Abstract
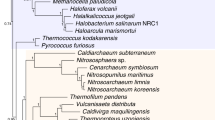
HOW many primary lineages of life exist and what are their evolutionary relationships? These are fundamental but highly controversial issues1. Woese and co-workers2–4 propose that archaebacteria, eubacteria and eukaryotes are the three primary lines of descent and their relationships can be represented by Fig. 1a (the 'archaebacterial tree') if one neglects the root of the tree. In contrast, Lake5,6 claims that archaebacteria are paraphyletic, and he groups eocytes (extremely thermophilic, sulphur-dependent bacteria) with eukaryotes, and halobacteria with eubacteria (the 'eocyte tree', Fig. 1b). Lake's view has gained considerable support as a result of an analysis6 of small subunit ribosomal RNA sequence data by a new approach, the evolutionary parsimony method7. Here we report that analysis of small subunit data by the neighbour-joining and maximum parasimony methods8,9 favours the archaebacterial tree and that computer simulations using either the archaebacterial or the eocyte tree as a model tree show that the probability of recovering the model tree is very high (>90 per cent) for both the neighbour-joining and maximum parsimony methods but is relatively low for the evolutionary parsimony method. Moreover, analysis of large subunit rRNA sequences by all three methods strongly favours the archaebacterial tree.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Penny, D. Nature 331, 111–112 (1988).
Woese, C. R. & Fox, G. E. Proc. natn. Acad. Sci. U.S.A. 74, 5088–5090 (1977).
Woese, C. R. & Wolfe, R. S. in Archaebacteria (eds Woese, C. R. & Wolfe, R. S.) The Bacteria Volume VIII, 561–564 (Academic, New York, 1985).
Woese, C. R. Microbiol. Rev. 51, 221–271 (1987).
Lake, J. A., Henderson, E., Oakes, M. & Clark, M. W. Proc. natn. Acad. Sci. U.S.A. 81, 3786–3790 (1984).
Lake, J. A. Nature 331, 184–186 (1988).
Lake, J. A. Molec. Biol. Evol. 4, 167–191 (1987).
Saitou, N. & Nei, M. Molec. Biol. Evol. 4, 406–425 (1987).
Fitch, W. M. Am. Nat. 111, 223–257 (1977).
Dams, E. et al. Nucleic Acids Res. 16, r87–r173 (1988).
Leffers, H., Kjems, J., Østergaard, L., Larsen, N. & Garrett, R. A. J. molec. Biol. 195, 43–61 (1987).
Felsenstein, J. Evolution 39, 783–791 (1985).
Cedergren, R., Gray, M. W., Abel, Y. & Sankoff, D. J. molec. Evol. 28, 98–112 (1988).
Gutell, R. R. & Fox, G. E. Nucleic Acids Res. 16, r175–r269 (1988).
Higgins, D. G. & Sharp, P. M. Gene 73, 237–244 (1988).
Li, W.-H., Wolfe, K. H., Sourdis, J. & Sharp, P. M. Symp. quant. Biol. 52, 847–856 (1987).
Olsen, G. J. Symp. quant. Biol. 52, 825–837 (1987).
Tautz, D., Hancock, J. M., Webb, D. A., Tautz, C. & Dover, G. A. Molec. Biol. Evol. 5, 366–376 (1988).
Lake, J. A. Nature 321, 257–258 (1986).
Earth's Earliest Biosphere (ed. Schopf, J. W.) (Princeton Univ. Press, Princeton, 1983).
Cavalier-Smith, T. in Endocytobiology III (eds Lee, J. J. & Fredrick, J. F.) Ann. New York Acad. Sci. 503, 17–54 (1987).
Berghöfer, B. et al. Nucleic Acids Res. 16, 8113–8128 (1988).
Lake, J. A. J. molec. Evol. 26, 59–73 (1987).
Kimura, M. J. molec. Evol. 16, 111–120 (1980).
Li, W.-H. Molec. Biol. Evol. (in the press).
Johansen, T., Johansen, S. & Haugli, F. B. Current Genet. 14, 265–273 (1988).
Höpfl, P., Ulrich, N., Hartmann, R. K., Ludwig, W. & Schleifer, K. H. Nucleic Acids Res. 16, 9043 (1988).
Murzina, N. V., Vorozheykina, D. P. & Matvienko, N. I. Nucleic Acids Res. 16, 8172 (1988).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Gouy, M., Li, WH. Phylogenetic analysis based on rRNA sequences supports the archaebacterial rather than the eocyte tree. Nature 339, 145–147 (1989). https://doi.org/10.1038/339145a0
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/339145a0
This article is cited by
-
Archaea and the origin of eukaryotes
Nature Reviews Microbiology (2017)
-
The hybrid nature of the Eukaryota and a consilient view of life on Earth
Nature Reviews Microbiology (2014)
-
An archaeal origin of eukaryotes supports only two primary domains of life
Nature (2013)
-
Some comments on calibration of molecular evolutionary rates
Immunogenetics (1992)
-
Early evolutionary relationships among known life forms inferred from elongation factor EF-2/EF-G sequences: Phylogenetic coherence and structure of the archaeal domain
Journal of Molecular Evolution (1992)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.