Abstract
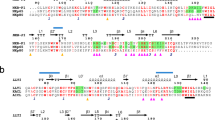
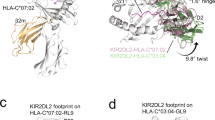
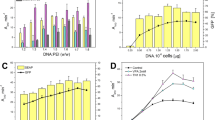
NKG2D, a homodimeric lectin-like receptor, is a unique stimulatory molecule that is found on natural killer cells, T cells and activated macrophages. The natural ligands for murine NKG2D are distant major histocompatibility complex homologs, retinoic acid early transcript (Rae1) and H-60 minor histocompatibility antigen. The crystal structure of the extracellular region of murine NKG2D reveals close homology with other C-type lectin receptors such as CD94, Ly49A, rat MBP-A and CD69. However, the precise mode of dimeric assembly varies among these natural killer receptors, as well as their surface topography and electrostatic properties. The NKG2D structure provides the first structural insights into the role and ligand specificity of this stimulatory receptor in the innate and adaptive immune system.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Lanier, L. L. On guard—activating NK cell receptors. Nature Immunol. 2, 23–27 (2001).
Karre, K., Ljunggren, H. G., Piontek, G. & Kiessling, R. Selective rejection of H-2-deficient lymphoma variants suggests alternative immune defence strategy. Nature 319, 675–678 (1986).
Lanier, L. L. NK cell receptors. Annu. Rev. Immunol. 16, 359–393 (1998).
Wagtmann, N. et al. Molecular clones of the p58 NK cell receptor reveal immunoglobulin- related molecules with diversity in both the extra- and intracellular domains. Immunity 2, 439–449 (1995).
Colonna, M. & Samaridis, J. Cloning of immunoglobulin-superfamily members associated with HLA-C and HLA-B recognition by human natural killer cells. Science 268, 405–408 (1995).
Yokoyama, W. M., Jacobs, L. B., Kanagawa, O., Shevach, E. M. & Cohen, D. I. A murine T lymphocyte antigen belongs to a supergene family of type II integral membrane proteins. J. Immunol. 143, 1379–1386 (1989).
Maenaka, K., Juji, T., Stuart, D. I. & Jones, E. Y. Crystal structure of the human p58 killer cell inhibitory receptor (KIR2DL3) specific for HLA-Cw3-related MHC class I. Struct. Fold Des. 7, 391–398 (1999).
Fan, Q. R. et al. Structure of the inhibitory receptor for human natural killer cells resembles haematopoietic receptors. Nature 389, 96–100 (1997).
Snyder, G. A., Brooks, A. G. & Sun, P. D. Crystal structure of the HLA-Cw3 allotype-specific killer cell inhibitory receptor KIR2DL2. Proc. Natl Acad. Sci. USA 96, 3864–3869 (1999).
Chapman, T. L., Heikeman, A. P. & Bjorkman, P. J. The inhibitory receptor LIR-1 uses a common binding interaction to recognize class I MHC molecules and the viral homolog UL18. Immunity 11, 603–613 (1999).
Boyington, J. C., Motyka, S. A., Schuck, P., Brooks, A. G. & Sun, P. D. Crystal structure of an NK cell immunoglobulin-like receptor in complex with its class I MHC ligand. Nature 405, 537–543 (2000).
Renedo, M. et al. A sequence-ready physical map of the region containing the human natural killer gene complex on chromosome 12p12.3-p13.2. Genomics 65, 129–136 (2000).
Ho, E. L. et al. Murine Nkg2d and Cd94 are clustered within the natural killer complex and are expressed independently in natural killer cells. Proc. Natl Acad. Sci. USA 95, 6320–6325 (1998).
Boyington, J. C. et al. Structure of CD94 reveals a novel C-type lectin fold: implications for the NK cell-associated CD94/NKG2 receptors. Immunity 10, 75–82 (1999).
Llera, A. S., Viedma, F., Sanchez-Madrid, F. & Tormo, J. Crystal structure of the C-type lectin-like domain from the human hematopoietic cell receptor CD69. J. Biol. Chem. (in the press, 2001).
Natarajan, K., Sawicki, M. W., Margulies, D. H. & Mariuzza, R. A. Crystal structure of human CD69: A C-type lectin-like activation marker of hematopoietic cells. Biochemistry 39, 14779–14786 (2000).
Tormo, J., Natarajan, K., Margulies, D. H. & Mariuzza, R. A. Crystal structure of a lectin-like natural killer cell receptor bound to its MHC class I ligand. Nature 402, 623–631 (1999).
Moretta, A., Biassoni, R., Bottino, C., Mingari, M. C. & Moretta, L. Natural cytotoxicity receptors that trigger human NK-cell-mediated cytolysis. Immunol. Today 21, 228–234 (2000).
Lanier, L. L. Turning on natural killer cells. J. Exp. Med. 191, 1259–1262 (2000).
Lanier, L. L., Corliss, B., Wu, J. & Phillips, J. H. Association of DAP12 with activating CD94/NKG2C NK cell receptors. Immunity 8, 693–701 (1998).
Lanier, L. L., Corliss, B. C., Wu, J., Leong, C. & Phillips, J.H. Immunoreceptor DAP12 bearing a tyrosine-based activation motif is involved in activating NK cells. Nature 391, 703–707 (1998).
Wu, J. et al. An activating immunoreceptor complex formed by NKG2D and DAP10. Science 285, 730–732 (1999).
Bauer, S. et al. Activation of NK cells and T cells by NKG2D, a receptor for stress-inducible MICA. Science 285, 727–729 (1999).
Diefenbach, A., Jamieson, A. M., Liu, S. D., Shastri, N. & Raulet, D. H. Ligands for the murine NKG2D receptor: expression by tumor cells and activation of NK cells and macrophages. Nature Immunol. 1, 119–126 (2000).
Vance, R. E., Jamieson, A. M. & Raulet, D. H. Recognition of the class Ib molecule Qa-1(b) by putative activating receptors CD94/NKG2C and CD94/NKG2E on mouse natural killer cells. J. Exp. Med. 190, 1801–1812 (1999).
Cosman, D. et al. The human cytomegalovirus (HCMV) glycoprotein, UL16, binds to the MHC class I-related protein, MICB/PERB11, and to two novel, MHC class I-related molecules, ULBP1 and ULBP2. FASEB J. 14, 1018 (2000).
Chalupny, J. et al. Soluble forms of the novel MHC class I-related molecules, ULBP1 and ULBP2, bind to, and functionally activate NK cells. FASEB J. 14, 1018 (2000).
Cerwenka, A. et al. Retinoic acid early inducible genes define a ligand family for the activating NKG2D receptor in mice. Immunity 12, 721–727 (2000).
Groh, V. et al. Broad tumor-associated expression and recognition by tumor-derived γδ T cells of MICA and MICB. Proc. Natl Acad. Sci. USA 96, 6879–6884 (1999).
Day, A. J. The C-type carbohydrate recognition domain (CRD) superfamily. Biochem. Soc. Trans. 22, 83–88 (1994).
Kolatkar, A. R. et al. Mechanism of N-acetylgalactosamine binding to a C-type animal lectin carbohydrate-recognition domain. J. Biol. Chem. 273, 19502–19508 (1998).
Holm, L. & Sander, C. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 233, 123–138 (1993).
Lopez-Botet, M., Llano, M., Navarro, F. & Bellon, T. NK cell recognition of non-classical HLA class I molecules. Semin. Immunol. 12, 109–119 (2000).
Richardson, J. S. The anatomy and taxonomy of protein structure. Adv. Protein Chem. 34, 167–339 (1981).
Lawrence, M. C. & Colman, P. M. Shape complementarity at protein-protein interfaces. J. Mol. Biol. 234, 946–950 (1993).
Connolly, M. L. Analytical molecular surface calculation. J. Appl. Cryst. 16, 548–558 (1983).
CCP4. The Collaborative Computational Project Number 4, suite programs for protein crystallography. Acta Cryst. 50, 760–763 (1994).
Brooks, A. G., Posch, P. E., Scorzelli, C. J., Borrego, F. & Coligan, J. E. NKG2A complexed with CD94 defines a novel inhibitory natural killer cell receptor. J. Exp. Med. 185, 795–800 (1997).
Colman, P. M. et al. Three-dimensional structure of a complex of antibody with influenza virus neuraminidase. Nature 326, 358–363 (1987).
Li, P. et al. Crystal structure of the MHC class I homolog MIC-A, a γδ T cell ligand. Immunity 10, 577–584 (1999).
Garboczi, D. N., Hung, D. T. & Wiley, D. C. HLA-A2-peptide complexes: refolding and crystallization of molecules expressed in Escherichia coli and complexed with single antigenic peptides. Proc. Natl Acad. Sci. USA 89, 3429–3433 (1992).
Busch, D. H., Pilip, I. M., Vijh, S. & Pamer, E. G. Coordinate regulation of complex T cell populations responding to bacterial infection. Immunity 8, 353–362 (1998).
Otwinowski, Z. & Minor, W. Processing of x-ray diffraction data collected in oscillation mode. Meth. Enzymol. 276, 307–326 (1997).
Pannu, N. S. & Read, R. J. Improved structure refinement through maximum likelyhood. Acta Cryst. A52, 659–668 (1996).
Brünger, A. T. et al. Crystallography and NMR system (CNS): A new software system for macromolecular structure determination. Acta Cryst. 54, 905–921 (1998).
Read, R. J. Improved fourier coefficients for maps using phases from partial structures with errors. Acta Cryst. 42, 140–149 (1986).
Jones, T. A., Cowan, S., Zou, J. Y. & Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Cryst. 47, 110–119 (1991).
Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Cryst. 26, 283–291 (1993).
Hooft, R. W., Vriend, G., Sander, C. & Abola, E. E. Errors in protein structures. Nature 381, 272 (1996).
Sheriff, S., Hendrickson, W. A. & Smith, J. L. Structure of myohemerythrin in the azidomet state at 1.7/1.3 Å resolution. J. Mol. Biol. 197, 273–296 (1987).
Esnouf, R. M. An extensively modified version of MOLSCRIPT that includes greatly enhanced coloring capabilities. J. Mol. Graph. 15, 132–134 (1997).
Merritt, E. A. & Murphy, M. E. P. Raster3D Version 2.0 - A program for photorealistic molecular graphics. Acta Cryst. 50, 869–873 (1994).
Berman, H. M. et al. The Protein Data Bank. Nucleic Acids Res . 28, 235–242 (2000).
Acknowledgements
We thank S. Greasley, A. Heine, N. Larsen and X. Dai for data collection and processing; S. Greasley, A. Heine and J. Speir for helpful suggestions; A. Heine for invaluable advice with molecular replacement; R. Stanfield and J. Stevens for critical comments; R. Stanfield for help with computational calculations; M. Elsliger for computational assistance; and the staff of SSRL beamline 9-2. Supported by National Institutes of Health grants CA58896 and AI42266 (to I. A. W.); DK55037 and AI62267 (to L. T.); a Gerhard Hess research fellowship of the Deutsche Forschungsgemeinschaft (to D. H. B); a post-doctoral fellowship of the German Academic Exchange Service (to M. G. R.); and a National Science Foundation predoctoral fellowship (to D. W. W.).
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Wolan, D., Teyton, L., Rudolph, M. et al. Crystal structure of the murine NK cell–activating receptor NKG2D at 1.95 Å. Nat Immunol 2, 248–254 (2001). https://doi.org/10.1038/85311
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/85311
This article is cited by
-
Regulation of immune cell function and differentiation by the NKG2D receptor
Cellular and Molecular Life Sciences (2011)
-
Characteristics of Epstein–Barr virus envelope protein gp42
Virus Genes (2010)
-
NKG2D
AfCS-Nature Molecule Pages (2009)
-
The NKG2D receptor: immunobiology and clinical implications
Immunologic Research (2008)