Abstract
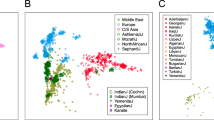
GENETIC variation at hypervariable loci is being used extensively for linkage analysis1 and individual identification2, and may be useful for inter-population studies2–5. Here we show that polymorphic microsatellites (primarily CA repeats) allow trees of human individuals to be constructed that reflect their geographic origin with remarkable accuracy. This is achieved by the analysis of a large number of loci for each individual, in spite of the small variations in allele frequencies existing between populations6,7. Reliable evolutionary relationships could also be established in comparisons among human populations but not among great ape species, probably because of constraints on allele length variation. Among human populations, diversity of microsatellites is highest in Africa, which is in contrast to other nuclear markers and supports the hypothesis of an African origin for humans.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Weissenbach, J. et al. Nature 359, 794–801 (1992).
Pena, S. D. J., Chakraborty, R., Epplen, J. T. & Jeffreys, A. J. (eds) DNA Fingerprinting: State of the Science (Birkhauser, Basel, 1993).
Gilbert, D. A., Lehman, N., O'Brien, S. J. & Wayne, R. K. Nature 344, 764–767 (1990).
Edwards, A., Hammond, H. A., Jin, L., Caskey, C. T. & Chakraborty, R. Genomics 12, 241–253 (1992).
Chakraborty, R., Deka, R., Jin, L. & Ferrell, R. E. Am. J. hum. Biol. 4, 387–397 (1992).
Smouse, P. E., Spielman, R. S. & Park, M. H. Am. Nat. 119, 445–463 (1982).
Mitton, J. B. Am. Nat. 111, 203–212 (1977).
Cann, R. L., Stoneking, M. & Wilson, A. C. Nature 325, 31–36 (1987).
Vigilant, L., Stoneking, M., Harpending, H., Hawkes, K. & Wilson, A. C. Science 253, 1503–1507 (1991).
Lewontin, R. Evol. Biol. 6, 381–398 (1972).
Nei, M. & Roychoudhury, A. Science 177, 434–436 (1972).
Cavalli-Sforza, L. L., Piazza, A., Menozzi, P. & Mountain, J. Proc. natn. Acad. Sci. U.S.A. 85, 6002–6006 (1988).
Bowcock, A. M. et al. Proc. natn. Acad. Sci. U.S.A. 88, 839–843 (1991).
Nei, M. & Roychoudhury, A. Molec. Biol. Evol. 10, 927–943 (1993).
Cavalli-Sforza, L. L., Menozzi, P. & Piazza, A. History and Geography of Human Genes (Princeton Univ. Press, Princeton, in the press).
Karlin, S. A First Course in Stochastic Processes (Academic, New York, 1969).
Walsh, J. B. Genetics 115, 553–567 (1987).
Tachida, H. & Iizuka, M. Genetics 131, 471–478 (1992).
Templeton, A. R. Am. Anthr. 95, 51–72 (1993).
Barbujani, G. & Sokal, R. R. Proc. natn. Acad. Sci. U.S.A. 87, 1816–1819 (1990).
Zei, G. et al. Ann. hum. Genet. 57, 123–140 (1993).
Cavalli-Sforza, L. L., Menozzi, P. & Piazza, A. Science 259, 639–646 (1993).
Bowcock, A. M. et al. Gene Geog. 5, 151–173 (1991).
Horai, S. et al. Molec. Biol. Evol. 10, 23–47 (1993).
Saitou, N. & Nei, M. Molec. Biol. Evol. 4, 406–425 (1987).
Bowcock, A. et al. Genomics 15, 376–386 (1993).
Felsenstein, J. PHYLIP (Phylogeny Inference Package) Version 3.5c (Department of Genetics, Univ. Washington, Seattle, 1993).
Cavalli-Sforza, L. L. & Edwards, A. W. F. Evolution 32, 550–570 (1967).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Bowcock, A., Ruiz-Linares, A., Tomfohrde, J. et al. High resolution of human evolutionary trees with polymorphic microsatellites. Nature 368, 455–457 (1994). https://doi.org/10.1038/368455a0
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/368455a0
This article is cited by
-
Disease-driven top predator decline affects mesopredator population genomic structure
Nature Ecology & Evolution (2024)
-
Testing hypotheses of invasion pathways of the ladybird Harmonia axyridis into and across South Africa
Biological Invasions (2024)
-
Sampling through space and time: multi-year analysis reveals dynamic population genetic patterns for an amphibian metapopulation
Conservation Genetics (2024)
-
Landscape structure does not hinder the dispersal of an invasive herbivorous mammal in the New Caledonian biodiversity hotspot
European Journal of Wildlife Research (2024)
-
Urban environment determines population genetics in the green toad, Bufotes viridis
European Journal of Wildlife Research (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.