Abstract
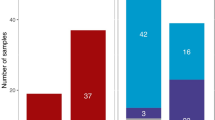
A sample of central Argentina (Córdoba) was genotyped for the first hypervariable region (HVS-I) plus a set of coding region mitochondrial DNA (mtDNA) single nucleotide polymorphisms (SNPs) (N = 102) and compared with a data set of Y-chromosome short tandem repeats (Y-STRs; N = 100) previously genotyped in the same individuals. We additionally compiled a database containing more than 4,000, 6,800, and 12,000 HVS-I sequences of Native American, sub-Saharan African, and European origin, respectively. The Y-Chromosome Haplotype Reference Database (YHRD) was used as a reference for the Y-STR profiles from Córdoba. The Native American component is highly prevalent on the maternal side (~41%) in contrast to the Y-chromosome paternal contribution (~2%), indicating a strong gender bias in the colonization and admixture processes that occurred in the recent history of Argentina, in agreement with historical records. The demographic input of African slaves in Córdoba was very high in the eighteenth century (~40% of the total population) but decreased dramatically after a few decades; therefore, the minor traces of sub-Saharan Y-chromosome and mtDNA lineages observed in our sample fit well with these historical records. The European Y-chromosome component of Córdoba (~97%; in contrast to the 57% observed in the mtDNA side) also mirrors the substantial immigration experienced by Argentina during the beginning of the last century, predominantly from Italy and Spain.
Similar content being viewed by others
Introduction
The present population of central Argentina (Córdoba) is the result of a complex amalgamation of different cultures and populations with different genetic ancestries. Today, various Native American groups exist that inhabit different regions of Córdoba. A small population of Comechingones (actual population size of ~5,000) still live in a relatively isolated region of the mountain ranges of Córdoba, whereas the Olongastas, considered a subgroup of the Diaguitas (~6,000 individuals), live at lower altitudes in the northwest of the province. Other groups, such as the Sanavirones in the northeast and the Pampeanos (formerly occupying the low flat areas of the Argentinean humid pampa), have since become extinct. Although some of these populations have preserved their folklore and other cultural traditions relatively well, none have retained their original languages and now speak Spanish exclusively.
During colonial times, the use of slave labor for agricultural development became an economic necessity for Spanish colonialists. The relatively low density of Native Americans coupled with the resistance of these population groups to Spanish acculturation and slavery led to the introduction of very large numbers of sub-Saharan Africans to the region. Although the demographic impact of Africans on the autochthonous population in other Latin American countries such as Colombia and Brazil was extremely high and still persists in modern populations (Alves-Silva et al. 2000; Beleza et al. 2005; Ely et al. 2006; Parra et al. 2001; Salas et al. 2005b, c; 2004), their real demographic impact in Argentina is a debatable topic. Some scholars maintain that a very significant number of African slaves were forcedly moved to Argentina, in particular to regions of intense agricultural activity: according to Victoria-Gomes (2002), in 1778 the population of Africans in Córdoba was ~44%, whereas in 1887 the official percentage of Africans was reduced to ~1.8%. The reasons for such a dramatic decrease remain uncertain; Victoria-Gomes (2002) pointed to epidemic causes (e.g., yellow fever) and colonial fights against neighboring populations (the number of Africans recruited in the army was disproportionately large compared with other ethnic groups).
During the nineteenth century, Argentina experienced a large-scale immigration of Europeans, notably from Italy and Spain (but including UK and Germany), which dramatically changed the demography of the country, mainly in urban areas. There are other well-known minorities in this region, including Jews (escaping persecution in the Second World War), Arabs, Armenians, and Japanese. Since Córdoba is one of the most important industrial centers of the country, over the past 50 years, it has attracted numerous immigrants from all over the country and from neighboring countries (i.e., Bolivia, Paraguay, etc.).
Finally, none of the existing Argentinean Native American groups live a completely isolated existence, and the degree of admixture, in particular with individuals of primary European ancestry, seems to be high in some populations. However, the magnitude of this admixture and the final demographic impact of African slaves in the region has not been properly evaluated.
We collected a sample of central Argentina (Córdoba) to estimate the different main ancestries that contributed to the present population. We analyzed mtDNA variation by sequencing the first hypervariable region (HVS-I) and a set of mtDNA coding region single nucleotide polymorphisms (SNPs). The degree of admixture on the maternal side was also contrasted with analysis of a set of Y-chromosome short tandem repeats (Y-STR) markers genotyped in the same individuals (Fondevila et al. 2003), firstly by inferring the haplogroup status of these Y-STR profiles, and secondly by searching these profiles in the worldwide Y Chromosome Haplotype Reference Database (YHRD).
Material and methods
Samples
We collected 102 healthy unrelated individuals from the province of Córdoba in central Argentina. Informed consent was given by all participants. The protocol and procedures employed were reviewed and approved by the review committee of the University of Santiago de Compostela, Spain, where genotyping was carried out. All persons gave their informed consent prior to inclusion.
PCR amplification and sequencing
All samples were amplified and sequenced (forward and reverse) for the HVS-I, analyzing the sequence range 16024–16400. Polymerase chain reaction (PCR) amplification was performed with GeneAmp 9700 thermocyclers, and sequencing analysis was performed as previously described (Álvarez-Iglesias et al. 2007). All samples were additionally genotyped for a set of ten SNPs following (Quintáns et al. 2004). Mutations are referred to the revised Cambridge Reference Sequence (rCRS) (Andrews et al. 1999). We followed a standardized forensic framework for nomenclature, as indicated in Carracedo et al. (2000) but with slight modifications considered in Salas et al. (2005a) concerning insertions. Data were checked following the phylogenetic principles described previously (Bandelt 1994; Bandelt et al. 2004a, b; Salas et al. 2007) to avoid sequence artifacts as much as possible.
Databases and statistical analysis
For phylogeographic purposes, we collected different available population data sets from the literature. Thus, for African haplotypes, we used roughly the same database reported in Černý et al. (2007), which consists of 6,856 profiles from different regions of the African continent. For Native American lineages, an HVS-I database that consists of 4,086 available sequences in the literature was employed, and for the western European database, we considered >12,000 publicly available profiles.
Only the HVS-I segment was used for population comparison, in particular, the 16090–16365 sequence range, as this is the common segment for the different populations used. Nomenclature of African haplotypes follows Salas et al. (2002, 2004), with updates in Černý et al. (2007), Kivisild et al. (2004), and Torroni et al. (2006). For Native American haplogroups, we use the most updated nomenclature from Bandelt et al. (2003) and Kong et al. (2006), and for the European profiles, Achilli et al. (2005, 2004), Loogväli et al. (2004), and Sun et al. (2006), among others. DnaSP 4.10.3 software (Rozas et al. 2003) was used for computation of different diversity indices. Principal component analysis was performed using Stata 9.1 (http://www.stata.com/).
Y-chromosome haplogroup inference
The number of occurrences of particular profiles in a worldwide database roughly indicates their most natural geographical origin. Minimal Y-chromosome haplotypes for 100 individuals were reported in Fondevila et al. (2003) and submitted to the YHRD (http://www.yhrd.org/index.html). We searched the YHRD for each of these profiles (YHRD; release 22) with particular focus on the main European source populations of Spain and Italy together with the major continental regions (e.g., Europe, Latin America, etc.), keeping the population grouping scheme provided by the YHRD. Haplogroup Predictor (https://home.comcast.net/~hapest5/index.html) was used for inferring haplogroup status of Y-STR profiles using flat a priori probabilities.
Results and discussion
Phylogeography of mtDNA HVS-I profiles
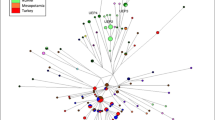
The Central Argentinean population has two well-differentiated mtDNA ancestral components: ~57% of the lineages are of European origin (predominantly from western Europe), and ~41% are typically Native American (Fig. 1). Only two individuals carried mtDNA of sub-Saharan provenance. About 31% of European lineages belong to haplogroup H, but there are representatives of other common European haplogroups (HV0, I, J, etc.), broadly reflecting the haplogroup spectra of a typical European population (Table 1). At the haplotypic level, the degree of molecular resolution does not allow the phylogeographic allocation of these European lineages to particular regions of Europe; in fact, most of these HVS-I profiles in Córdoba occur across the whole of Europe. There are only few outlined exceptions; for instance, we did not find matches for the haplogroup H profile C16111T T16209C C16270T. Some lineages have some more restricted geographical occurrence. For example, there are seven matches for the haplogroup W sequence C16173T C16223T C16292T T16325C T16352C; curiously, all of them are in Romania (Brandstätter et al. 2007) and Georgia (Quintana-Murci et al. 2004).
With regard to the Native American component, we observed representatives of the four main haplogroups, A2 (~8%), B2 (~5%), C1 (~14%), and D1 (~14%). Five out of nine of the A2 haplotypes match with the basal A2 haplotype, which is common across the American continent. There are four instances of the basal C1 haplotype in Córdoba, also common in populations living at different continental latitudes. In contrast, the basal HVS-I haplotype of D1 (T16362C T16325C) was not detected in our sample. However, we found 11 identical matches of the most common D1 haplotype C16223T C16242T T16311C T16325C T16362C, ten of them in other Argentinean populations: four matches in the Mapuches (Ginther et al. 1993) and the rest in the Pilagá and Wichí (Cabana et al. 2006) and in the Coyas (Álvarez-Iglesias et al. 2007). Two other matches were also observed in the Genographic database (https://www3.nationalgeographic.com/genographic/resources.html). There are other interesting matches for the D1 haplotypes of Córdoba. Haplotype C16187T T16189C T16209C C16223T T16325C T16362C appears four times in the database, three of them in the aboriginal and in the general Chilean population (Horai et al. 1993; Moraga et al. 2000) and one in the ancient Kaweskar DNA samples (Patagonia-Tierra del Fuego; South of Argentina, and Chile) studied by García-Bour et al. (2004).
As shown in Table 2, for the sequence range 16090–16385, there are 24 (out of 42) different Native American haplotypes in Córdoba. Most of them (N = 17) are only observed once, four sequences appear twice, two sequences occur five times, and one haplotype appears seven times (C16223T C16242T T16311C T16325C T16362C). Sequence (S) and nucleotide (π) diversities and average number of nucleotide diversity (M) are quite high for the Native American component (S = 0.948; π = 0.01767; M = 6.04) in contrast to the average values for the continental Native American component (S = 0.945; π = 0.01519; M = 3.43), indicating that the admixture process with Europeans was relatively gradual, allowing the preservation of a significant part of the Native American original gene pool in today’s general population of Córdoba. There are 13 identical matches between the Native American lineages and South America: nine in central America and ten in North America. However, six haplotypes are found across America (North, Central, and South) (Table 2). Moreover, there are nine haplotypes with only one representative in Córdoba that were still not observed in our large database.
We only detected two sequences of sub-Saharan origin belonging to haplogroups L1b1 and L1c1. For the L1b1 sequence (see Table 1), we did not find exact matches, but there are some one-step-mutation neighbors—such as A16166G on top of T16126C C16187T T16189C C16193T C16223T C16264T C16270T C16278T A16293G T16311C, which is found in Cabinda (Beleza et al. 2005)—in the Bakaka of south Cameroon (Coia et al. 2005) but also in the Tacuarembó from Uruguay. The typical central African L1c1 lineage G16129A A16163G C16187T T16189C T16209C C16223T C16278T A16293G C16294T T16311C C16360T also appears at moderate frequencies in Cabinda (Beleza et al. 2005) and other American countries.
Principal component analysis
We carried out a principal component analysis (PCA) based on mtDNA haplogroup frequencies and using several European, sub-Saharan African and two Native American data sets as population references (see Fig. 2 legend). PC1 (which accounts for 21% of the variability) primarily separates African from non-African populations, whereas PC2 (18%) basically splits European from Native American samples (see nested plot in Fig. 1). The plot also shows a clear-cut heterogeneity pattern in the African samples. In contrast, the Native American groups are tightly grouped in a single cluster, reflecting the fact that most of them essentially carry Native American lineages. A second round of PC analysis (the two first PCs accounting for 61% of the variability) was carried out excluding the African samples (main plot in Fig. 2). PC1 (32%) shows in one pole the European samples and in the other extreme the Native Americans ones. The European component of the Córdoba sample (57%) is clearly reflected in the PC2 (29%), as it is also the case for the Mexican data set (Green et al. 2000), which also has an important European mtDNA background (~15%).
Principle component analysis (PCA) based on mtDNA haplogroup frequencies. The main PCA plot includes several European and Native American samples plus the one from Córdoba analyzed in this study, whereas the nested small PCA plot includes three additional sub-Saharan African samples. References for population samples are as follows: a Africa (green dots): Mozambique (Salas et al. 2002), Angola (Plaza et al. 2004), Cabinda (Beleza et al. 2005); b Europe (blue dots): Italy (Bini et al. 2003), Galicia from Spain (Salas et al. 1998), and Germany (Lutz et al. 1998); and c America (red dots): Mexico (Green et al. 2000), Mapuche from Chile (Moraga et al. 2000), Guarani Kaiowá from Brazil (Marrero et al. 2007), Coya (Álvarez-Iglesias et al. 2007), Toba and Wichí from Argentina (Cabana et al. 2006), Navajo (Monson et al. 2002), Ayoreo from Bolivia and Paraguay (Dornelles et al. 2004), Ngobe from Panamá (Kolman et al. 1995), Córdoba (this study)
The most likely ancestry of Y-STR profiles
Inference of the haplogroup status based on the Bayes approach implemented in Haplogroup Predictor showed that ~70% of the profiles yielded a posteriori probabilities >0.98 in a unique and well-defined haplogroup (Table 3). In contrast to the mtDNA variation, most Y-STR lineages (~97%) can be allocated to typical European haplogroups, whereas only a small fraction correspond to lineages of likely Native American (~2%) and African ancestry (~1%) (Fig. 1). The most common European haplogroup is R1b (42%). This lineage is supposed to have spread into the rest of Europe from Iberian and other southern European refugia after the Last Glacial Maximum and today is the most frequent Y-chromosome lineage in Europe. Some other typical Mediterranean clades are also present in our central Argentinean sample. For instance, J2 is of near-Eastern origin but today it is prevalent across Mediterranean coastal regions, including the Iberian Peninsula. Its sister clade, J1, is probably of a more southern European origin and it is now common in the Mediterranean coastal regions. G2 is the most common G lineage in western Europe and makes up 8–10% of several Mediterranean populations (Spain, Italy, Greece, and Turkey).
Y-STRs profiles were also searched in the YHRD to investigate the number of times a particular profile was previously observed in other worldwide populations. This procedure provides additional indication regarding their most natural geographical origin. Strikingly, 25% of the Y-STR profiles do not find a single match in the YHRD. In agreement with their inferred haplogroup status, most haplotypes are more frequently observed in Europe or in populations with important European ancestry (e.g., USA). In agreement with historical documentation, a substantial number of profiles, 29 and 28%, have a higher frequency in Spain and Italy, respectively. For instance, profile 14/11–14/13/30/24/11/13/13 (inferred haplogroup status = R1b) appears three times in Córdoba with 25 matches in Spain (1.5%). The number of profiles that match certain central European samples is relatively high; however, this may just reflect the fact that the YHRD is substantially enriched with profiles obtained from specific areas (the YHRD European data set comprises one third of profiles obtained from Germany and Poland).
Haplotype 27 is of Native American ancestry (a posteriori probability for its haplogroup Q status = 0.94). Profile 5 has a more ambiguous haplogroup allocation (e.g., a posteriori probability of belonging to haplogroup Q = 0.17). When searching the YHRD, we did not find a single match in the whole database; however, a total of seven one-step mutation derivatives are only observed in Americans. Finally, haplotype 55 belongs to the sub-Saharan haplotype E3a (a posteriori probability for its haplogroup status = 0.96), and there are neither matches nor one-step mutation profiles in the YHRD (which could again reflect sampling bias due to the limited presence of sub-Saharan samples in the database).
Final remarks
There is a clear gender bias in the mtDNA and Y-chromosome composition of central Argentina. PCA, based on mtDNA haplogroup frequencies, with the two first principal components accounting for 61% of the variability), together with phylogeographic inferences, clearly indicates the halfway position of the Córdoba population between Europeans and Native Americans. In contrast, most Y-chromosomes in the general population of Córdoba are of European origin, with evidence for an important input from Italy and Spain. This is in agreement with historical records indicating that between 1869 and 1991, the average contribution of Italians and Spanish was 34% and 22% of total newcomers, respectively (source: INDEC, Instituto Nacional de Estadísticas y Censo from Argentina; http://www.indec.gov.ar). The presence of mtDNA and Y-chromosome lineages of sub-Saharan origin in Córdoba is low (<2%) but fits well with the demographic inferences of another study (Victoria-Gomes 2002).
Our results contrast with those obtained for other Argentinean populations. For instance, Dipierri et al. (1998) show that the Native American component in two northwestern Argentinean populations is ~65%, the introgression being more evident on the Y-chromosome side with frequencies of ~28% in Quebrada de Humahuaca and ~64% in San Salvador de Jujuy. On the other hand, the population of La Plata (Argentina) shows a Native American component of ~46% and a paternal contribution of ~11% (Martinez-Marignac et al. 2004). In the very isolated Argentinean village of Acuña (Bailliet et al. 2001), mtDNA is mainly Native American, whereas the Y-chromosome part is essentially European. As in Córdoba, all the above-mentioned studies clearly indicate directional mating in Argentina. Moreover, all these studies indicate Argentina shows a clear pattern of population substructure on the specific maternal and paternal genomes, which also corroborates the findings of previous studies based on autosomal STR markers (Toscanini et al. 2006). The forensic field and medical genetic studies will benefit from population studies across the Argentinean territory that would allow detailed knowledge of population structure and its consequences when estimating the weight of forensic haploid evidence (Egeland and Salas 2008) or evaluating the possibility of spurious positive results in medical genetic studies (Salas and Carracedo 2007).
References
Achilli A, Rengo C, Magri C, Battaglia V, Olivieri A, Scozzari R, Cruciani F, Zeviani M, Briem E, Carelli V, Moral P, Dugoujon JM, Roostalu U, Loogväli E-L, Kivisild T, Bandelt H-J, Richards M, Villems R, Santachiara-Benerecetti AS, Semino O, Torroni A (2004) The molecular dissection of mtDNA haplogroup H confirms that the Franco-Cantabrian glacial refuge was a major source for the European gene pool. Am J Hum Genet 75:910–918
Achilli A, Rengo C, Battaglia V, Pala M, Olivieri A, Fornarino S, Magri C, Scozzari R, Babudri N, Santachiara-Benerecetti AS, Bandelt H-J, Semino O, Torroni A (2005) Saami and Berbers—an unexpected mitochondrial DNA link. Am J Hum Genet 76:883–886
Álvarez-Iglesias V, Jaime JC, Carracedo Á, Salas A (2007) Coding region mitochondrial DNA SNPs: targeting East Asian and Native American haplogroups. Forensic Sci Int Genet 1:44–55
Alves-Silva J, da Silva Santos M, Guimaraes PE, Ferreira AC, Bandelt H-J, Pena SD, Prado VF (2000) The ancestry of Brazilian mtDNA lineages. Am J Hum Genet 67:444–461
Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N (1999) Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet 23:147
Bailliet G, Castilla EE, Adams JP, Orioli IM, Martinez-Marignac VL, Richard SM, Bianchi NO (2001) Correlation between molecular and conventional genealogies in Aicuña: a rural population from Northwestern Argentina. Hum Hered 51:150–159
Bandelt H-J (1994) Phylogenetic networks. Verhandlungen des Naturwissenschaftlichen Vereins Hamburg 34:51–71
Bandelt H-J, Herrnstadt C, Yao Y-G, Kong Q-P, Kivisild T, Rengo C, Scozzari R, Richards M, Villems R, Macaulay V, Howell N, Torroni A, Zhang Y-P (2003) Identification of Native American founder mtDNAs through the analysis of complete mtDNA sequences: some caveats. Ann Hum Genet 67:512–524
Bandelt H-J, Salas A, Bravi CM (2004a) Problems in FBI mtDNA database. Science 305:1402–1404
Bandelt H-J, Salas A, Lutz-Bonengel S (2004b) Artificial recombination in forensic mtDNA population databases. Int J Legal Med 118:267–273
Beleza S, Gusmão L, Amorim A, Carracedo Á, Salas A (2005) The genetic legacy of western Bantu migrations. Hum Genet 117:366–375
Bini C, Ceccardi S, Luiselli D, Ferri G, Pelotti S, Colalongo C, Falconi M, Pappalardo G (2003) Different informativeness of the three hypervariable mitochondrial DNA regions in the population of Bologna (Italy). Forensic Sci Int 135:48–52
Brandstätter A, Egyed B, Zimmermann B, Duftner N, Padar Z, Parson W (2007) Migration rates and genetic structure of two Hungarian Ethnic Groups in Transylvania, Romania. Ann Hum Genet 71:791–803
Cabana GS, Merriwether DA, Hunley K, Demarchi DA (2006) Is the genetic structure of Gran Chaco populations unique? Interregional perspectives on native South American mitochondrial DNA variation. Am J Phys Anthropol 131:108–119
Carracedo Á, Bar W, Lincoln P, Mayr W, Morling N, Olaisen B, Schneider P, Budowle B, Brinkmann B, Gill P, Holland M, Tully G, Wilson M (2000) DNA commission of the international society for forensic genetics: guidelines for mitochondrial DNA typing. Forensic Sci Int 110:79–85
Černý V, Salas A, Hájek M, Žaloudková M, Brdička R (2007) A bidirectional corridor in the Sahel-Sudan belt and the distinctive features of the Chad Basin populations: a history revealed by the mitochondrial DNA genome. Ann Hum Genet 71(Pt 4):433–452
Coia V, Destro-Bisol G, Verginelli F, Battaggia C, Boschi I, Cruciani F, Spedini G, Comas D, Calafell F (2005) Brief communication: mtDNA variation in North Cameroon: lack of Asian lineages and implications for back migration from Asia to sub-Saharan Africa. Am J Phys Anthropol 128:678–681
Dipierri JE, Alfaro E, Martínez-Marignac VL, Bailliet G, Bravi CM, Cejas S, Bianchi NO (1998) Paternal directional mating in two Amerindian subpopulations located at different altitudes in northwestern Argentina. Hum Biol 70:1001–1010
Dornelles CL, Battilana J, Fagundes NJ, Freitas LB, Bonatto SL, Salzano FM (2004) Mitochondrial DNA and Alu insertions in a genetically peculiar population: the Ayoreo Indians of Bolivia and Paraguay. Am J Hum Biol 16:479–488
Egeland T, Salas A (2008) Statistical evaluation of haploid genetic evidence. TOForensicSJ 1(1):4-11
Ely B, Wilson JL, Jackson F, Jackson BA (2006) African-American mitochondrial DNAs often match mtDNAs found in multiple African ethnic groups. BMC Biol 4:34
Fondevila M, Jaime JC, Salas A, Lareu MV, Carracedo A (2003) Y-chromosome STR haplotypes in Cordoba (Argentina). Forensic Sci Int 137:217–220
García-Bour J, Pérez-Pérez A, Álvarez S, Fernández E, López-Parra AM, Arroyo-Pardo E, Turbón D (2004) Early population differentiation in extinct aborigines from Tierra del Fuego-Patagonia: ancient mtDNA sequences and Y-chromosome STR characterization. Am J Phys Anthropol 123:361–370
Ginther C, Corach D, Penacino GA, Rey JA, Carnese FR, Hutz MH, Anderson A, Just J, Salzano FM, King MC (1993) Genetic variation among the Mapuche Indians from the Patagonian region of Argentina: mitochondrial DNA sequence variation and allele frequencies of several nuclear genes. Exs 67:211–219
Green LD, Derr JN, Knight A (2000) mtDNA affinities of the peoples of North-Central Mexico. Am J Hum Genet 66:989–998
Horai S, Kondo R, Nakagawa-Hattori Y, Hayashi S, Sonoda S, Tajima K (1993) Peopling of the Americas, founded by four major lineages of mitochondrial DNA. Mol Biol Evol 10:23–47
Kivisild T, Reidla M, Metspalu E, Rosa A, Brehm A, Pennarun E, Parik J, Geberhiwot T, Usanga E, Villems R (2004) Ethiopian mitochondrial DNA heritage: tracking gene flow across and around the gate of tears. Am J Hum Genet 75:752–770
Kolman CJ, Bermingham E, Cooke R, Ward RH, Arias TD, Guionneau-Sinclair F (1995) Reduced mtDNA diversity in the Ngöbé Amerinds of Panamá. Genetics 140:275–283
Kong Q-P, Bandelt H-J, Sun C, Yao Y-G, Salas A, Achilli A, Wang CY, Zhong L, Zhu CL, Wu SF, Torroni A, Zhang Y-P (2006) Updating the East Asian mtDNA phylogeny: a prerequisite for the identification of pathogenic mutations. Hum Mol Genet 15:2076–2086
Loogväli E-L, Roostalu U, Malyarchuk BA, Derenko MV, Kivisild T, Metspalu E, Tambets K, Reidla M, Tolk H-V, Parik J, Pennarun E, Laos S, Lunkina A, Golubenko M, Barać L, Peričić M, Balanovsky OP, Gusar V, Khusnutdinova EK, Stepanov V, Puzyrev V, Rudan P, Balanovska EV, Grechanina E, Richard C, Moisan JP, Chaventré A, Anagnou NP, Pappa KI, Michalodimitrakis EN, Claustres M, Gölge M, Mikerezi I, Usanga E, Villems R (2004) Disuniting uniformity: a pied cladistic canvas of mtDNA haplogroup H in Eurasia. Mol Biol Evol 21:2012–2021
Lutz S, Weisser HJ, Heizmann J, Pollak S (1998) Location and frequency of polymorphic positions in the mtDNA control region of individuals from Germany. Int J Legal Med 111:67–77
Marrero AR, Silva-Junior WA, Bravi CM, Hutz MH, Petzl-Erler ML, Ruiz-Linares A, Salzano FM, Bortolini MC (2007) Demographic and evolutionary trajectories of the Guarani and Kaingang natives of Brazil. Am J Phys Anthropol 132:301–310
Martinez-Marignac VL, Bertoni B, Parra EJ, Bianchi NO (2004) Characterization of admixture in an urban sample from Buenos Aires, Argentina, using uniparentally and biparentally inherited genetic markers. Hum Biol 76:543–557
Monson KL, Miller KWP, Wilson MR, DiZinno JA, Budowle B (2002) The mtDNA population database: an integrated software and database resource for forensic comparison. Forensic Sci Commun 4(2)
Moraga ML, Rocco P, Miquel JF, Nervi F, Llop E, Chakraborty R, Rothhammer F, Carvallo P (2000) Mitochondrial DNA polymorphisms in Chilean aboriginal populations: implications for the peopling of the southern cone of the continent. Am J Phys Anthropol 113:19–29
Parra EJ, Kittles RA, Argyropoulos G, Pfaff CL, Hiester K, Bonilla C, Sylvester N, Parrish-Gause D, Garvey WT, Jin L, McKeigue PM, Kamboh MI, Ferrell RE, Pollitzer WS, Shriver MD (2001) Ancestral proportions and admixture dynamics in geographically defined African Americans living in South Carolina. Am J Phys Anthropol 114:18–29
Plaza S, Salas A, Calafell F, Corte-Real F, Bertranpetit J, Carracedo Á, Comas D (2004) Insights into the western Bantu dispersal: mtDNA lineage analysis in Angola. Hum Genet 115:439–447
Quintana-Murci L, Chaix R, Wells RS, Behar DM, Sayar H, Scozzari R, Rengo C, Al-Zahery N, Semino O, Santachiara-Benerecetti AS, Coppa A, Ayub Q, Mohyuddin A, Tyler-Smith C, Qasim Mehdi S, Torroni A, McElreavey K (2004) Where west meets east: the complex mtDNA landscape of the southwest and Central Asian corridor. Am J Hum Genet 74:827–845
Quintáns B, Álvarez-Iglesias V, Salas A, Phillips C, Lareu MV, Carracedo Á (2004) Typing of mitochondrial DNA coding region SNPs of forensic and anthropological interest using SNaPshot minisequencing. Forensic Sci Int 140:251–257
Rozas J, Sánchez-DelBarrio JC, Messeguer X, Rozas R (2003) DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19:2496–2497
Salas A, Bandelt H-J, Macaulay V, Richards MB (2007) Phylogeographic investigations: the role of trees in forensic genetics. Forensic Sci Int 168:1–13
Salas A, Carracedo Á (2007) Studies of association in complex diseases: statistical problems related to the analysis of genetic polymorphisms. Rev Clin Esp 207:563–565
Salas A, Carracedo Á, Macaulay V, Richards M, Bandelt H-J (2005a) A practical guide to mitochondrial DNA error prevention in clinical, forensic, and population genetics. Biochem Biophys Res Commun 335:891–899
Salas A, Carracedo Á, Richards M, Macaulay V (2005b) Charting the ancestry of African Americans. Am J Hum Genet 77:676–680
Salas A, Comas D, Lareu MV, Bertranpetit J, Carracedo Á (1998) mtDNA analysis of the Galician population: a genetic edge of European variation. Eur J Hum Genet 6:365–375
Salas A, Richards M, De la Fé T, Lareu MV, Sobrino B, Sánchez-Diz P, Macaulay V, Carracedo Á (2002) The making of the African mtDNA landscape. Am J Hum Genet 71:1082–1111
Salas A, Richards M, Lareu MV, Scozzari R, Coppa A, Torroni A, Macaulay V, Carracedo Á (2004) The African diaspora: mitochondrial DNA and the Atlantic slave trade. Am J Hum Genet 74:454–465
Salas A, Richards M, Lareu MV, Sobrino B, Silva S, Matamoros M, Macaulay V, Carracedo Á (2005c) Shipwrecks and founder effects: divergent demographic histories reflected in Caribbean mtDNA. Am J Phys Anthropol 128:855–860
Sun C, Kong QP, Palanichamy M, Agrawal S, Bandelt H-J, Yao YG, Khan F, Zhu CL, Chaudhuri TK, Zhang YP (2006) The dazzling array of basal branches in the mtDNA macrohaplogroup M from India as inferred from complete genomes. Mol Biol Evol 23:683–690
Torroni A, Achilli A, Macaulay V, Richards M, Bandelt H-J (2006) Harvesting the fruit of the human mtDNA tree. Trends Genet 22:339–345
Toscanini UF, Gusmão L, Berardi G, Amorim A, Carracedo Á, Salas A, Raimondi EH (2006) Testing for genetic structure in different urban Argentinian populations. Forensic Sci Int 165(1):35–40
Victoria-Gomes M (2002) La presencia negroafricana en al Argentina. Pasado y permanencia. Boletín digital de la Biblioteca de la Nación Argentina 9:2
Acknowledgments
We thank Chris Phillips for critically reading the manuscript and making useful suggestions. The ‘Ramón y Cajal’ Spanish programme from the Ministerio de Educación y Ciencia (RYC2005-3). Grants from the Xunta de Galicia (PGIDIT06PXIB208079PR) and Fundación de Investigación Médica Mutua Madrileña awarded to AS partially supported this project.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Salas, A., Jaime, J.C., Álvarez-Iglesias, V. et al. Gender bias in the multiethnic genetic composition of central Argentina. J Hum Genet 53, 662–674 (2008). https://doi.org/10.1007/s10038-008-0297-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10038-008-0297-8
Keywords
This article is cited by
-
Genetic variation in populations from central Argentina based on mitochondrial and Y chromosome DNA evidence
Journal of Human Genetics (2018)
-
Association of candidate gene polymorphisms with clinical subtypes of preterm birth in a Latin American population
Pediatric Research (2017)
-
Charting the Y-chromosome ancestry of present-day Argentinean Mennonites
Journal of Human Genetics (2016)
-
Genomic insights on the ethno-history of the Maya and the ‘Ladinos’ from Guatemala
BMC Genomics (2015)
-
Evaluating the accuracy of AIM panels at quantifying genome ancestry
BMC Genomics (2014)