Abstract
The Han Chinese is the largest single ethnic group in the world, consisting of ten Chinese branches. With the exception of the Pinghua branch, the genetic structure of this group has been studied extensively, and Y chromosome and mitochondrial (mt)DNA data have demonstrated a coherent genetic structure of all Han Chinese. It is therefore believed that the Pinghua branch, being members of an old branch of the Han Chinese, despite being scattered in and around Guangxi Province where members of the Daic and Hmong-Mien are more prevalent than Han Chinese, is no exception. We have studied 470 individual samples (including 195 males) from Pinghua populations and other ethnic groups (Zhuang, Kam, Mulam, Laka, and Mien) from six areas (Hezhou, Fuchuan, Luocheng, Jinxiu, Sanjiang, and Wuxuan) in the north of the Guangxi Zhuang Autonomous Region of China. Both mtDNA and the Y chromosomes were typed in these samples. High frequencies of the Y chromosome haplogroups O2a* and O*, which always present at a high frequency among the populations of the southern minorities, were found in Pinghua populations. Only Pinghua populations in Luocheng and Jinxiu maintain the Han frequent haplogroup O3a5a. mtDNA lineages B4a, B5a, M*, F1a, M7b1, and N* were found in Pinghua populations, exhibiting a pattern similar to the neighboring indigenous populations, especially the Daic populations. Cluster analyses (dendrograms, principal component analyses, and networks) of Pinghua populations, the other Han branches, and other ethnic groups in East Asia indicated that Pinghua populations are much closer to the southern minorities than to the other Han branches. Admixture analyses confirmed this result. In conclusion, we argue that Pinghua populations did not descend from Han Chinese, but from southern minorities. The ancestral populations of Pinghua people were assimilated by the Han Chinese in terms of language, culture, and self-identification and, consequently, the Pinghua people became an exceptional branch of Han Chinese’s coherent genetic structure.
Similar content being viewed by others
Introduction
The Han Chinese are the largest ethnic group in the world, with more than 1.3 billion people—19% of the world population (Wen et al. 2004a). Most Han Chinese populations live in the agricultural region of East China; however, some are scattered in border areas among the minorities or in other countries. The origin, development, and expansion of the Han Chinese is an ongoing process with a long history. Han culture has greatly influenced the cultures of neighboring populations, contributing to the development of world civilization (Xu 1992). The ancestors of the Sino-Tibetan (an ethnic/linguistic phylum including Han Chinese and Tibeto-Burman) migrated from Southeast Asia to the upriver area basin of the Yellow River and became the proto-Sino-Tibetan population 10,000–40,000 years ago. Around 5000–6000 years ago, the Huaxia, who were the ancestors of Han Chinese, descended from the proto-Sino-Tibetan population (Su et al. 2000) and started their expansion based on their advanced agriculture, technology, and culture. As a large nation with fairly clear history, the genetic structure of Han Chinese is therefore a good prototype for studying the expansion of large ethnic groups.
Han populations and Han culture have expanded to South China during the past 2000 years. The indigenous populations in South China belong to the Daic, Hmong-Mien, and Austro-Asiatic phyla. Southern Han populations were then formed by interactions between Han Chinese and the southern minorities. Although some differences can be found between Southern Han and Northern Han in terms of language and culture, relative coherence is still maintained. In earlier genetics studies, we observed that the genetic structures of most Southern Han populations are still quite similar to that of the Northern Han, without much influence of the southern minorities. However, the Han population in the Guangxi province exhibits a rather different structure from the other Han populations, with a rather low proportion of Northern Han in admixture analyses (Wen et al. 2004a).
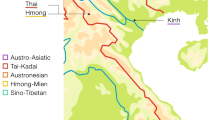
The language of Han Chinese is called Chinese or the Sinitic language, which is classified into ten major dialects: Mandarin, Jin, Cantonese (Yue), Wu, Hui, Xiang, Hakka, Gan, Min, and Pinghua (Fig. 1). Han Chinese populations are then classified into ten groups according to the linguistic classification. All nine Han branches have been investigated in previous molecular anthropological studies except for the Pinghua population. The Han populations in Guangxi belong to the Cantonese, Mandarin, Hakka, and Pinghua groups. These four branches may have been included in our previously studied Guangxi mixed sample. Since none of the nine previously studied branches exhibited any genetic particularity, the special structure of the Guangxi mixed sample may result from the uninvestigated Pinghua population proportion.
Branches of Han Chinese, sub-branches of Pinghua Han, and the sample distribution. The digitally marked areas on the map are the sampled regions: 1 Luocheng, 2 Sanjiang, 3 Hezhou, 4 Jinxiu, 5 Fuchuan, 6 Wuxuan. Han Chinese is classified into three subfamilies: Chin, Chi, and Chu. These three names are derived from the three strongest kingdoms during the Warring States Period (475–221 BC) when the Han Chinese started the colonization in East and South China and the dialects diversified into Western (Chin), Eastern (Chi), and Southern (Chu) groups. The upper map shows the distribution of all the Han Chinese branches, and only the majority branch is shown in the areas with more than one branch. Therefore, the Pinghua area in the upper map is a little different from the lower map with the distribution of Pinghua population alone
The word Pinghua means the Plebeian Dialect. The Pinghua Chinese is an old branch of Han Chinese, with a population of three to four million. Most of the Pinghua populations are located in the Guangxi province; however, some are also scattered in the Hunan, Yunnan, and Guangdong provinces adjacent to Guangxi. It has been suggested that the history of the Pinghua Chinese is older than those of the Hakka, Cantonese, and Min Chinese. Pinghua people maintain the typical Chinese language and culture, while they also display some cultural characteristics of the southern minorities, such as accent, raiment, and favorite foods. Within the Pinghua branch, the populations are classified into four subgroups (Xu 1999a): the North Guangxi, South Guangxi, Wuling, and Yulin (Fig. 1). The North Guangxi subgroup is the most typical Pinghua branch.
Alternative names of the Pinghua people are diverse in different areas (Zhan et al. 2003). The name Pinghua is used in the suburbs of Nanning (the capital of Guangxi), Yongning, and Lingui; Pingsheng (Plebeian Tone) in Yangshuo; Zheyuanhua (Canefield Dialect) in Youjiang; Tuguaihua (Native Chatter) in Rongshui and Rongan; Tubaihua (Native Colloquialism) in Guigang; Tuhua (Native Dialect) in Pingle; Cunhua (Rustic Dialect) in Hengxian; Kehua (Visitor Dialect—the same meaning as Hakkanese but not the same language) in Zuojiang; Baixinghua (Common Dialect) in Yongfu.
None of these Pinghua populations have been studied genetically. Here we report on our study of the genetic structure of Pinghua Chinese from different areas of the Guangxi province, both in the paternal and maternal lineages. The Pinghua branch was found to be the most different from the other Han branches, indicating that the formation of Han Chinese occurred not only by demic diffusion but also by the assimilation of minorities. We have also attempted to summarize the genetic relationships among the Han branches.
Materials and methods
Population samples
We collected 197 individual samples of Pinghua populations from the Hezhou, Fuchuan, Luocheng, Jinxiu, and Wuxuan Counties, Guangxi Zhuang Autonomous Region. Blood samples were collected randomly from healthy and unrelated individuals, including 101 males and 96 females. The individual samples were taken from individuals residing in different towns and villages to avoid the sampling bias. Neighboring control samples were also collected in the Guangxi indigenous minorities, including the Laka, Zhuang, Mulam, Kam, and Mien. The control samples were also from healthy and unrelated individuals, with a sample size of 273 (94 males and 179 females). All subjects signed informed consent forms. DNA was extracted by the classical phenol–chloroform method from the blood samples.
Y chromosome markers
Thirteen single nucleotide polymorphisms (SNPs) in the Y chromosome non-recombining portion were typed in the 195 male samples by PCR-restriction fragment length polymorphism (RFLP) (M130, M89, M9, M119, M110, M101, M268, M95, M88, M122, M164, M159, and M7). Seven SNPs (Yap, M15, M175, M111, M134, M117, M121), and six short tandem repeat polymorphisms (STRPs, DYS19, DYS389I, DYS390, DYS391, DYS392, DYS393) were typed by using fluorescently labeled primers for PCR amplification. The denatured products were electrophoresed on an acrylamide gel using an ABI 3100 genetic analyzer (Applied Biosciences, Foster City, CA). These markers are all highly informative in East Asia (Su et al. 2000; Li 2005; Shi et al. 2005). Y chromosome haplogroups were determined according to the classification of the Y-DNA Haplogroup Tree 2007 provided by International Society of Genetic Genealogy (ISOGG).
Mitochondrial DNA markers
Sequencing of the mtDNA HVS-1 region was performed for all 470 samples. Primers for HVS-1 sequencing were L15974 and H16488 (Yao et al. 2002). Purified PCR products were sequenced using the BigDye terminator cycle sequencing kit and an ABI 3100 genetic analyzer (Wen et al. 2004a, b, c). Primers were designed for amplifying multiple fragments that contain haplogroup diagnostic polymorphisms in the coding regions. The PCR products were then digested by restriction enzymes: 663HaeIII, 5176AluI, 12406HpaI, 4831HhaI, 9824HinfI, 5417RsaI, 10310NlaIII, 13259HincII, 9bp indel, 3391HaeIII, 10397AluI, 4715HaeIII (Kivisild et al. 2002; Yao et al. 2002). Both the HVS-1 motif and the coding region variations were used to infer haplogroups according to the classification of Kivisild et al. (2002). The HVS-1 sequences of 470 individuals have been submitted to GenBank (accession numbers EU277025–EU277489).
Statistical analysis
Population data on the Y chromosome (Li et al. 2003; Wen et al. 2004a, c; Li 2005; Chen et al. 2006; Feng 2007) and mtDNA ( Wen et al. 2004a, b, c; Li 2005; Li et al. 2007) were retrieved from the literature and included in our comparative analysis. Population dendrograms of Y haplogroups and mtDNA haplogroups were constructed separately by the furthest neighbor method using SPSS ver. 13.0 software (SPSS, Chicago, IL). The plots of principal component (PC) analysis of the East Asian populations were also constructed by SPSS ver. 13.0 using Y chromosome and mtDNA data separately. Admix ver. 2.0 (Dupanloup and Bertorelle 2001) and LEADMIX (Wang 2003) were used to evaluate the genetic contributions of Han Chinese and southern indigenous populations to the Pinghua Chinese population. The data of parent populations used in the admixture analyses were the sum of all the data collected from the literature. For Han Chinese, all of the data from the Han populations, with the exception of the Pinghua population, were included. For the southern minorities, we pooled the Hmong-Mien and Daic population data from four southern provinces (Guizhou, Guangxi, Hunan, and Guangdong). Both data from the literature and that obtained from our new samples were included. Three networks of the Y chromosome STRP haplotypes and eight networks of mtDNA HVS-1 motifs were constructed by the median-joining method (Bandelt et al. 1999) using Network ver. 4.201.
Results
Y chromosome haplogroup frequencies
Y chromosome haplogroups of all the 195 male samples were determined by the newest classification of ISOGG. Data derived from our newly collected samples are given in Table 1. The major haplogroups of Pinghua populations are O2a*, O3a5a, and O*, which is similar to the pattern (O2a*, O3*, K, and O*) of the neighboring minorities. However, the frequent haplogroup of Han Chinese, O3a5a, only appears at a high frequency in Luocheng and Jinxiu, two small population samples. O3a5a is rare in the southern indigenous populations, as determined in previous studies. The distributions of some haplogroups are ethnically associated: O1a and O2a* are frequent in Daic populations (Li 2005); O3* and O2a* are frequent in Hmong-Mien populations (Feng 2007); O3, O3a5, and O3a5a are frequent in Han Chinese (Su et al. 2000; Wen et al. 2004a; Shi et al. 2005). The presence of O2a* as the most frequent haplogroup of the Pinghua population indicates that the population is closer to southern minorities than to the other Han Chinese populations.
Mitochondrial DNA haplogroup frequencies
Forty-three mtDNA haplogroups were found in the 470 samples (Table 2). The mtDNA haplogroups present at high frequencies in the Pinghua populations are B4a, B5a, M*, F1a, M7b1, and N*; this is very similar to the frequencies in neighboring populations, especially Daic populations. The Pinghua population from Jinxiu, which has a high frequency of haplogroup F3, is different from the other populations. The frequency of haplogroup M7* in the Luocheng Pinghua population is also higher than those of other populations. Both mtDNA and Y chromosome haplogroup patterns of these two populations are different from those of the other Pinghua populations, but this may also result from the small sample size. The B, F, R9a, R9b, N9a, and M7 haplogroups are frequent haplogroups of southern indigenous populations. The Daic populations are characterized by B4a, B5a, F1a, M7b1, M7b*, M*, R9A, and R9b (Li et al. 2007); the Hmong-Mien populations by B4a, B5a, M*, M7b*, C, B4b1, M7b1, F1a, B4*, and R9b (Wen et al. 2004b); the Han Chinese by A, C, D, G, M8a, Y, and Z (Wen et al. 2004a). The mtDNA haplogroup pattern of Pinghua populations matches the pattern of the southern indigenous populations—not that of Han Chinese.
Dendrogram clustering
In the dendrograms of Fig. 2, we clustered the data of Y chromosome and mtDNA haplogroups from various populations in East Asia separately. The data of the populations in the same linguistic group were then pooled. Ten Han Chinese branches and six other linguistic phyla were pooled by averaging the population frequencies.
In the dendrogram of Y chromosome haplogroups, all of the Han branches, except for the Pinghua branch, cluster in one group, indicating the highly coherent paternal genetic structure of the Han Chinese ethnic group. The Tibeto-Burman falls into the Han cluster, matching the linguistic classification of the Sino-Tibetan phylum. The southern indigenous phyla form another cluster. The Pinghua Han branch falls into this clustering, exhibiting the southern feature of Pinghua populations. The northern-most phylum, Altaic, clusters with neither of the groups. Within the Sino-Tibetan cluster, the populations are clustered more geographically than linguistically. The geographically close branches of Xiang and Yue are also close in the dendrogram, though linguistically, Xiang belongs to the same branch of Wu, Hui, and Gan. The geographic influence is even clearer in mtDNA data clustering. The southernmost populations (Daic, Hmong-Mien, Pinghua, Yue, Min) fall into one cluster.
Principal component analyses
The Han Chinese and southern minority data were used in the principal component analyses. In the Y chromosome plot of Fig. 3a, southern and northern Han populations can be seen to cluster tightly in both PC 1 and PC 2. In the mtDNA plot of Fig. 3b, northern Han populations still cluster tightly, while some of the southern Han populations are scattered. Therefore, the highly coherent genetic structure of the Han Chinese is mirrored in the PC analyses, while the distribution of southern minority data is dispersed widely. The Pinghua populations are very far from the Han Chinese cluster in the Y chromosome plot, except for the Jinxiu and Luocheng populations. In the mtDNA plot, all of the Pinghua populations are scattered among the southern minorities, which may also reflect the sexual bias in the gene flow among populations. Maternal lineages are always more frequently floating by the paternal-dominant marriage. Therefore, the maternal lineages of Pinghua populations will be much closer to those of their neighbors, Daic and Hmong-Mien, than the paternal lineages.
Network analyses of the major haplogroups
The major Y chromosome haplogroups are O2a*, O*, and O3a5a. In the reference populations, we only found a high frequency of O* in the Zhuang population; in most of the data available on the Han ethnic population, O* is either rare or unexamined. Therefore, we did not analyze the network of O*. In Fig. 4, networks of O2a*, O3*, and O3a5a were analyzed using six STRPs. The STRP haplotypes of the Pinghua samples (Electronic Supplementary Material, Table 1) were compared with those of the Han, Daic, and Hmong-Mien (Chen et al. 2006; Feng 2007; Li 2005; Li 2007). There are at least two central haplotypes in the network of O2a*: one in the upper portion, and one towards the center. The upper center is occupied by Daic, Hmong-Mien, and Pinghua samples. Most of the other Han branch samples are in the upper part, and a Han-specific sub-clade can be observed in the terminal nodes with Min speakers from eastern Guangdong. This Han sub-clade is derived from the Hmong-Mien haplotype and matches the neighboring history of Han and She (an eastern Hmong-Mien population) in eastern Guangdong (Li 2007). Pure Daic or pure Hmong-Mien sub-clades can also be observed, indicating that the network is ethnically informative. Most of the Daic and Hmong-Mien haplotypes are in the center of the O2a* network. Most of the Pinghua and other Han haplotypes are likely derived haplotypes, as they are mostly at the terminals. The Pinghua samples share many more haplotypes with the southern minorities than with other Han Chinese. They also have more haplotypes connected to those of the southern minorities, indicating that most of the Pinghua people with the O2a* haplogroup are derived from the southern minorities. Compared with the two indigenous groups, the Pinghua haplotypes are more related to Hmong-Mien. In the network of O3*, Hmong-Mien haplotypes are closer to the center. The Han sub-clades are derived from the Hmong-Mien haplotypes, while the Daic haplotypes are on the periphery. Pinghua samples share more haplotypes in common with the Daic samples. In the network of O3a5a, Pinghua samples are more related to the other Han samples. These samples are from Luocheng, with highest frequency of O3a5a among the Pinghua population samples.
The mtDNA HVS-1 motif and the RFLP information are given in Electronic Supplementary Material Table 2. Based on the motifs, the networks of mtDNA haplogroups B4*, B4a, B5a, N9a, F*, F1a, R9b, and M7 were analyzed (Fig. 5). The reference data for the mtDNA HVS-1 motifs are from the literature (Wen et al. 2004b; Hill et al. 2007; Li et al. 2007). The mtDNA networks are always “sun-like” with a huge central haplotype and a great number of “sunlight” haplotypes around the center. Almost all of the ethnic phyla share the central haplotype, while fewer share the “sunlight” haplotypes. Interactions among the ethnic phyla can be frequently observed. In the networks of F*, M7, and N9a, the Pinghua samples are more related to the other Han samples, indicating that Pinghua people maintained more maternal lineages from Han Chinese than paternal lineages. Overall, however, the maternal lineages of the Pinghua populations are still closer to those of the southern minorities.
Admixture analyses
The Han Chinese originated in North China (around the Shaanxi Province) and spread to South China during the past two and a half thousand years (Wen et al. 2004a). Therefore, the possible genetic sources of southern Han can be northern Han and the southern minorities (mostly Daic and Hmong-Mien populations). The admixture ratio of these two parent populations in the southern Han populations can be estimated by admixture analyses. We used two programs (ADMIX2.0 and LEADMIX) to estimate the admixture ratio of the maternal and paternal lineages in Pinghua populations separately. We assumed that the admixture started 2500 years ago. As the other Han Chinese populations are all quite similar to each other, we pooled those samples (1693 individuals for Y chromosome and 2159 individuals for mtDNA; data are provided in Electronic Supplementary Material Table 3) as one of the potential parent populations. For another potential parent population, southern minorities, we pooled all of the Daic and Hmong-Mien samples from South China (1677 for Y chromosome and 2374 for mtDNA). Table 3 clearly showns that the results of the two programs are almost the same. The southern minority contribution to the Pinghua population is much higher than that of the Han Chinese, in both the maternal and paternal lineages. The negative proportions for the Han Chinese, shown in Table 3, may mean that the Han contributed more to the southern minorities than to the Pinghua populations. The relationships between the Pinghua population and the two potential parent populations in the networks are also included in Table 3. In the Y chromosome networks, the Pinghua samples share, or are connected with, significantly more haplotypes of the southern minorities than with the Han Chinese (P < 10−12). In the mtDNA networks, the Pinghua populations are not significantly closer to the southern minorities than to the other Han Chinese (P = 0.326). In conclusion, we argue that Pinghua populations are paternally derived from the southern minorities, and maternally influenced by other Han Chinese populations.
Discussion
Expansion principle of large nations represented by the coherent genetic structure of Han Chinese
Ethnically and linguistically, there are ten major branches of the Han Chinese. In our genetics analyses, the Han Chinese have shown a coherent genetic structure, with the exception of the Pinghua branch. The genetic structure of Han Chinese cannot only reveal the population history of Han itself but also shed light on some principles of the formation of large nations. It is widely accepted that the Han have a history of more than 5000 years. Evidence from archaeology, history, linguistics, and recent molecular anthropology studies suggest that the Huaxia, the predecessors of the Han, evolved from the ancient Sino-Tibetan population of western China and lived in the upriver basin of the Yellow River about 5000–6000 ago. The Huaxia flourished during the three earliest dynasties (Xia, Shang, and Zhou) and grew and spread out as agriculture improved; first eastward, then southward as the Han Chinese. In the past 2000 years, there have been at least three recorded waves of southward migration: (1) in the Eastern Jin Dynasty (317–420 AD), a population of more than 900,000 moved to the south; (2) more populations moved in the late Tang Dynasty, after the An-Shi Rebellion (755–762 A.D.); (3) in the Southern Song Dynasty (1127–1297 A.D.), a population of around 5 million moved to the south (Ge et al. 1997). When the Han migrants arrived in new regions, they were assimilated by or assimilated some of the indigenous populations, a population process that was common in Han Chinese history. In the Zhou Dynasty, Eastern Yi was assimilated; in the Qin and Han Dynasties, Chu and Yue; in the Southern and Northern Dynasties, five northern groups (Xiongnu, Xianbei, Jie, Di, Qiang); in the Tang Dynasty, Gok-Turks; in the Yuan Dynasty, Khitan and Jurchens. However, these striking events have not resulted in high proportions of exotic genetic contributions to the Han Chinese, as the population of Han Chinese was always much larger than those of the other ethnic groups; therefore, the coherent genetic structure can still be observed (Wen et al. 2004a). In conclusion, we suggest that a larger population must have been the precondition for assimilating the exotic populations. The formations of large nations were therefore mainly demic diffusions. If an exotic population had a preponderant population, it was almost impossible to be assimilated in ancient times, such as the Uighur and Mongol, as the cultures and languages diffused slowly without the intervention of modern media technology.
Social background of the Pinghua populations’ specific genetic structure
Our analyses have demonstrated that the genetic structure of Pinghua populations is much closer to that of the southern minorities. Therefore, they are not branches formed by the demic diffusion of Han Chinese, but are instead formed by the indigenous minorities who have accepted the Han culture. In Guangxi province, the population mainly belongs to the Daic phylum; some Hmong-Mien populations are also scattered among the Daic areas. Southern minorities are the dominant population in Guangxi and generally would not have been assimilated. However, Guangxi, which has been a province of China for the last 2500 years, has always been under the influence of Han culture, the dominant culture of China. People could easily travel from the neighboring Han-dominant provinces, Guangdong and Hunan, to Guangxi, thereby facilitating the spread of Han culture. Therefore, some populations would have accepted the Han culture, becoming this special branch of Han Chinese. Pinghua people are Han Chinese only in language, culture, and self-identification—not in genetic structure. This form of assimilation, in the absence of a Han-dominant population, was uncommon in the formation history of the Han.
Historically, the diffusion of Han culture in Guangxi began very early; the Zhuang, Kam, and Mien were all deeply influenced by the Han culture, and a large proportion of Han vocabulary was assimilated in their languages. Even the Thai people, who left Guangxi at least 1500 years ago, still have a large Han vocabulary as well as other influences from Han culture. Pinghua populations are located along the route used by the first group of Han migrants into Guangxi and Guangdong. The First Emperor of the Qin Dynasty (221–206 B.C.) dredged the Lingqu canal in north Guangxi, led his army through the region of Pinghua populations, and ultimately arrived in Guangdong. North Guangxi was not the final destination for the first group of Han migrants, but the Guangxi minorities along the way were deeply influenced by the culture, ultimately becoming a branch of Han.
However, we cannot prove that the genetic structure of present Pinghua population was inherited from the ancient Pinghua population. The Pinghua population might also begin with the early Han migrants and still have the core genetic proportion of Han origin. For example, in the network of the O3a5a haplogroup, a frequent Y-haplogroup of Han Chinese (Fig. 4), Pinghua samples are more related to other Han samples than to the southern minorities, and some are at the terminals of the network. This implies that the paternal lineage has independently evolved in other Hans and the Pinghua for some generations since the divergence of the two groups. Thus, it is equally plausible that an ancient Han (defined genetically and culturally) settled the Guangxi province, and then an extensive gene flow from the neighboring indigenous groups to the Han migrants has markedly reduced genetic signatures of the ancient Hans at the population level. That is, the Pinghua might be “genetically switched” descendants of the Han migrants.
Southern indigenous characters of Pinghua populations
Since the Qin Dynasty, there have been several waves of military migrations from North China to Guangxi. In 214 BC, the Qin army arrived in Guangxi; in 1053 AD, the Song army, led by General Di Qing, arrived for a counterinsurgency; in 1368 AD, the Ming army, led by General Liao Yongzhong, reoccupied Guangxi (Yuan and Huang 2005). However, the Han migrants in Guangxi were never dominant in terms of population numbers and were ultimately diluted in the indigenous populations. The dominant Han culture and language, however, was introduced with the migrants and influenced the indigenous populations. Genetically, Pinghua populations are not descendants of Han migrants, but of the culturally switched minorities. Therefore, Pinghua populations exhibit not only obvious characteristics of Han culture, but those of the southern minority culture as well, such as raiment, wedding, and exequies customs (Xu 1999a, b). Linguistically, Pinghua also maintain some southern minority features. For example, the alveolar lateral fricative consonant [ ] is not a remnant of the ancient Chinese, but an accent of Daic. In Southern Pinghua, the quartering of the entering tone is also a characteristic of Daic languages. Alternatively, Pinghua exhibit more typical Chinese phonology and vocabulary (Li 2000). Hereby, we assume that Pinghua was formed while the Guangxi minorities were learning the Chinese language based on a native language background, resulting in the dual traits of the Pinghua dialect (Pan 2002). The language and culture were not formed at one time, but were the result of Han migrants of different periods mixing with the native cultures.
] is not a remnant of the ancient Chinese, but an accent of Daic. In Southern Pinghua, the quartering of the entering tone is also a characteristic of Daic languages. Alternatively, Pinghua exhibit more typical Chinese phonology and vocabulary (Li 2000). Hereby, we assume that Pinghua was formed while the Guangxi minorities were learning the Chinese language based on a native language background, resulting in the dual traits of the Pinghua dialect (Pan 2002). The language and culture were not formed at one time, but were the result of Han migrants of different periods mixing with the native cultures.
Reference
Bandelt HJ, Forster P, Rohl A (1999) Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16:37–48
Chen J, Li H, Qin ZD, Liu WH, Lin WX, Yin RX, Jin L, Pan SL (2006) Y-chromosome genotyping and genetic structure of Zhuang populations. Acta Genet Sin 33:1060–1072
Dupanloup I, Bertorelle G (2001) Inferring admixture proportions from molecular data: extension to any number or parental populations. Mol Biol Evol 18:672–675
Feng H (2007) General history of Yao Nationality (Yaozu Tongshi). Minzu Press, Beijing
Ge JX, Wu SD, Cao SD (1997) The migration history of China. Fujian People’s Press, Fuzhou
Hill C, Soares P, Mormina M, Macaulay V, Clarke D, Blumbach PB, Vizuete-Forster M, Forster P, Bulbeck D, Oppenheimer S, Richards M (2007) A mitochondrial stratigraphy for island southeast Asia. Am J Hum Genet 80:29–43
Kivisild T, Tolk HV, Parik J, Wang Y, Papiha SS, Bandelt HJ, Villems R (2002) The emerging limbs and twigs of the East Asian mtDNA tree. Mol Biol Evol 19:1737–1751
Li H (2005) Genetic structure of Austro-Tai populations. PhD Dissertation of human biology, Fudan University, Shanghai
Li H (2007) Abscondence of Min-Yue Ethnic group revealed by molecular anthropology. J Guangxi Univ Nationalities (Phil Soc Sci) 29:42–47
Li H, Pan W, Wen B, Yang N, Jin J, Jin L, Lu D (2003) Origin of Hakka and Hakkanese: a genetics analysis. Acta Genet Sin 30:873–880
Li H, Cai XY, Winograd-Cort ER, Wen B, Cheng X, Qin ZD, Liu WH (2007) Mitochondrial DNA diversity and population differentiation in Southern East Asia. Am J Phys Anthropol 134:481–488
Li LJ (2000) The history of PingHua. Chin Lang Literat (Zhongguo Yuwen) 6:24–30
Pan WY (2002) Language contact and the formation of Southeast Chinese Dialects. In: Zou JY (ed) Proc Hong Kong Language Contact Round Table. City University of Hong Kong Press, Hong Kong
Shi H, Dong YL, Wen B, Xiao CJ, Peter A. Underhill, Shen PD, Chakraborty R, Jin L, Su B (2005) Y-chromosome evidence of Southern Origin of the East Asian-Specific Haplogroup O3-M122. Am J Hum Genet 77:408–419
Su B, Xiao C, Deka R, Seielstad MT, Kangwanpong D, Xiao J, Lu D, Underhill P, Cavalli-Sforza L, Chakraborty R, Jin L (2000) Y chromosome haplotypes reveal prehistorical migrations to the Himalayas. Hum Genet 107:582–590
Wang J (2003) Maximum-likelihood estimation of admixture proportions from genetic data. Genetics 164:747–765
Wen B, Li H, Lu D, Song X, Zhang F, He Y, Li F, Gao Y, Mao X, Zhang L, Qian J, Tan J, Jin J, Huang W, Deka R, Su B, Chakraborty R, Jin L (2004a) Genetic evidence supports demic diffusion of Han culture. Nature 431:302–305
Wen B, Li H, Gao S, Mao X, Gao Y, Li F, Zhang F, He Y, Dong Y, Zhang Y, Huang W, Jin J, Xiao C, Lu D, Chakraborty R, Su B, Deka R, Jin L (2004b) Genetic structure of Hmong-Mien speaking populations in East Asia as revealed by mtDNA lineages. Mol Biol Evol 22:725–733
Wen B, Xie XH, Gao S, Li H, Shi H, Song XF, Qian TZ, Xiao CJ, Jin JZ, Su B, Lu D, Chakraborty Ranajit, Jin L (2004c) Analyses of Genetic Structure of Tibeto-Burman populations reveals sex-biased admixture in Southern Tibeto-Burmans. Am J Hum Genet 74: 856–865
Xu JS (1992) The development history of the Han Nationality. Sichuan Ethnic Press, Chengdu
Xu JS (1999a) Formation and culture traits of PingHua Population. J Guangxi Univ Nationalities (Phil Soc Sci) 21:103–108
Xu JS (1999b) Formation and culture traits of PingHua Population (continuation). J Guangxi Univ Nationalities (Phil Soc Sci) 21:93–97
Yao YG, Kong QP, Bandelt HJ, Kivisild T, Zhang YP (2002) Phylogeographic differentiation of mitochondrial DNA in Han Chinese. Am J Hum Genet 70:635–651
Yuan SL, Huang NJ (2005) Alternations and the external reasons for their prosperity of Guangxi dominating languages (including Local Chinese Dialects and Putonghua). J Liuzhou Teach Coll 20:64–68
Zhan BH, Chui SH, Liu XZ, Yang W (2003) The issues about the adscription of GuangXi PingHua. Chin Lang Res (Yuwen Yanjiu) 3:47–52
Acknowledgments
We especially thank all of the donors of our samples. The sample collectors include Yunping Lei of Fudan University, Zhe Wei, Shuo Lian, and Lisha Wang of Guangxi Medical University. Weixiong Lin and Xiaoqiu Luo also took part in the DNA extraction work. Our research was supported by the National Social Science Fund projects of China (2004 No.05XMZ015) to JSX, the National Natural Science Foundation of China (No.39993420) and the Genographic Project of National Geography to LJ, and Guangxi Natrural Science Foundation (No.GSN0728160, GSY0542044) to SLP.
Author information
Authors and Affiliations
Consortia
Corresponding author
Additional information
Rui-Jing Gan and Shang-Ling Pan contributed equally to this work.
Website:
Y chromosome haplogroup nomenclature of International Society of Genetic Genealogy: http://www.isogg.org/tree/ISOGG_YDNATreeTrunk07.html
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Gan, RJ., Pan, SL., Mustavich, L.F. et al. Pinghua population as an exception of Han Chinese’s coherent genetic structure. J Hum Genet 53, 303–313 (2008). https://doi.org/10.1007/s10038-008-0250-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10038-008-0250-x
Keywords
This article is cited by
-
Differentiated adaptative genetic architecture and language-related demographical history in South China inferred from 619 genomes from 56 populations
BMC Biology (2024)
-
Analysis of maternal genetic structure of mitochondrial DNA control region from Tai-Kadai-speaking Buyei population in southwestern China
BMC Genomics (2024)
-
Y-chromosome diversity suggests southern origin and Paleolithic backwave migration of Austro-Asiatic speakers from eastern Asia to the Indian subcontinent
Scientific Reports (2015)
-
A genetic contribution from the Far East into Ashkenazi Jews via the ancient Silk Road
Scientific Reports (2015)
-
Tracing the legacy of the early Hainan Islanders - a perspective from mitochondrial DNA
BMC Evolutionary Biology (2011)