Abstract
Senescence is a cellular response to damage and stress. The senescence response prevents cancer by suppressing the proliferation of cells with a compromised genome and contributes to optimal wound healing in normal tissues. Persistent senescent cells are also thought to drive aging and age-associated pathologies through their secretion of inflammatory factors that modify the tissue microenvironment and alter the function of nearby normal or transformed cells. Understanding how senescent cells alter the microenvironment would be aided by the ability to induce or eliminate senescent cells at will in vivo. Here, we combine the use of the synthetic nucleoside analog ganciclovir (GCV) with herpes simplex virus thymidine kinase (HSVtk) activity to create or eliminate senescent human cells. We show that low concentrations of GCV induce senescence through the accumulation of nuclear DNA damage while higher concentrations of GCV, similar to those used in vivo, kill non-dividing senescent cells via mitochondrial DNA (mtDNA) damage and caspase-dependent apoptosis. Using this system, we effectively eliminated xenografted normal human senescent fibroblasts or induced senescence in human breast cancer cells in vivo. Thus, cellular senescence and mtDNA damage are outcomes of synthetic nucleoside analog treatment, indicating that the GCV–HSVtk combination can be used effectively to promote the targeted formation or eradication of senescent cells.
Similar content being viewed by others
Main
Cellular senescence suppresses cancer by halting the proliferation of damaged cells that are at risk for malignant progression.1, 2, 3, 4 First formally described in the 1960s,5 cellular senescence has been linked to two important phenomena in complex organisms: tumor suppression and aging. It is now clear that cellular senescence is an essential tumor-suppressive mechanism, rivaling apoptosis in importance.1, 2 More recently, the senescence response was shown to participate in tissue repair and occurs after DNA-damaging cancer therapies, potentially contributing to the success and side effects of these treatments.4, 6, 7, 8
The senescence growth arrest is tumor suppressive, because it prevents the expansion of cells that are potentially oncogenic. How cellular senescence contributes to aging or tissue repair remains somewhat unclear. The senescence response might drive aging by depleting progenitor cell pools. Additionally, the ability of senescent cells to secrete inflammatory molecules might contribute to tissue repair, but persistent senescent cells might drive chronic inflammation, a major risk factor for age-related pathologies.6 Most senescent cells express the p16INK4a tumor-suppressor protein, which also increases with age in mouse and human tissues.9, 10, 11 p16INK4a expression correlates well with other senescence markers, such as senescence-associated beta galactosidase (SA-βgal) activity.12 The expression of both biomarkers often coincides in vivo in mice and humans.9, 11 Based on the expression of these two biomarkers, there is substantial evidence that senescent cells accumulate with age in vertebrate tissues; moreover, senescent cells are evident at sites of numerous age-related pathologies, such as atherosclerosis, osteoarthritis and precancerous lesions.13, 14, 15, 16, 17, 18
Recent data show that most senescent cells eventually disappear in mice, with evidence of tissue infiltration by innate immune cells, suggesting that senescent cells are cleared by the immune system.19, 20, 21 Similarly, senescent cells induced during wound repair appear to be cleared by the immune system.22, 23 Nonetheless, senescent cells accumulate with age and pathology and are presumed to resist immune clearance by as yet unknown mechanisms. Importantly, in a mouse model of accelerated aging, elimination of senescent cells prevented certain late-life pathologies, lending strong credibility to the hypothesis that senescent cells contribute to aging phenotypes.24
Understanding the nature and functions of senescent cells during wound repair, cancer therapy and aging requires new tools to modulate and visualize the senescence response in tissues. When targeted to senescent cells, for example, using senescence-associated promoters, these tools could allow the detection of senescent cells in tissues and living mice and/or their specific induction or elimination in a regulated fashion. Recently, mouse models were developed in which the p16INK4a promoter drove expression of a transgene that caused apoptosis or luciferase expression in senescent cells in vivo.24, 25
As another step toward developing approaches to study/target senescent cells, we show here that ganciclovir C9H5N13O4 (GCV) and the herpes simplex virus thymidine kinase (HSVtk) can be used to both induce and kill senescent cells in vivo. GCV is a synthetic nucleoside analog widely used to treat herpes viral infections in humans.26 The viral thymidine kinase binds GCV with high affinity and converts it to a toxic nucleotide after incorporation into the viral and nuclear DNA of infected cells, thereby promoting apoptotic cell death.27 The usefulness of the GCV–HSVtk combination has a long history in cancer gene therapy28 and is currently a potential therapeutic tool when incorporated into oncolytic viruses.29, 30, 31
Here we show that, in response to low doses of GCV, normal and cancer cells expressing HSVtk undergo senescence due to nuclear DNA damage. In response to higher GCV doses, normal and senescent cells expressing HSVtk undergo apoptosis as a consequence of mitochondrial DNA (mtDNA) damage. Thus, the GCV–HSVtk combination can be used as a multifunctional inducer or killer of senescent cells.
Results
GCV induces nuclear DNA damage and cell death in human fibroblasts
We used a lentiviral vector to constitutively express the trimodal reporter-killer fusion protein (3MR) in primary human foreskin fibroblasts (HCA2). 3MR contains monomeric red fluorescent protein (mRFP), Renilla luciferase (rLUC), and a functional fragment of the HSVtk.32 Following infection and selection, 3MR expression was confirmed by mRFP fluorescence and rLUC immunofluorescence (Supplementary Figure S1).
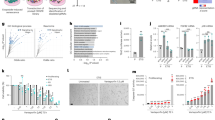
With increasing concentrations of GCV, 3MR-expressing cells reduced proliferation or cell number, proportional to the dose of GCV (0–1 μg/ml) (Figure 1a). At low doses (0.001–0.01 μg/ml), GCV retarded or arrested proliferation. At higher doses (≥0.1 μg/ml), GCV caused cell loss (Figure 1a). Cell loss entailed an accumulation of cells with a sub-G1 DNA content (Figure 1b), suggesting apoptotic cell death. We confirmed apoptosis using Annexin V—DAPI live-dead staining, which showed increased numbers of early and late apoptotic cells (Figure 1c). Intermediate GCV doses (0.01–0.1 μg/ml) increased DNA double-strand breaks, as measured by nuclear 53BP1 DNA damage foci, detectable by immunofluorescence (Figure 1d).33, 34 53BP1 foci increased from low levels, typical of proliferating primary cells, to ∼50% of cells harboring at least three 53BP1 foci (Figure 1d). 53BP1 foci and apoptosis caused by GCV occurred specifically in 3MR-expressing cells but not in control cells in which GCV was non-toxic up to ∼10 μg/ml (Figures 1d–f and Supplementary Figure S2).
GCV-induced nuclear DNA damage triggers cellular senescence or death. (a) HCA2 cells expressing 3MR were given the indicated concentrations of GCV 48 h after plating; media+or −GCV was replenished every 2 days. Cell number was determined 5 and 12 days later and is plotted as the log of the number of cell divisions. (b) HCA2-3MR cells were treated as in (a) and collected 7 days after GCV addition for cell-cycle analysis by flow cytometry. Cell-cycle distribution is plotted as the percentage of cells in each phase of the cell cycle for each concentration of GCV. (c) HCA2-3MR cells were treated as in (a) and collected 4 days after GCV treatment for live-dead apoptosis analysis using flow cytometry. Representative samples are shown and the data are plotted in the right graph as the percentage of cells in the live (Q4), early apoptotic (Q3) or late apoptotic/necrotic (Q1 and Q2) quadrants for each concentration of GCV. (d) HCA2 or HCA2-3MR cells were plated on glass slides and fixed for immunofluorescence following 48 h treatment with the indicated doses of GCV. Left panels: 53BP1 (green) shows both diffuse nuclear staining and punctate focal staining (DNA damage foci). Nuclear DNA was stained with DAPI (4,6-diamidino-2-phenylindole; blue). The red arrowheads show cells with ≥3 53BP1 foci. Right panel: The percentage of nuclei with ≥3 53BP1 foci was determined in at least 200 cells per GCV concentration. Light gray: HCA2-control cells; dark gray: HCA2-3MR cells. (e) HCA2 or HCA2-3MR cells were plated and counted 24 h later (day 0) and 7 days following GCV addition at the indicated concentrations (media+or −GCV were replenished every 2 days). (f) HCA2 cells were treated as in (a) and collected 4 days after GCV treatment for live-dead apoptosis analysis by flow cytometry. Scatter plots are shown in Supplementary Figure S2A. The data is plotted as percentage of cells in the live (Q4), early apoptotic (Q3) or late apoptotic/necrotic (Q1 and Q2) quadrants for each concentration of GCV
GCV induces senescence and persistent DNA damage foci
Low concentrations of GCV (0.01 μg/ml) induced cellular senescence in nearly all 3MR-expressing fibroblasts, as determined by an absence of cell loss (Figure 1a) and a dose-dependent increase in the percentage of cells expressing SA-βgal activity (Figure 2a). At higher concentrations (0.1–1 μg/ml), most cells detached from the culture dish (Figures 1a and b), although the few cells that remained attached were almost all SA-βgal-positive (Figure 2a). Moreover, 3MR-expressing cells treated with GCV failed to replicate DNA (Figure 2b), as expected of growth-arrested senescent cells. Further, 53BP1 foci induced by sustained GCV treatment of 3MR-expressing cells were persistent, a feature of DNA-SCARS (DNA segments with chromatin alterations reinforcing senescence) and a hallmark of senescent cells.34 Thus, removal of GCV for 3 days following an initial 9-day treatment did not reduce the number of 53BP1 foci, supporting the conclusion that GCV can induce senescence in 3MR-expressing cells, with accompanying persistent DNA-SCARS34 (Figure 2c). Finally, GCV-induced senescence in 3MR-expressing cells entailed increased levels of SA-βgal, as well as increased levels of the p53-regulated cell-cycle inhibitor p21WAF1 (Supplementary Figures S3A and B).
GCV induces senescence through DNA damage accumulation. (a) Top panel: HCA2-3MR cells were given the indicated concentrations of GCV 48 h after plating; media+or −GCV were replenished every 2 days. Cells were fixed and SA-βGal staining (blue) was performed 10 days later. Lower panel: The fraction of SA-βGal-positive cells was quantified and plotted as the percentage of positive cells per GCV concentration. (b) EdU (5-ethynyl-2′-deoxyuridine) DNA labeling indexes were determined in cells treated with GCV. Light gray: HCA2-control cells; dark gray: HCA2-3MR cells. Cells were seeded on glass slides and given the indicated concentrations of GCV 48 h after plating for 7 days (media+or −GCV were replenished every 2 days). EdU was added from day 7 to day 8 (24 h pulse) and the percentage of EdU-positive cells was determined in at least 200 cells per GCV concentration. (c) Persistent DNA damage foci were determined in cells treated with GCV. Light gray: HCA2-3MR cells treated with GCV for 9 days; dark gray: HCA2-3MR cells treated similarly and chased for 3 days in GCV-free media. Cells on glass slides were given the indicated concentrations of GCV 48 h after plating for 9 or 12 days (media+or −GCV were replenished every 2 days). The percentage of cells with ≥3 persistent 53BP1 nuclear foci (DNA-SCARS) was determined as described in Figure 1d
GCV induces caspase-dependent apoptosis in senescent cells
Upon phosphorylation by HSVtk, GCV is known to kill dividing cells by incorporating into nuclear DNA, causing replication-dependent double strand breaks.35, 36 Although doses of GCV between 0.1 and 1 μg/ml induced senescence (growth arrest) of surviving 3MR-expressing cells (Figures 1 and 2), these growth-arrested cells continued to die over longer intervals (Figure 1a). To directly test whether GCV could kill non-dividing senescent cells, we induced senescence in control and 3MR-expressing cells using X-irradiation (10 Gy; Supplementary Figure S3),18, 33, 34 and measured the number of senescent cells that remained attached to the culture dish following GCV treatment (Figure 3a). Despite their non-dividing status, senescent 3MR-expressing cells showed substantial detachment between 4 and 9 days following GCV treatment. Similarly to proliferating cells, GCV killed these senescent cells by apoptosis, as determined by Annexin V—DAPI live-dead staining (Figures 3b and c).
GCV induces caspase-dependent apoptosis in senescent cells. (a) HCA2 and HCA2-3MR cells were X-irradiated to induce senescence. Three days later, cells were given the indicated concentrations of GCV and counted 4 and 9 days following GCV addition. (b and c) HCA2-3MR cells were rendered senescent as in (a). Eight days after irradiation, cells were treated with the indicated GCV doses and collected 4 days later for live-dead apoptosis analysis by flow cytometry. Representative scatter plots are displayed in (b). The data are plotted as the percentage cells in the live (Q4), early apoptotic (Q3) or late apoptotic/necrotic (Q1 and Q2) quadrants for each concentration of GCV in (c). (d) HCA2-3MR cells were made senescent as in (a). Seven days after irradiation, cells were treated with 10 μg/ml of GCV in the presence of a caspase inhibitor. Cells were immunostained for cytochrome c (green) and 53BP1 2 and 4 days later. Nuclei were counterstained for 53BP1 (blue; contrast was increased to highlight nucleoplasmic 53BP1). Red arrowheads denote cells with diffuse cytoplasmic cytochrome c staining. (e) Quantitation of the percentage of cells with diffuse cytoplasmic cytochrome c staining 2, 3 and 4 days after GCV treatment. FITC, fluorescein isothiocyanate; GCV, ganciclovir
To confirm apoptotic cell death, we used the sensitive CICR (Caspase-Independent Cytochrome c Release) assay that prevents late-stage caspase-dependent apoptosis progression, thus allowing apoptotic cells to accumulate over several days.37 Following simultaneous treatment with GCV and a broad caspase inhibitor, accumulated cells attempting to undergo apoptosis were scored by immunofluorescence for diffuse cytoplasmic cytochrome c released from the mitochondria (as opposed to punctate mitochondrial cytochrome c in non-apoptotic cells) (Figure 3d). CICR quantification showed that 10 μg/ml GCV induced apoptosis in ∼50% of senescent cells over a 4-day interval following GCV exposure (Figure 3e). Thus, GCV induced caspase-dependent cell death in senescent cells.
GCV incorporates into the mtDNA and causes mtDNA damage
Senescent fibroblasts do not synthesize nuclear DNA but have an increase in mitochondrial mass.38 We reasoned that GCV might kill senescent cells by S-phase-independent incorporation into mtDNA, as suggested by anti-cancer strategies using HSVtk.39, 40, 41 To test this possibility, we incubated senescent and non-senescent cells, either control or 3MR-expressing, with 0.1 μg/ml tritiated GCV for 48 h. Radioactivity was detected in the mtDNA of 3MR-expressing cells, whether dividing (non-senescent) or non-dividing (senescent) (Figure 4a). To determine whether GCV damaged mtDNA in 3MR-expressing cells, we analyzed mtDNA by agarose gel electrophoresis. We detected intact mtDNA (16.5 kb) in control and 3MR-expressing senescent cells. However, only in GCV-treated 3MR-expressing senescent cells, we detected smaller (damaged) molecules that migrated faster (Figure 4b).
GCV induces apoptosis in senescent (SEN) cells through mtDNA fragmentation. (a) HCA2 and HCA2-3MR were rendered senescent using x-ray irradiation as in Figure 3 and were treated with 0.1 μg/ml of tritiated GCV. mtDNA was isolated and radioactivity measured. Incorporated GCV molecules per molecule of mtDNA is depicted. (b) mtDNA from control or irradiation-induced senescent cells treated or not with 10 μg/ml GCV were analyzed by agarose gel electrophoresis
GCV eliminates senescent 3MR-expressing cells in vivo
To determine whether the GCV–HSVtk combination could eliminate senescent cells in vivo, we took advantage of the rLUC in 3MR to track the fate of 3MR-expressing cells in mouse xenografts. We induced senescence in 3MR-expressing HCA2 cells by X-irradiation (10 Gy) and, 3 days later, injected 5 × 106 cells into the right flank of nu/nu mice. One day later, we administered PBS (vehicle control) or GCV (25 mg/kg) for 10 days and measured luminescence at regular intervals thereafter (Figure 5a). In the absence of GCV (vehicle alone), luminescence declined between 6 and 10 days, presumably due to clearance by the innate immune system, as reported.22, 42 GCV, however, eliminated senescent cells significantly faster, with 50% clearance by 6 days and complete clearance by 8 days (Figure 5a).
GCV induces cell death and senescence in vivo. (a) HCA2-3MR cells were injected subcutaneously into nu/nu mice, and mice were treated with GCV for 5 days. In vivo luminescence was measured every 2 days starting from day 1 after cell inoculation. (b) Control or 3MR-expressing MDA-MB231-D3H2LN cells were treated with GCV at the indicated concentrations, and cell viability evaluated by Trypan blue staining after 5 days. (c) Control or 3MR-expressing MDA-MB231-D3H2LN cells were injected into nu/nu mouse mammary fat pad, and mice were treated for 5 days with phosphate-buffered saline (PBS) or GCV starting on the day of cell injection (d0). Another group was injected with 3MR cells and treated with GCV at 10 days after injection. In vivo luminescence was measured at the indicated days after cancer cell injection. (d) Tumors from mice treated with PBS or GCV were collected 21 days after cell injection and stained for SA-βgal. The percentage of SA-βgal-positive cells was determined. Lower panels: Representative images of the SA-βgal staining (senescent cells are blue). WT, wild type
GCV induces senescence in 3MR-expressing breast cancer cells in vivo
We used a lentivirus to express 3MR in a human breast cancer cell line, MDA-MB231-D3H2LN, which also expresses firefly luciferase. In culture, high-dose GCV (2 and 20 μg/ml) eliminated >95% of cells within 5 days (Figure 5b). GCV was shown to efficiently reduce HSVtk-harboring cancer cells in vivo.43 Similarly, GCV treatment at the time of injection of 3MR-expressing MDA-MB231-D3H2LN cells (GCV d0) prevented tumor formation (Figure 5c). Moreover, GCV treatment 10 days after cancer cell injection (GCV d10) slowly shrank established tumors (Figure 5c), suggesting not all cancer cells underwent rapid apoptosis but rather some underwent senescence. Indeed, tumors treated with GCV at d10 and collected 21 days after injection had a significant percentage of senescent cells (Figure 5d). Thus, the GCV–HSVtk combination can also trigger cell senescence in vivo.
Discussion
Nucleoside analogs such as GCV are used to target cytomegaloviruses and herpes viruses.26 Drug specificity is achieved because the viral TK has a much higher affinity for GCV and similar analogs than the cellular TK. TK phosphorylates GCV, and, upon further phosphorylation by cellular enzymes, the drug competes with deoxyguanosine triphosphate for incorporation into nascent DNA chains, leading to DNA synthesis termination. In the presence of HSVtk, GCV not only prevents viral DNA replication but also causes nuclear DNA damage during S-phase, followed by apoptosis. Our finding that GCV can trigger cellular senescence or cell death (in dividing cells), and can trigger cell death in non-dividing cells via mtDNA damage, explains some of the clinical efficacy of these compounds. Because senescent cells can be targeted by the immune system in vivo, inducing senescence in virally infected cells could accelerate their clearance.19, 20, 21, 22, 23 Alternatively, the senescence induced by GCV can be converted to cell death due to inhibition of mtDNA synthesis, leading to the further elimination of infected cells. Interestingly, the HSVtk-GCV combination is currently being tested in combination with herpes virus-derived oncolytic viruses to enhance tumor cell death in targeted cancer therapies.29, 30, 31 Our findings that GCV induces cell death and senescence even in cancer cells could explain the enhanced tumor cell elimination, possibly through an enhanced immune response against senescent cancer cells.
Senescent cells accumulate with age and are thought to cause or contribute to a variety of age-associated diseases.13, 14, 15, 16, 17, 24, 44 One striking characteristic of senescent cells is the secretion of inflammatory cytokines, proteases and other molecules that can alter the tissue microenvironment.18, 33, 45, 46, 47 We and others have shown that these secreted factors can disrupt normal tissue structure and function, induce malignant phenotypes in pre-malignant and non-aggressive cells and stimulate malignant tumorigenesis in vivo through inflammation and vascularization.13, 14, 18, 45, 46, 48 Senescent cells are resistant to apoptosis, and, as the persistent presence of these cells can be harmful, it is important to find ways to eliminate them, preferably by causing apoptotic cell death, which occurs with minimal inflammation.
HSVtk and GCV have been used to induce apoptotic cell death in cell culture or tissues.35, 41, 49, 50 We show that cells harboring HSVtk additionally undergo senescence in response to low GCV doses, whereas higher doses cause senescent cells to die by apoptosis owing to mtDNA damage. Because senescent cells increase their mitochondrial mass,38 they may be even more sensitive to GCV-mediated mitochondrial toxicity than non-senescent cells.40, 41 Of note, as apoptosis was shown to be induced by persistent single-strand DNA breaks in the mitochondrial genome,51 the same mechanism could explain the effects of GCV we observed in senescent cells.
In addition to enhancing the potential effects of HSVtk-GCV based therapies, the fact that GCV can kill non-dividing senescent cells could be used to selectively eliminate senescent HSVtk-harboring cells in vivo—for example, if HSVtk were driven by a senescence-sensitive promoter, as recently described for another inducible killing strategy.24 Additionally, the multifunctional 3MR protein has the added advantage of providing imaging by bioluminescence and fluorescence.36, 52, 53
A genetically engineered mouse model was recently developed to kill senescent cells in vivo using a drug that causes caspase activation.24 This study was pivotal in implicating a role for senescent cells in certain degenerative pathologies associated with aging. Here, our findings suggest killing senescent cells might also retard cancer progression. Using the HSVtk-GCV system described here, we are in the process of testing the idea that 3MR transgenic mouse models might allow tracking and eliminating senescent cells in vivo.
Materials and Methods
Cell culture
Human diploid HCA2 (foreskin) fibroblasts and human breast carcinoma cells (MDA-MB-231-D3H2LN Bioware expressing firefly luciferase; PerkinElmer, Waltham, MA, USA) were cultured in Dulbecco’s modified Eagle’s media plus 10% fetal bovine serum and 100 U/ml streptomycin and penicillin.
Antibodies and reagents
Antibodies against 53BP1 (1 : 2000) and cytochrome c (1 : 800) were from Bethyl (Montgomery, TX, USA; BL182) and Pharmingen (No. 556432), respectively. The secondary antibody was Alexa Fluor 488 goat anti-rabbit IgG (No. A11008, 1:750; Invitrogen, Carlsbad, CA, USA). Mouse monoclonal antibody against rLUC (1 : 200) was from Chemicon (Millipore, Billerica, MA, USA; MAB4400), and ganciclovir (GCV) was from Sigma (St Louis, MO, USA). Radio-labeled GCV was from Moravek Biochemicals Inc. (Brea, CA, USA).
Constructs
Stable expression of 3MR was achieved using lentiviruses generated as described.54 We cloned the 3MR sequence into a Gateway entry vector and transferred it to a lentiviral destination vector (670–1 puromycin resistance) under the control of the CMV/TO promoter.55
Irradiation and SA-βgal staining
To induce senescence, cells were cultured to confluence, X-irradiated (10 Gy), allowed to recover overnight and re-seeded; 10 days later, cells displayed the enlarged senescent morphology and expressed SA-βgal, as described.12 For SA-βgal staining in tissues, 21 days following tumor inoculation, mice were euthanized and tumors collected, embedded in OCT, flash frozen in liquid nitrogen and stored at −80 °C. Embedded tumors were cryosectioned (10 μm), mounted on slides and stained for SA-βgal using fast red as a counterstain.
Immunofluorescence
Cells were cultured in four- or eight-well chamber slides (Nunc, Penfield, NY, USA), fixed in 4% formaldehyde (Sigma) for 10 min at 4 °C and permeabilized in PBS-0.5% Triton for 10 min at 4 °C. Slides were blocked for 30 min in blocking buffer (1% IgG-free BSA, 4% donkey serum (Jackson ImmunoResearch, West Grove, PA, USA) in PBS) and incubated with primary antibodies diluted in blocking buffer for 1 h at room temperature or overnight at 4 °C. Cells were washed, incubated with secondary antibodies for 30 min at room temperature, washed and mounted with slow-fade gold (Molecular Probes, Invitrogen) or Vectashield containing DAPI (Vector Labs, Burlingame, CA, USA). Images were acquired using an Olympus BX20 fluorescence microscope with spotfire software (Diagnostics Instruments, Sterling Heights, MI, USA) and processed with Photoshop CS (Adobe, San Jose, CA, USA).
DNA synthesis detection
Cells in chamber slides were pulsed with the modified thymidine analogue EdU for 24 h before fixation and permeabilization, as described above. Cells were processed as per the manufacturer’s protocol to detect the incorporation of EdU into DNA using Click-iT chemistry (Invitrogen C10340). Slides were washed with PBS and mounted with Vectashield containing DAPI (Vector Labs) and analyzed for immunofluorescence.
Flow cytometry analysis for apoptosis detection
Live trypsinized cells and their supernatant were collected, centrifuged, rinsed with PBS and processed according to the manufacturer’s apoptosis detection kit protocol, except that propidium iodine was replaced by DAPI at a concentration of 800 ng/ml (BD, San Jose, CA, USA; No. 556547). The data was collected using BD LSRFortessa (BD) and analyzed using FlowJo 10.0.3 (FlowJo, Ashland, OR, USA).
Flow cytometry for cell-cycle analysis
Live trypsinized cells were collected, centrifuged, rinsed with PBS, centrifuged and resuspended in sodium citrate 0.2%, Triton X-100 0.1%, RNase A 100 μg/ml and DAPI 1 μg/ml. Samples were analyzed following 20 min of incubation at room temperature using BD LSRFortessa and the data analyzed using FlowJo 9.6.2 (Watson Pragmatic cell-cycle distribution).
Caspase-independent cytochrome c release assay
Cells in four-well chamber slides were given GCV for 2, 3 or 4 days in the presence Z-VAD-FMK, a pan-caspase inhibitor (Sigma). Cells were fixed, processed for cytochrome c immunofluorescence and scored as apoptotic if there was diffuse staining in both the cytoplasm and nucleus, and lack of reticulated or punctate mitochondrial staining in the cytoplasm, as described.37
Measurement of GCV incorporation and mtDNA damage
HCA2 and HCA2-3MR fibroblasts were X-irradiated (10 Gy) to induce senescence. Ten days later, cells were given 0.1 μg/ml tritiated GCV (3H-GCV; 3.9 Ci/mmol) (Moravek Biochemicals) for 48 h, and washed. Mitochondria and mtDNA were purified using Mitochondria Isolation (Pierce, Rockford, IL, USA) and Mitochondrial DNA Isolation kits (Abcam, San Francisco, CA, USA). Scintillation fluid was mixed with 100 ng of mtDNA (evaluated by densitometry on agarose gels) and radioactivity was quantified. For DNA damage analysis, cells were treated similarly but incubated with 10 μg/ml cold GCV for 48 h and loaded onto 0.5% agarose gels. DNA was stained with SYBR Gold (Invitrogen) and detection performed using a Typhoon apparatus (GE Healthcare Biosciences, Pittsburgh, PA, USA).
Tumor growth assays
Tumor growth was followed as described.13, 45, 56 Briefly, we injected into the mammary fat pads of 5-week-old nu/nu mice a 100-μl suspension of 106 MDA-MB231-D3H2LN (control) or MDA-MB231-D3H2LN-3MR cells and monitored tumor growth by firefly luminescence using a IVIS Spectrum II Biophotonic imager (Xenogen, PerkinElmer) and caliper measurements over a 21-day period. The clearance of HCA2-3MR primary fibroblasts after injection of 5 × 106 cells was performed similarly, except luminescence from rLUC in 3MR was detected over 10 days. Mice were treated with 25 mg/kg of GCV by daily intraperitoneal injections over 5 days.
Statistical analysis
Error bars on all graphs are the S.D. of three independent measurements. Statistical analyses were performed using Graphpad Prism software (GraphPad, La Jolla, CA, USA).
Abbreviations
- 3MR:
-
trimodal reporter-killer fusion protein
- CICR:
-
Caspase-Independent Cytochrome c Release
- DNA-SCARS:
-
DNA segments with chromatin alterations reinforcing senescence
- GCV:
-
ganciclovir
- HSVtk:
-
herpes simplex virus thymidine kinase
- mtDNA:
-
mitochondrial DNA
- RFP:
-
red fluorescent protein
- rLUC:
-
Renilla luciferase
- SA-βgal:
-
senescence-associated beta galactosidase
References
Campisi J, d’Adda di Fagagna F . Cellular senescence: when bad things happen to good cells. Nature Rev Mol Cell Biol 2007; 8: 729–740.
Braig M, Schmitt CA . Oncogene-induced senescence: putting the brakes on tumor development. Cancer Res 2006; 66: 2881–2884.
Prieur A, PD. S . Cellular senescence in vivo: a barrier to tumorigenesis. Curr Opin Cell Biol 2008; 20: 150–155.
Collado M, Serrano M . Senescence in tumours: evidence from mice and humans. Nat Rev Cancer 2010; 10: 51–57.
Hayflick L, Moorhead PS . The serial cultivation of human diploid cell strains. Exp Cell Res 1961; 25: 585–621.
Rodier F, Campisi J . Four faces of cellular senescence. J Cell Biol 2011; 192: 547–556.
Coppe JP, Desprez PY, Krtolica A, Campisi J . The senescence-associated secretory phenotype: the dark side of tumor suppression. Annu Rev Pathol 2010; 5: 99–118.
Le ON, Rodier F, Fontaine F, Coppe JP, Campisi J, DeGregori J et al. Ionizing radiation-induced long-term expression of senescence markers in mice is independent of p53 and immune status. Aging Cell 2010; 9: 398–409.
Krishnamurthy J, Torrice C, Ramsey MR, Kovalev GI, Al-Regaiey K, Su L et al. Ink4a/Arf expression is a biomarker of aging. J Clin Invest 2004; 114: 1299–1307.
Zindy F, Quelle DE, Roussel MF, Sherr CJ . Expression of the p16INK4a tumor suppressor versus other INK4 family members during mouse development and aging. Oncogene 1997; 15: 203–211.
Ressler S, Bartkova J, Niederegger H, Bartek J, Scharffetter-Kochanek K, Jansen-Durr P et al. p16INK4A is a robust in vivo biomarker of cellular aging in human skin. Aging Cell 2006; 5: 379–389.
Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C et al. A novel biomarker identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci USA 1995; 92: 9363–9367.
Krtolica A, Parrinello S, Lockett S, Desprez P, Campisi J . Senescent fibroblasts promote epithelial cell growth and tumorigenesis: A link between cancer and aging. Proc Natl Acad Sci USA 2001; 98: 12072–12077.
Parrinello S, Coppe JP, Krtolica A, Campisi J . Stromal-epithelial interactions in aging and cancer: senescent fibroblasts alter epithelial cell differentiation. J Cell Sci 2005; 118 (Pt 3): 485–496.
Martin JA, Buckwalter JA . The role of chondrocyte senescence in the pathogenesis of osteoarthritis and in limiting cartilage repair. J Bone Joint Surg Am 2003; 85-A Suppl 2: 106–110.
Matthews C, Gorenne I, Scott S, Figg N, Kirkpatrick P, Ritchie A et al. Vascular smooth muscle cells undergo telomere-based senescence in human atherosclerosis: effects of telomerase and oxidative stress. Circ Res 2006; 99: 156–164.
Erusalimsky JD, Kurz DJ . Cellular senescence in vivo: its relevance in ageing and cardiovascular disease. Exp Gerontol 2005; 40: 634–642.
Coppe JP, Patil CK, Rodier F, Sun Y, Munoz DP, Goldstein J et al. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol 2008; 6: 2853–2868.
Ventura A, Kirsch DG, McLaughlin ME, Tuveson DA, Grimm J, Lintault L et al. Restoration of p53 function leads to tumour regression in vivo. Nature 2007; 445: 661–665.
Xue W, Zender L, Miething C, Dickins RA, Hernando E, Krizhanovsky V et al. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature 2007; 445: 656–660.
Kang TW, Yevsa T, Woller N, Hoenicke L, Wuestefeld T, Dauch D et al. Senescence surveillance of pre-malignant hepatocytes limits liver cancer development. Nature 2011; 479: 547–551.
Krizhanovsky V, Yon M, Dickins RA, Hearn S, Simon J, Miething C et al. Senescence of activated stellate cells limits liver fibrosis. Cell 2008; 134: 657–667.
Jun JI, Lau LF . The matricellular protein CCN1 induces fibroblast senescence and restricts fibrosis in cutaneous wound healing. Nat Cell Biol 2010; 12: 676–685.
Baker DJ, Wijshake T, Tchkonia T, LeBrasseur NK, Childs BG, van de Sluis B et al. Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. Nature 2011; 479: 232–236.
Yamakoshi K, Takahashi A, Hirota F, Nakayama R, Ishimaru N, Kubo Y et al. Real-time in vivo imaging of p16Ink4a reveals cross talk with p53. J Cell Biol 2009; 186: 393–407.
De Clercq E . Antiviral drugs in current clinical use. J Clin Virol 2004; 30: 115–133.
Kaina B . DNA damage-triggered apoptosis: critical role of DNA repair, double-strand breaks, cell proliferation and signaling. Biochem Pharmacol 2003; 66: 1547–1554.
Nasu Y, Kusaka N, Saika T, Tsushima T, Kumon H . Suicide gene therapy for urogenital cancer: current outcome and prospects. Mol Urol 2000 Summer 4: 67–71.
Abate-Daga D, Andreu N, Camacho-Sanchez J, Alemany R, Herance R, Millan O et al. Oncolytic adenoviruses armed with thymidine kinase can be traced by PET imaging and show potent antitumoural effects by ganciclovir dosing. PLoS One 2011; 6: e26142.
Agarwalla PK, Aghi MK . Oncolytic herpes simplex virus engineering and preparation. Methods Mol Biol 2012; 797: 1–19.
Raty JK, Pikkarainen JT, Wirth T, Yla-Herttuala S . Gene therapy: the first approved gene-based medicines, molecular mechanisms and clinical indications. Curr Mol Pharmacol 2008; 1: 13–23.
Ray P, De A, Min JJ, Tsien RY, Gambhir SS . Imaging tri-fusion multimodality reporter gene expression in living subjects. Cancer Res 2004; 64: 1323–1330.
Rodier F, Coppe JP, Patil CK, Hoeijmakers WA, Munoz DP, Raza SR et al. Persistent DNA damage signalling triggers senescence-associated inflammatory cytokine secretion. Nat Cell Biol 2009; 11: 973–979.
Rodier F, Munoz DP, Teachenor R, Chu V, Le O, Bhaumik D et al. DNA-SCARS: distinct nuclear structures that sustain damage-induced senescence growth arrest and inflammatory cytokine secretion. J Cell Sci 2011; 124 (Pt 1): 68–81.
Furman PA, McGuirt PV, Keller PM, Fyfe JA, Elion GB . Inhibition by acyclovir of cell growth and DNA synthesis of cells biochemically transformed with herpesvirus genetic information. Virology 1980; 102: 420–430.
Rubsam LZ, Davidson BL, Shewach DS . Superior cytotoxicity with ganciclovir compared with acyclovir and 1-beta-D-arabinofuranosylthymine in herpes simplex virus-thymidine kinase-expressing cells: a novel paradigm for cell killing. Cancer Res 1998; 58: 3873–3882.
Goldstein J, Rodier F, Garbe JC, Stampfer MR, Campisi J . Caspase-independent cytochrome c release is a sensitive measure of low level apoptosis in cell culture models. Aging Cell 2005; 4: 217–222.
Passos JF, Nelson G, Wang C, Richter T, Simillion C, Proctor CJ et al. Feedback between p21 and reactive oxygen production is necessary for cell senescence. Mol Syst Biol 2010; 6: 347.
Beltinger C, Fulda S, Kammertoens T, Uckert W, Debatin KM . Mitochondrial amplification of death signals determines thymidine kinase/ganciclovir-triggered activation of apoptosis. Cancer Res 2000; 60: 3212–3217.
van der Eb MM, Geutskens SB, van Kuilenburg AB, van Lenthe H, van Dierendonck JH, Kuppen PJ et al. Ganciclovir nucleotides accumulate in mitochondria of rat liver cells expressing the herpes simplex virus thymidine kinase gene. J Gene Med 2003; 5: 1018–1027.
Herraiz M, Beraza N, Solano A, Sangro B, Montoya J, Qian C et al. Liver failure caused by herpes simplex virus thymidine kinase plus ganciclovir therapy is associated with mitochondrial dysfunction and mitochondrial DNA depletion. Hum Gene Ther 2003; 14: 463–472.
Xue W, Zender L, Miething C, Dickins RA, Hernando E, Krizhanovsky V et al. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature 2007; 445: 656–650.
Moolten FL . Tumor chemosensitivity conferred by inserted herpes thymidine kinase genes: paradigm for a prospective cancer control strategy. Cancer Res 1986; 46: 5276–5281.
Paradis V, Youssef N, Dargere D, Ba N, Bonvoust F, Deschatrette J et al. Replicative senescence in normal liver, chronic hepatitis C, and hepatocellular carcinomas. Hum Pathol 2001; 32: 327–332.
Coppe JP, Patil CK, Rodier F, Krtolica A, Beausejour CM, Parrinello S et al. A human-like senescence-associated secretory phenotype is conserved in mouse cells dependent on physiological oxygen. PLoS One 2010; 5: e9188.
Coppe JP, Kauser K, Campisi J, Beausejour CM . Secretion of vascular endothelial growth factor by primary human fibroblasts at senescence. J Biol Chem 2006; 281: 29568–29574.
Freund A, Orjalo AV, Desprez PY, Campisi J . Inflammatory networks during cellular senescence: causes and consequences. Trends Mol Med 2010; 16: 238–246.
Krtolica A, Larocque N, Genbacev O, Ilic D, Coppe JP, Patil CK et al. GROalpha regulates human embryonic stem cell self-renewal or adoption of a neuronal fate. Differentiation 2011; 81: 222–232.
Nishiyama Y, Rapp F . Anticellular effects of 9-(2-hydroxyethoxymethyl) guanine against herpes simplex virus-transformed cells. J Gen Virol 1979; 45: 227–230.
Davidson RL, Kaufman ER, Crumpacker CS, Schnipper LE . Inhibition of herpes simplex virus transformed and nontransformed cells by acycloguanosine: mechanisms of uptake and toxicity. Virology 1981; 113: 9–19.
Tann AW, Boldogh I, Meiss G, Qian W, Van Houten B, Mitra S et al. Apoptosis induced by persistent single-strand breaks in mitochondrial genome: critical role of EXOG (5′-EXO/endonuclease) in their repair. J Biol Chem 2011; 286: 31975–31983.
Ferbeyre G, de Stanchina E, Lin AW, Querido E, McCurrach ME, Hannon GJ et al. Oncogenic ras and p53 cooperate to induce cellular senescence. Mol Cell Biol 2002; 22: 3497–3508.
Parrinello S, Samper E, Krtolica A, Goldstein J, Melov S, Campisi J . Oxygen sensitivity severely limits the replicative lifespan of murine fibroblasts. Nat Cell Biol 2003; 5: 741–747.
Beausejour CM, Krtolica A, Galimi F, Narita M, Lowe SW, Yaswen P et al. Reversal of human cellular senescence: roles of the p53 and p16 pathways. EMBO J 2003; 22: 4212–4222.
Campeau E, Ruhl VE, Rodier F, Smith CL, Rahmberg BL, Fuss JO et al. A versatile viral system for expression and depletion of proteins in mammalian cells. PLoS One 2009; 4: e6529.
Liu D, Hornsby PJ . Senescent human fibroblasts increase the early growth of xenograft tumors via matrix metalloproteinase secretion. Cancer Res 2007; 67: 3117–3126.
Acknowledgements
We thank Dr. Sanjiv Gambhir (Stanford University) for the 3MR plasmid and members of the Rodier and Campisi laboratories for valuable comments and discussions. This work was supported by grants from the USA National Institutes of Health (AG09909, AG017242) to JC and the Canadian Institute for Health Research (MOP114962) and a “Fonds de Recherche Québec Santé” junior I career award (22624) to FR.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by M Agostini
Supplementary Information accompanies this paper on Cell Death and Disease website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Laberge, RM., Adler, D., DeMaria, M. et al. Mitochondrial DNA damage induces apoptosis in senescent cells. Cell Death Dis 4, e727 (2013). https://doi.org/10.1038/cddis.2013.199
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cddis.2013.199
Keywords
This article is cited by
-
Association between single nucleotide polymorphism of rs1937 in TFAM gene and longevity among the elderly Chinese population: based on the CLHLS study
BMC Geriatrics (2022)
-
Elimination of senescent cells by β-galactosidase-targeted prodrug attenuates inflammation and restores physical function in aged mice
Cell Research (2020)
-
Osteocyte Cellular Senescence
Current Osteoporosis Reports (2020)
-
Senescence in the pathogenesis of age-related macular degeneration
Cellular and Molecular Life Sciences (2020)
-
RACK1 promotes tumorigenicity of colon cancer by inducing cell autophagy
Cell Death & Disease (2018)