Abstract
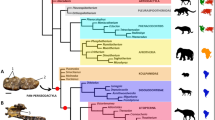
Higher level relationships among placental mammals, as well as the historical biogeography and morphological diversification of this group, remain unclear1,2,3. Here we analyse independent molecular data sets, having aligned lengths of DNA of 5,708 and 2,947 base pairs, respectively, for all orders of placental mammals. Phylogenetic analyses resolve placental orders into four groups: Xenarthra, Afrotheria, Laurasiatheria, and Euarchonta plus Glires. The first three groups are consistently monophyletic with different methods of analysis. Euarchonta plus Glires is monophyletic or paraphyletic depending on the phylogenetic method. A unique nine-base-pair deletion in exon 11 of the BRCA1 gene provides additional support for the monophyly of Afrotheria, which includes proboscideans, sirenians, hyracoids, tubulidentates, macroscelideans, chrysochlorids and tenrecids. Laurasiatheria contains cetartiodactyls, perissodactyls, carnivores, pangolins, bats and eulipotyphlan insectivores. Parallel adaptive radiations have occurred within Laurasiatheria and Afrotheria. In each group, there are aquatic, ungulate and insectivore-like forms.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Novacek, M. J. Mammalian phylogeny: shaking the tree. Nature 356, 121–125 (1992).
Shoshani, J. & McKenna, M. C. Higher taxonomic relationships among extant mammals based on morphology, with selected comparisons of results from molecular data. Mol. Phylogenet. Evol. 9, 572–584 (1998).
de Jong, W. W. Molecules remodel the mammalian tree. Trends Ecol. Evol. 13, 270–275 (1998).
Springer, M. S., Burk, A., Kavanagh, J. R., Waddell, V. G. & Stanhope, M. J. The interphotoreceptor retinoid binding protein gene in therian mammals: implications for higher level relationships and evidence for loss of function in the marsupial mole. Proc. Natl Acad. Sci. USA 94, 13754–13759 (1997).
Springer, M. S. et al. Endemic African mammals shake the phylogenetic tree. Nature 388, 61–64 (1997).
Stanhope, M. J. et al. Molecular evidence for multiple origins of Insectivora and for a new order of endemic African insectivore mammals. Proc. Natl Acad. Sci. USA 95, 9967–9972 (1998).
Teeling, E. C. et al. Molecular evidence regarding the origin of echolocation and flight in bats. Nature 403, 188–192 (2000).
Asher, R. J. A morphological basis for assessing the phylogeny of the ‘Tenrecoidea’ (Mammalia, Lipotyphla). Cladistics 15, 231–252 (1999).
Luckett, W. P. & Hartenberger, J.-L. Monophyly or polyphyly of the order Rodentia: Possible conflict between morphological and molecular interpretations. J. Mammal. Evol. 1, 127–147 (1993).
Waddell, P. J., Cao, Y., Hauf, J. & Hasegawa, M. Using novel phylogenetic methods to evaluate mammalian mtDNA, including amino acid-invariant sites-logdet plus site stripping, to detect internal conflicts in the data, with special reference to the positions of hedgehog, armadillo and elephant. Syst. Biol. 48, 31–53 (1999).
Penny, D., Masegawa, M., Waddell, P. J. & Hendy, M. D. Mammalian evolution: Timing and implications from using the logdeterminant transform for proteins of differing amino acid composition. Syst. Biol. 48, 76–93 (1999).
McKenna, M. C. & Bell, S. K. Classification of Mammals Above the Species Level (Columbia Univ. Press, New York, 1997).
Rainger, R. Agenda for Antiquity: Henry Fairfield Osborn and Vertebrate Paleontology at the American Museum of Natural History, 1890–1935 (Univ. Alabama Press, Tuscaloosa, Alabama, 1991).
Kumar, S. & Hedges, S. B. A molecular timescale for vertebrate evolution. Nature 392, 917–920 (1998).
Foote, M., Hunter, J. P., Janis, C. M. & Sepkoski, J. J. Jr Evolutionary and preservational constraints on origins of biologic groups: divergence times of eutherian mammals. Science 283, 1310–1314 (1999).
Rich, T. H. et al. A tribosphenic mammal from the Mesozoic of Australia. Science 278, 1438–1442 (1997).
Mouchaty, S. K., Gullberg, A., Janke, A. & Arnason, U. The phylogenetic position of the Talpidae within Eutheria based on analysis of complete mitochondrial sequences. Mol. Biol. Evol. 17, 60–67 (2000).
Waddell, P. J., Okada, N. & Hasegawa, M. Towards resolving the interordinal relationships of placental mammals. Syst. Biol. 48, 1–5 (1999).
Rasmussen, D. T. in The Evolution of Perissodactyls (eds Prothero, D. R. & Schoch, R. M.) 57–78 (Oxford Univ. Press, Oxford, 1989).
Matthew, W. D. The Carnivora and Insectivora of the Bridger basin, middle Eocene. Mem. Am. Nat. Hist. 9, 291–567 (1909).
Novacek, M. J. The skull of leptictid insectivorans and the higher-level classification of eutherian mammals. Bull. Am. Mus. Nat. Hist. 183, 1–111 (1986).
Easteal, S. Molecular evidence for the early divergence of placental mammals. BioEssays 21, 1052–1058 (1999).
Thompson, J. D., Higgins, G. D. & Gibson, T. J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 (1994).
Swofford, D. L., Olsen, G. P., Waddell, P. J. & Hillis, D. M. in Molecular Systematics (eds Hillis, D. M., Moritz, C. & Mable, B. K.) 407–492 (Sinauer, Sunderland, Massachusetts, 1996).
Krajewski, C., Blacket, M., Buckley, L. & Westerman, M. A multigene assessment of phylogenetic relationships within the dasyurid marsupial subfamily Sminthopsinae. Mol. Phylogenet. Evol. 8, 236–248 (1997).
Swofford, D. L. PAUP*. Phylogenetic Analysis Using Parsimony (* and Other Methods) Version 4, (Sinauer, Sunderland, Massachusetts, 1998).
Rambaut, A. & Grassly, N. C. Seq-Gen: An application for the Monte Carlo simulation of DNA sequence evolution along phylogenetic trees. Comput. Appl. Biosci. 13, 303–306 (1997).
Rambaut, A. & Bromham, L. Estimating divergence dates from molecular sequences. Mol. Biol. Evol. 15, 442–448 (1998).
Acknowledgements
We thank F. Catzeflis for tissue samples. This work was supported by the NSF (M.S.S.) and the TMR program of the European Commission (W.W.d.J.; M.J.S.).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Madsen, O., Scally, M., Douady, C. et al. Parallel adaptive radiations in two major clades of placental mammals. Nature 409, 610–614 (2001). https://doi.org/10.1038/35054544
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/35054544
This article is cited by
-
Geometric morphometrics as a tool to identify Dasypodini osteoderms: Implications for the oldest records of Dasypus
Journal of Mammalian Evolution (2023)
-
Odontogenic ameloblast-associated (ODAM) is inactivated in toothless/enamelless placental mammals and toothed whales
BMC Evolutionary Biology (2019)
-
Rooting phylogenetic trees under the coalescent model using site pattern probabilities
BMC Evolutionary Biology (2017)
-
Phylogeny analysis from gene-order data with massive duplications
BMC Genomics (2017)
-
Mandibulate convergence in an armoured Cambrian stem chelicerate
BMC Evolutionary Biology (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.