brown, adipose tissue. Proc Natl Acad Sci USA 2001; 98: 12532–12537.
Wang ND, Finegold MJ, Bradley A, Ou CN, Abdelsayed SV, Wilde MD et al. Impaired energy homeostasis in C/EBP alpha knockout mice. Science 1995; 269: 1108–1112.
Tanaka T, Yoshida N, Kishimoto T, Akira S . Defective adipocyte differentiation in mice lacking the C/EBPbeta and/or C/EBPdelta gene. EMBO J 1997; 16: 7432–7443.
Smink JJ, Leutz A . Instruction of mesenchymal cell fate by the transcription factor C/EBPbeta. Gene 2012; 497: 10–17.
Lee EK, Lee MJ, Abdelmohsen K, Kim W, Kim MM, Srikantan S et al. miR-130 suppresses adipogenesis by inhibiting peroxisome proliferator-activated receptor gamma expression. Mol Cell Biol 2011; 31: 626–638.
Tang YF, Zhang Y, Li XY, Li C, Tian W, Liu L . Expression of miR-31, miR-125b-5p, and miR-326 in the adipogenic differentiation process of adipose-derived stem cells. OMICS 2009; 13: 331–336.
Sakaue H, Ogawa W, Matsumoto M, Kuroda S, Takata M, Sugimoto T et al. Posttranscriptional control of adipocyte differentiation through activation of phosphoinositide 3-kinase. J Biol Chem 1998; 273: 28945–28952.
Bartel DP . MicroRNAs: target recognition and regulatory functions. Cell 2009; 136: 215–233.
Skarn M, Namlos HM, Noordhuis P, Wang MY, Meza-Zepeda LA, Myklebost O . Adipocyte differentiation of human bone marrow-derived stromal cells is modulated by microRNA-155, microRNA-221, and microRNA-222. Stem Cells Dev 2012; 21: 873–883.
Zhang JF, Fu WM, He ML, Wang H, Wang WM, Yu SC et al. MiR-637 maintains the balance between adipocytes and osteoblasts by directly targeting Osterix. Mol Biol Cell 2011; 22: 3955–3961.
Qin L, Chen Y, Niu Y, Chen W, Wang Q, Xiao S et al. A deep investigation into the adipogenesis mechanism: profile of microRNAs regulating adipogenesis by modulating the canonical Wnt/beta-catenin signaling pathway. BMC Genomics 2010; 11: 320.
Huang J, Zhao L, Xing L, Chen D . MicroRNA-204 regulates Runx2 protein expression and mesenchymal progenitor cell differentiation. Stem Cells 2010; 28: 357–364.
Ouzounova M, Vuong T, Ancey PB, Ferrand M, Durand G, Le-Calvez Kelm F et al. MicroRNA miR-30 family regulates non-attachment growth of breast cancer cells. BMC Genomics 2013; 14: 139.
Bridge G, Monteiro R, Henderson S, Emuss V, Lagos D, Georgopoulou D et al. The microRNA-30 family targets DLL4 to modulate endothelial cell behavior during angiogenesis. Blood 2012; 120: 5063–5072.
Amar L, Benoit C, Beaumont G, Vacher CM, Crepin D, Taouis M et al. MicroRNA expression profiling of hypothalamic arcuate and paraventricular nuclei from single rats using Illumina sequencing technology. J Neurosci Methods 2012; 209: 134–143.
Wu T, Zhou H, Hong Y, Li J, Jiang X, Huang H . miR-30 family members negatively regulate osteoblast differentiation. J Biol Chem 2012; 287: 7503–7511.
Eguchi T, Watanabe K, Hara ES, Ono M, Kuboki T, Calderwood SK . OstemiR: a novel panel of microRNA biomarkers in osteoblastic and osteocytic differentiation from mesencymal stem cells. PLoS One 2013; 8: e58796.
Zaragosi LE, Wdziekonski B, Brigand KL, Villageois P, Mari B, Waldmann R et al. Small RNA sequencing reveals miR-642a-3p as a novel adipocyte-specific microRNA and miR-30 as a key regulator of human adipogenesis. Genome Biol 2011; 12: R64.
Li XH, Ha CT, Fu D, Xiao M . Micro-RNA30c negatively regulates REDD1 expression in human hematopoietic and osteoblast cells after gamma-irradiation. PLoS One 2012; 7: e48700.
Hu R, Liu W, Li H, Yang L, Chen C, Xia ZY et al. A Runx2/miR-3960/miR-2861 regulatory feedback loop during mouse osteoblast differentiation. J Biol Chem 2011; 286: 12328–12339.
Kim EJ, Kang IH, Lee JW, Jang WG, Koh JT . MiR-433 mediates ERRgamma-suppressed osteoblast differentiation via direct targeting to Runx2 mRNA in C3H10T1/2 cells. Life Sci 2013; 92: 562–568.
Hassan MQ, Maeda Y, Taipaleenmaki H, Zhang W, Jafferji M, Gordon JA et al. miR-218 directs a Wnt signaling circuit to promote differentiation of osteoblasts and osteomimicry of metastatic cancer cells. J Biol Chem 2012; 287: 42084–42092.
Zhang Y, Xie RL, Gordon J, LeBlanc K, Stein JL, Lian JB et al. Control of mesenchymal lineage progression by microRNAs targeting skeletal gene regulators Trps1 and Runx2. J Biol Chem 2012; 287: 21926–21935.
Kapinas K, Kessler C, Ricks T, Gronowicz G, Delany AM . miR-29 modulates Wnt signaling in human osteoblasts through a positive feedback loop. J Biol Chem 2010; 285: 25221–25231.
Luzi E, Marini F, Sala SC, Tognarini I, Galli G, Brandi ML . Osteogenic differentiation of human adipose tissue-derived stem cells is modulated by the miR-26a targeting of the SMAD1 transcription factor. J Bone Miner Res 2008; 23: 287–295.
Li Z, Hassan MQ, Volinia S, van Wijnen AJ, Stein JL, Croce CM et al. A microRNA signature for a BMP2-induced osteoblast lineage commitment program. Proc Natl Acad Sci USA 2008; 105: 13906–13911.
Holmen SL, Giambernardi TA, Zylstra CR, Buckner-Berghuis BD, Resau JH, Hess JF et al. Decreased BMD and limb deformities in mice carrying mutations in both Lrp5 and Lrp6. J Bone Miner Res 2004; 19: 2033–2040.
Mani A, Radhakrishnan J, Wang H, Mani MA, Nelson-Williams C, Carew KS et al. LRP6 mutation in a family with early coronary disease and metabolic risk factors. Science 2007; 315: 1278–1282.
Anjos-Afonso F, Bonnet D . Isolation, culture, and differentiation potential of mouse marrow stromal cells. Curr Protoc Stem Cell Biol 2008; Chapter 2: Unit 2B 3.
Tsang WP, Kwok TT . The miR-18a* microRNA functions as a potential tumor suppressor by targeting on K-Ras. Carcinogenesis 2009; 30: 953–959.
Zhao M, Qiao M, Harris SE, Oyajobi BO, Mundy GR, Chen D . Smurf1 inhibits osteoblast differentiation and bone formation in vitro and in vivo. J Biol Chem 2004; 279: 12854–12859.
Acknowledgements
This work was supported by grants81071436 and 81271977 to Baoli Wang from Natural Science Foundation of China and by grant 11JCZDJC16500 to Baoli Wang from Tianjin Municipal Natural Science Foundation, China. Xiaoxia Li was partially supported by grant 81101596 from Natural Science Foundation of China.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by A Stephanou
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Wang, J., Guan, X., Guo, F. et al. miR-30e reciprocally regulates the differentiation of adipocytes and osteoblasts by directly targeting low-density lipoprotein receptor-related protein 6. Cell Death Dis 4, e845 (2013). https://doi.org/10.1038/cddis.2013.356
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cddis.2013.356
Keywords
This article is cited by
-
The role of wnt signaling in diabetes-induced osteoporosis
Diabetology & Metabolic Syndrome (2023)
-
miRNAs as the important regulators of myasthenia gravis: involvement of major cytokines and immune cells
Immunologic Research (2023)
-
MiR-422a promotes adipogenesis via MeCP2 downregulation in human bone marrow mesenchymal stem cells
Cellular and Molecular Life Sciences (2023)
-
Regulatory of miRNAs in tri-lineage differentiation of C3H10T1/2
Stem Cell Research & Therapy (2022)
-
miR-34a regulates adipogenesis in porcine intramuscular adipocytes by targeting ACSL4
BMC Genetics (2020)