Abstract
NPY is a 36-aminoacid peptide expressed in several areas of the nervous system. Neuropeptide Y (NPY) receptors represent a widely diffused system that is involved in the regulation of multiple biological functions. The human NPY gene is located in chromosome 7. The functional significance of coding Leu7Pro polymorphism in the signal peptide of preproNPY is known. Six hundred and fifty four individuals of 14 ethnic Indian populations were screened for three mutations in the NPY gene, including Leu7Pro. We found that the Pro7 frequencies among the studied populations were much higher than in previous studies from other parts of the world. The highest allele frequency of Pro7 was detected in the Kota population in the Nilgiri Hill region of south India, and this may reflect a founder event in the past or genetic drift. All populations followed the Hardy–Weinberg equilibrium for the assayed markers. A total of five haplotypes were observed, only two of which were found to occur with a high frequency in all populations. No linkage disequilibrium (LD) was observed across the tested alleles in any population with the exception of Leu7Pro and Ser50Ser in the Badaga population (χ 2 = 13.969; p = 0.0001).
Similar content being viewed by others
Introduction
Neuropeptide Y (NPY) is the most abundant neuropeptide in the brain and is a widely diffused system that is involved in the regulation of multiple biological functions. NPY is expressed in several regions of the nervous system and is a pleiotropic factor that participates in the control of multiple physiological processes, such as cognitive functions, eating behavior, circadian rhythms, neuroendocrine mechanisms, reproductive and cardiovascular functions (McDonald 1990; Gabriel et al. 1996; Gribkoff et al. 1998; Bray et al. 2000; Magni 2003). NPY is a 36-amino-acid peptide that consists of an alpha-helix folded underneath a proline helix with a tyrosine residue at the carboxy terminus. The gene is highly conserved with 92% amino acid sequence identity between the cartilaginous fish Torpedo marmorata and mammals, which are separated by an evolutionary distance of more than 400 million years (Larhammar 1996). This high level of conservation indicates that NPY presumably has a critical physiological function and that interindividual variation at this locus is likely to be minimal.
A thymidine (T) to cytosine (C) polymorphism is present at nucleotide 1,128 of the human NPY gene, resulting in a substitution of leucine to proline (Leu7–Pro7) in the signal part of preproNPY (Karvonen et al. 1998). Individuals with the Leu7/Pro7 genotype have an average 42% maximal increase in plasma NPY in response to physiological stress when compared with Leu7/Leu7 individuals (Kallio et al. 2001). The Leu7Pro polymorphism in the NPY gene has been shown to be implicated in lipid metabolism regulation. Presence of the Pro7 variant is a suspected cardiovascular risk factor in Caucasians (Karvonen et al. 1998, 2001; Niskanen et al. 2000a). The Leu7Pro polymorphism has been associated with accelerated progression of carotid atherosclerosis in both obese healthy subjects (Karvonen et al. 2001) and patients with type-2 diabetes (Niskanen et al. 2000a). Type-2 diabetic patients with the Pro7 substitution had a higher prevalence of diabetic retinopathy (Niskanen et al. 2000b; Pettersson-Fernholm et al. 2004).
In general, the frequency of the Pro7 allele has been reported to be 8% among Finns and 3% among Dutchmen (Karvonen et al. 1998). There was no report of the presence of this allele among Japanese (Drube et al. 2001; Makino et al. 2001; Itokawa et al. 2003) and Korean populations (Ding et al. 2002). Jia et al. (2005) reported an extremely low frequency of the Pro7 allele in China. Although the NPY gene has been studied in some European and Asian populations, no published studies exist on Indian populations, which comprise nearly 5,000 endogamous populations.
There are 4,635 well-defined endogamous populations in India, which are culturally stratified as tribes and non-tribes. The 532 tribal communities, who are assumed to be the aboriginal inhabitants of the subcontinent, constitute 7.76% of the total population (http://www.censusindia.net). A few studies reported a closer affinity of Indian castes with either European or Asian populations (Bamshad et al. 2001). Our previous study suggested that the vast majority of the Indian maternal gene pool (>98%), consisting of Indo-European and Dravidian speakers, is genetically more or less uniform (Thanseem et al. 2006). The populations selected in the present study encompass the entire range of Indian social structure. In this study we evaluate the allele frequency, haplotype frequency and intermarker linkage disequilibrium (LD) distributions for three single-nucleotide polymorphisms (SNPs) of NPY gene in 14 different Indian populations. We find important differences in the distributions of allele and haplotype frequencies among different populations. These differences in the allele frequency and haplotype frequencies among populations will be useful in association studies or linkage analysis to determine phenotypes such as obesity, hypertension, and alcoholism.
Materials and methods
Populations
Study populations included a total of 654 unrelated males (to allow future studies of both mitochondrial and Y chromosomal variations) belonging to 14 ethnic groups, who inhabit geographically diverse regions of India (Fig. 1, Table 1). All subjects were normal healthy volunteers. The study protocol was approved by the ethics committee of the Centre for Cellular and Molecular Biology, Hyderabad, India. After obtaining informed written consent, a blood sample was collected from each individual.
Genotyping
Genomic DNA was extracted from all participants using the published protocol (Thangaraj et al. 2002). Forward (NPE2F 5′-CCTGGGTTCTCTCTGCGGGACTG-3′) and reverse (NPE2R 5′-CCCATTTTGTGTAGAGTGTGCCCTGT-3′) primers were used to amplify 516 bp of exon 2 and its flanking region of NPY. Amplifications were performed in a 10- μl volume containing one unit of AmpliTaqGold (PerkinElmer, Wellesley, MA, USA), 1X PCR buffer, 1.5 mM MgCl2, 200 μM deoxynucleotide triphosphates, 1 pmol of forward primer, one pmol of reverse primer, and 40 ng of genomic DNA. PCR products were visually verified on 2% agarose gels and were directly sequenced using Big Dye Terminator cycle sequencing on an ABI PRISM 3730 DNA analyzer (Applied Biosystems, Foster City, CA, USA).
Statistical analysis
All NPY alleles were tested for Hardy–Weinberg equilibrium using χ 2tests with one degree of freedom and Monte Carlo simulation using the HWSIM program (Cubells et al. 1997). The relationship between the geographic latitude of the population and the associated Pro7 allele frequency was estimated by simple linear regression using SPSS software. F statistics were calculated according to the methods of Weir and Cockerham (1984) with FSTAT (Goudet 1995). Haplotypes were constructed by maximum likelihood with an eExpectation–maximization method. Within each population, the haplotype frequencies were obtained from three SNPs by summing the frequencies of all haplotypes with each specific combination of alleles at two sites. Haplotype frequencies were estimated by Arlequin (Schneider et al. 2000). Linkage disequilibrium (D′ and r 2) was estimated using HaploView 3.12 (Barrett et al. 2005).
Results
A total of three polymorphic sites were observed in exon 2 of NPY gene, only one of which resulted in an amino acid change (T1128C/Leu7Pro; A1198G/Ala28Ala; G1258A/Ser50Ser). We designated the wild and mutant alleles with their respective nucleotides for all three coding SNPs. For haplotypes, we used these designations in order from 5′ to 3′. For example, a haplotype described as TGG indicated the presence of all ancestral alleles at the three coding sites. A significant difference in allele and genotypic frequencies was observed among populations (Table 2). The Pro7 allele was observed in nine out of 14 populations at frequencies ranging from 0.014 (Kurumba) to 0.233 (Kota); Pro7 homozygotes were found only in the Kota (Table 2). To investigate geographical trends in Pro7 allelic variation, we plotted the frequency of Pro7 frequency against degrees of latitude. A weak tendency for the Pro7 frequency to decrease north-to-south was observed, although the Kota were an outlier (Fig. 2) with a correlation coefficient r = 0.250 (p > 0.05). Of the 15,937 individuals screened around the world thus far, including the present study, the average frequency of Pro7 was found to be 0.048.
The Ala28Ala marker was polymorphic only in the Siddi, with a minor allele frequency of 0.026. The Ser50Ser marker was polymorphic in all studied populations, with the minor allele (A) frequency ranging from 0.186 (Kota) to 0.682 (Onge). None of the populations deviated significantly from Hardy–Weinberg equilibrium for all the polymorphic loci.
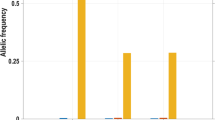
When haplotypes were constructed, a total of five haplotypes were observed out of which only two (TGG, TGA) were found at high frequency in all populations (combined frequency was 0.94–1.0) (Fig. 3). Haplotype TGG carried all ancestral alleles and haplotype TGA carried the derived allele at the third marker. Haplotype CGG was found at very low frequencies in several populations, but reached a frequency of 0.233 in the Kota because of the high frequency of the Pro7 allele in this population. In the Pro7 allele-bearing haplotypes, the “C” allele (Pro7) was found in association with the two wild-type alleles (CGG) with frequencies ranging from 1.7 to 23.3%, while in Kurumbas the Pro7 allele was associated with the mutant allele (A) of Ser50Ser locus (CGA). Haplotype CGA was found only in the Kurumba (0.014), while haplotype TAG was found only in the Siddi (0.026). Linkage disequilibrium was detected only between Leu7Pro and Ser50Ser (D’ = 1; r 2 = 0.069; χ 2 = 13.969; p = 0.001) loci in the Badaga.
Discussion
We report, for the first time, significant frequencies of the Pro7 allele of NPY in the majority of the Indian populations studied [0.014 (Kurumba) to 0.233 (Kota)]. The Pro7 allele has already been found in a few populations worldwide, such as Asians [Japanese (Drube et al. 2001; Makino et al. 2001; Okubo et al. 2001; Lappalainen et al. 2002; Itokawa et al. 2003); Koreans (Ding et al. 2002); Japanese in Tokyo and Han, Chinese in Beijing (http://www.hapmap.org)] and Africans [African–American (Lappalainen et al. 2002); Yoruba in Ibadan, Nigeria (http://www.hapmap.org)]. Jia et al. (2005) reported an extremely low frequency of Pro7 in a Chinese population, although Duan et al. (2005) failed to detect this allele in the same region. In our study, the highest frequency of this allele (0.23) was observed in the Kota (haplotype CGG frequency is 0.23) Since the Kota have been a small, isolated population for at least 2,000 years (Breeks 1873; Saha 1976; Roychoudhury et al. 2001), the high frequency of Pro7 is likely due to a founder effect or genetic drift. The Pro7 allele is also found in the Siddi, who are a recent migrant group from Africa (haplotype CGG frequency is 0.017). Ding et al. (2003) compiled data on 6,626 individuals from several studies that found the Pro7Pro genotype in only seven samples (six Finns and one European American). They proposed that the Pro7 allele might have originated in the north of Europe and spread to neighboring groups in varying frequencies due to the effects of genetic drift and isolation by distance. Lappalainen et al. (2002) also found the Pro7 allele at significant frequencies in Bedouin populations from the Middle East (0.078), and at lower frequencies in north and northeast African populations (0.022 in Moroccans and 0.011 in Ethiopian Jews), suggesting a possible African/Middle East origin for the Pro7 allele. The presence of the Pro7 allele in the Siddi, an African immigrant population, would support this interpretation. However, Pro7 is not found among populations thought to be the first migrants out of Africa, such as the Onge (Thangaraj et al. 2005), suggesting that its origin may postdate the first migration out of Africa.
Previous studies suggested that the “A” allele Ala28Ala locus was an “African-specific” polymorphism (http://www.hapmap.org). The frequency of this “A” allele in African populations is 0.05 (in Harare, Zimbabwe), 0.025 (Nigeria) and 0.045 (African American) (http://www.hapmap.org). The fact that this allele is found only in the Siddi is in accord with the African ancestry of the Siddi. In addition to the two major common haplotypes (TGG and TGA), three others (CGG, CGA and TAG) were observed in at least one of the populations studied and have an average frequency of >1%. Lack of significant LD between pairs of loci in the NPY gene region may be attributed to the low frequency of the alleles, since all measures of LD show some allele frequency dependence in finite sample sizes (Zapata 2000; Abecasis et al. 2001; Mueller 2004). The observation that the TGG haplotype is both common and highly conserved suggests that other factors may be acting to maintain the haplotype at high frequency. One possibility is positive selection that is so recent there has been little opportunity for recombination to break up the haplotype. This may result in LD around a specified allele that is disproportionately long-range given its population frequency. Alternatively, epistatic selection for combinations of alleles at two or more loci on a chromosome may influence LD patterns across haplotypes.
Understanding the evolutionary history of the NPY locus may provide insight into the evolutionary history of the gene in these populations and help to identify the haplotype blocks (The International HapMap Consortium 2003) that are most informative when identifying potential functional variation in this locus, which is associated with many physiological mechanisms.
References
Abecasis GR, Noguchi E, Heinzmann A, Traherne JA, Bhattacharyya S, Leaves NI, Anderson GG, Zhang Y, Lench NJ, Carey A, Cardon LR, Moffatt MF, Cookson WO (2001) Extent and distribution of linkage disequilibrium in three genomic regions. Am J Hum Genet 68:191–197
Bamshad M, Kivisild T, Watkins WS, Dixon ME, Ricker CE, Rao BB, Naidu JM, Prasad BV, Reddy PG, Rasanayagam A, Papiha SS, Villems R, Redd AJ, Hammer MF, Nguyen SV, Carroll ML, Batzer MA, Jorde LB (2001) Genetic evidence on the origins of Indian caste populations. Genome Res 11:994–1004
Barrett JC, Fry B, Maller J, Daly MJ (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21:263–265
Bray MS, Boerwinkle E, Hanis CL (2000) Sequence variation within the neuropeptide Y gene and obesity in Mexican Americans. Obes Res 8:219–226
Breeks JW (ed)(1873) An account of the primitive tribes and monuments of the Nilgiris. India Museum, London
Cubells JF, Kobayashi K, Nagatsu T, Kidd KK, Kidd JR, Calafell F, Kranzler HR, Ichinose H, Gelernter J (1997) Population genetics of a functional variant of the dopamine β-hydroxylase gene (DBH). Am J Med Genet 74:374–379
Ding B (2003) Distribution of the NPY 1128C allele frequency in different populations. J Neural Transm 110:1199–1204
Ding B, Bertilsson L, Wahlestedt C (2002) The single nucleotide polymorphism T1128C in the signal peptide of neuropeptide Y (NPY) was not identified in a Korean population. J Clin Pharm Ther 27:211–212
Drube J, Kawamura N, Nakamura A, Ando T, Komaki G, Inada T (2001) No leucine(7)-to-proline(7) polymorphism in the signal peptide of neuropeptide Y in Japanese population or Japanese with alcoholism. Psychiatr Genet 11:53–55
Duan S, Gao R, Xing Q, Du J, Liu Z, Chen Q, Wang H, Feng G, He L (2005) A family-based association study of schizophrenia with polymorphisms at three candidate genes. Neurosci Lett 379:32–36
Gabriel SM, Davidson M, Haroutunian V, Powchik P, Bierer LM, Purohit DP, Perl DP, Davis KL (1996) Neuropeptide deficits in schizophrenia vs Alzheimer’s disease cerebral cortex. Biol Psychiatry 39:82–91
Goudet J (1995) FSTAT (Version 1.2): a computer program to calculate F-statistics. J Hered 86:485–486
Gribkoff VK, Pieschl RL, Wisialowski TA, van den Pol AN, Yocca FD (1998) Phase shifting of circadian rhythms and depression of neuronal activity in the rat suprachiasmatic nucleus by neuropeptide Y: mediation by different receptor subtypes. J Neurosci 18:3014–3022
Itokawa M, Arai M, Kato S, Ogata Y, Furukawa A, Haga S, Ujike H, Sora I., Ikeda K, Yoshikawa T (2003) Association between a novel polymorphism in the promoter region of the neuropeptide Y gene and schizophrenia in humans. Neurosci Lett 347:202–204
Jia C, Liu Z, Liu T, Ning Y (2005) The T1128C polymorphism of neuropeptide Y gene in a Chinese population. Arch Med Res 36:175–177
Kallio J, Pesonen U, Karvonen MK, Kojima M, Hosoda H, Kangawa K, Koulu M (2001) Enhanced exercise-induced GH secretion in subjects with Pro7 substitution in the prepro-NPY. J Clin Endocrinol Metab 86:5348–5352
Karvonen MK, Pesonen U, Koulu M, Niskanen L, Laakso M, Rissanen A, Dekker J, Hart LM, Valve R, Uusitupa MIJ (1998) Association of a leucine(7)-to-proline(7) polymorphism in the signal peptide of neuropeptide Y with high serum cholesterol and LDL cholesterol levels. Nat Med 4:1434–1437
Karvonen MK, Valkonen VP, Lakka TA, Salonen R, Koulu M, Pesonen U, Tuomainen TP, Kauhanen J, Nyyssonen K, Lakka HM, Uusitupa MI, Salonen JT (2001) Leucine7 to proline7 polymorphism in the preproneuropeptide Y is associated with the progression of carotid atherosclerosis, blood pressure and serum lipids in Finnish men. Atherosclerosis 159:145–151
Lappalainen J, Kranzler HR, Malison R, Price LH, Van Dyck C, Rosenheck RA, Cramer J, Southwick S, Charney D, Krystal J, Gelernter J (2002) A functional neuropeptide Y Leu7Pro polymorphism associated with alcohol dependence in a large population sample from the United States. Arch Gen Psychiatry 59:825–831
Larhammar D (1996) Evolution of neuropeptide Y, peptide YY and pancreatic polypeptide. Regul Pept 62:1–11
Magni P (2003) Hormonal control of the neuropeptide Y system. Curr Protein Pept Sci 4:45–57
Makino K, Kataoka Y, Hirakawa Y, Ikeda A, Yamauchi A, Oishi AR (2001) A leucine(7)-to-proline(7) polymorphism in the signal peptide of neuropeptide Y was not identified in the Japanese population. J Clin Pharm Ther 26:201–203
McDonald JK (1990) Role of neuropeptide Y in reproductive function. Ann NY Acad Sci 611:258–272
Mueller JC (2004) Linkage disequilibrium for different scales and applications. Brief Bioinform 5:355–364
Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York
Niskanen L, Karvonen MK, Valve R, Koulu M, Pesonen U, Mercuri M, Rauramaa R, Toyry J, Laakso M, Uusitupa MIJ (2000a) Leucine 7 to proline 7 polymorphism in the neuropeptide Y gene is associated with enhanced carotid atherosclerosis in elderly patients with type 2 diabetes and control subjects. J Clin Endocr Metab 85:2266–2269
Niskanen L, Voutilainen-Kaunisto R, Terasvirta M, Karvonen MK, Valve R, Pesonen U, Laakso M, Uusitupa MI, Koulu M (2000b) Leucine 7 to proline 7 polymorphism in the neuropeptide y gene is associated with retinopathy in type 2 diabetes. Exp Clin Endocrinol Diabetes 108:235–236
Okubo T, Harada S (2001) Polymorphism of the neuropeptide Y gene: an association study with alcohol withdrawal. Alcohol Clin Exp Res 6:59S–62S
Pettersson-Fernholm K, Karvonen MK, Kallio J, Forsblom CM, Koulu M, Pesonen U, Fagerudd JA, Groop PH, FinnDiane Study Group (2004) Leucine 7 to proline 7 polymorphism in the preproneuropeptide Y is associated with proteinuria, coronary heart disease, and glycemic control in type 1 diabetic patients. Diabetes Care 27:503–509
Roychoudhury S, Roy S, Basu A, Banerjee R, Vishwanathan H, Usha Rani MV, Sil SK, Mitra M, Majumder PP (2001) Genomic structures and population histories of linguistically distincttribal groups of India. Hum Genet 109:339–350
Saha N (1976) Population genetic studies in Kerala and the Nilgiris. Hum Hered 26:175–197
Schneider S, Roessli D, Excoffier L (2000) Arlequin v.2.0: a software for population genetics data analysis. Genetics and Biometry Laboratory, University of Geneva, Geneva
Thangaraj K, Joshi MB, Reddy AG, Gupta NJ, Chakravarty B, Singh L (2002) CAG repeat expansion in the androgen receptor gene is not associated with male infertility in Indian populations. J Androl 23:815–818
Thangaraj K, Chaubey G, Kivisild T, Reddy AG, Singh VK, Rasalkar AA, Singh L (2005) Reconstructing the origin of Andaman Islanders. Science 308:996
Thanseem I, Thangaraj K, Chaubey G, Singh VK, Bhaskar LVKS, Reddy BM, Reddy AG, Singh L (2006) Genetic affinities among the lower castes and tribal groups of India: inference from Y chromosome and mitochondrial DNA. BMC Genet 7:42
The International HapMap Consortium (2003) The International HapMap Project. Nature 426:789–796
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370
Zapata C (2000) The D′ measure of overall gametic disequilibrium between pairs of multiallelic loci. Evolution Int J Org Evolution 54:1809–1812
Acknowledgments
This project was supported by a grant from Department of Biotechnology, Government of India (Grant no. BT/PR3607/SPD/09/261/2003).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Bhaskar, L.V.K.S., Thangaraj, K., Shah, A.M. et al. Allelic variation in the NPY gene in 14 Indian populations. J Hum Genet 52, 592–598 (2007). https://doi.org/10.1007/s10038-007-0158-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10038-007-0158-x
Keywords
This article is cited by
-
Association between neuropeptide Y receptor 2 polymorphism and the smoking behavior of elderly Japanese
Journal of Human Genetics (2010)
-
Neuropeptide Y gene polymorphisms are not associated with obesity in a South Indian population
European Journal of Clinical Nutrition (2010)
-
Neuropeptide Y gene functional polymorphism influences susceptibility to hypertension in Indian population
Journal of Human Hypertension (2010)