Abstract
In vertebrates, embryonic hematopoietic stem and progenitor cells (HSPCs) are derived from a subset of endothelial cells, the hemogenic endothelium (HE), through the endothelial-to-hematopoietic transition (EHT). Notch signaling is essential for HSPC development during embryogenesis across vertebrates. However, whether and how it regulates EHT remains unclear. Here, we show that G protein-coupled receptor 183 (Gpr183) signaling serves as an indispensable switch for HSPC emergence by repressing Notch signaling before the onset of EHT. Inhibition of Gpr183 significantly upregulates Notch signaling and abolishes HSPC emergence. Upon activation by its ligand 7α-25-OHC, Gpr183 recruits β-arrestin1 and the E3 ligase Nedd4 to degrade Notch1 in specified HE cells and then facilitates the subsequent EHT. Importantly, 7α-25-OHC stimulation promotes HSPC emergence in vivo and in vitro, providing an attractive strategy for enhancing the in vitro generation of functional HSPCs.
Similar content being viewed by others
Introduction
Hematopoietic stem cells (HSCs) produce all lineages of blood cells and maintain lifelong hematopoiesis in vertebrates. Although HSC transplantation is used to treat hematopoietic malignancies including leukemia, its application has been limited mainly due to the severe shortage of compatible donors to supply functional hematopoietic stem and progenitor cells (HSPCs). Therefore, understanding the mechanism underlying HSC or HSPC generation and development is of great clinical importance for regenerative medicine. Previous studies have demonstrated that embryonic HSPCs in vertebrates are generated from a group of specialized endothelial cells, named hemogenic endothelium (HE)1,2,3,4, through the endothelial-to-hematopoietic transition (EHT)5,6,7. Several important regulators or signaling pathways have been identified as playing roles in this process8,9,10. However, how these intrinsic and extrinsic regulators and signaling pathways spatially and temporally regulate the development of HSPCs from hemogenic endothelial cells (HECs) still remains elusive.
Notch signaling is an important and conserved signaling pathway during embryogenesis, particularly neurogenesis and cardiovascular development. Previous studies revealed that Notch signaling is indispensable for embryonic HE specification11, as well as artery development12,13, and the function of Notch signaling in promoting definitive haematopoiesis could be uncoupled from that in artery development11,14. However, recent studies reported that elevated levels of Notch signaling impede HSPC generation in chick and mouse embryos15,16, suggesting unexpected complexities in the role of Notch signaling during hematopoiesis.
G protein-coupled receptors (GPCRs) constitute the largest receptor family; they are involved in a variety of physiological processes. In most cases, GPCRs are responsible for sensing external signals including odors, taste, neurotransmitters or inflammation signals17. The contribution of GPCRs to embryonic hematopoiesis has only recently been realized. Thrombin receptor (f2r) inhibits EHT by interacting with RhoA/ROCK signaling18. Prostaglandin E2 (PGE2), which signals through a GPCR, can expand HSPC population19. Moreover, Gpr56 is highly enriched in hematopoietic clusters and is essential for HSPC generation20. However, how GPCR signaling is integrated with other hematopoiesis-related signaling pathways during embryonic HSPC development is still poorly characterized.
Here, we report a novel regulator of HSPC development: GPCR gpr183, identified through a transcriptome analysis using a newly generated Tg(runx1:en-GFP) line. Functional analysis indicates that gpr183 is indispensable for HSPC emergence but not HE specification in both zebrafish and mouse. Unexpectedly, we find that Notch signaling is not continuously required during HSPC development: after HE specification, Notch signaling is downregulated to allow HSPC emergence. The downregulation of Notch signaling requires Gpr183, which, upon activation by its endogenous ligand 7α-25-OHC, recruits β-arrestin1 (Arrb1) and E3 ligase Nedd4 to degrade Notch1 receptor in HE, thus facilitating the transition from endothelial to hematopoietic lineage. Importantly, activation of the Gpr183-Arrb1-Notch1 signaling cascade can promote HSPC development in both zebrafish and mouse, suggesting conservation of this mechanism and potential therapeutic value of Gpr183 ligands in regenerative medicine.
Results
Establishment of Tg(runx1:en-GFP) line and RNA sequencing
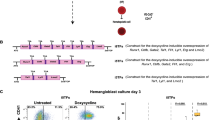
Using a mouse runx1+24 enhancer21, we generated a new zebrafish transgenic line Tg(runx1:en-GFP) that recapitulated runx1 endogenous expression during HE specification and HSPC emergence (Figure 1A, Supplementary information, Figure S1A, S1B and Movie S1). runx1 knockdown greatly attenuated the population of GFP+ cells in the aorta-gonad-mesonephros (AGM), the caudal hematopoietic tissue (CHT) and the thymus (Supplementary information, Figure S1B), supporting the notion that these GFP+ cells are runx1-dependent HSPCs22. To explore the mechanism governing HE specification and transformation into HSPCs through EHT, non-hemogenic endothelial cells (ECs, flk1+runx1−), specified HECs (flk1+runx1+), and potential HSPCs (flk1−runx1+) were sorted from Tg(flk1:mCherry/runx1:en-GFP) embryos at 28 h post fertilization (hpf; before the emergence of HSPCs) for RNA deep sequencing analysis (Figure 1A and 1B). RNA-Seq analysis showed a number of differentially expressed genes between two populations in three pairwise comparisons (Figure 1C). Compared with the EC population, many genes known to be involved in hematopoiesis were enriched in HEC or HSPC population, validating reliability of the RNA-Seq data (Supplementary information, Figure S1C and Table S1, RPKM > 2). Heatmap analysis revealed a number of genes highly expressed in HE or HSPC population but were previously less characterized in the context of hematopoiesis (Figure 1D). Expression profile screening by whole mount in situ hybridization confirmed that most of these genes were expressed in the AGM region (Supplementary information, Figure S2A). To functionally test these genes, we designed antisense morpholinos (MOs) targeting 11 different genes for knockdown experiments. After knockdown of seven genes, expression of HSPC markers runx1 and cmyb was severely attenuated at 36 hpf (Supplementary information, Figure S2B and S2C), indicating their important roles in definitive hematopoiesis. To determine the conservation of these genes during HE development across species, we compared zebrafish dataset with embryonic day 10.5 (E10.5) mouse HE sequencing results (data not shown) and found that gpr183 was the most enriched gene in both species.
RNA-deep sequencing analysis of gene expression in ECs, HE and HSPCs from Tg(flk1:mCherry/runx1:en-GFP) transgenic embryos. (A) Fluorescence signals in AGM and thymus of the indicated transgeneic lines showing that Tg(runx1:en-GFP) is able to label HE and HSPCs. White arrowheads denote HE or emerging HSPCs in AGM. (B) Flowchart of sorting and RNA-Seq analysis of the indicated cells from 28 hpf Tg(flk1:mCherry/runx1:en-GFP) embryos. (C) RNA-Seq analysis results showing the numbers of differentially expressed genes in sorted ECs, HECs and HSPCs from 28 hpf Tg(flk1:mCherry/runx1:en-GFP) embryos. (D) Heat-map analysis of selected genes based on RNA-Seq data.
Gpr183 is essential for HSPC emergence
Gpr183, also called Epstein-Barr virus-induced gene 2 or EBI2, is known for its role in guiding follicular B cell migration after activation by its endogenous ligand, 7-25-OHC23,24,25. However, it was unknown whether and how gpr183 is involved in HSPC development. The expression profile showed that gpr183 is expressed in the zebrafish AGM region starting at 26 hpf (Supplementary information, Figure S3A and S3B). Moreover, heatmap analysis confirmed that gpr183 is highly enriched in the HE population (Figure 1D), which was verified by the qPCR results in sorted ECs, HECs and HSPCs (Supplementary information, Figure S3C). Double fluorescence in situ hybridization using 30 hpf Tg(fli1a:eGFP) embryos confirmed that gpr183 is expressed in some endothelial cells in the AGM region (Supplementary information, Figure S3D).
To determine the function of gpr183 in zebrafish, we generated a gpr183-null mutant by removing 11 base pairs in the second exon (292 base pairs after the start codon) using the CRISPR/Cas9 technique (Figure 2A and Supplementary information, Figure S3E). We found that both cmyb expression at 36 hpf and T lymphocyte marker rag1 expression at 5 days post fertilization (dpf) were significantly downregulated in gpr183 mutant (Figure 2B). 7α-25-OHC is synthesized by Ch25h and Cyp7b1 sequentially in vivo26,27. Clotrimazole, an inhibitor of Cyp7b126, was used to block 7α-25-OHC synthesis, and this treatment effectively attenuated runx1 expression in AGM and rag1 expression in thymus (Figure 2C). After crossing gpr183 mutant with the Tg(flk1:mCherry/runx1:en-GFP) reporter line, we found that the HEC population at 36 hpf was significantly disrupted in the absence of gpr183 signaling (Figure 2D and 2E). Moreover, gpr183 deficiency also attenuated EHT (Supplementary information, Movie S2). Kidney marrow, or head kidney, is a hematopoietic organ in adult zebrafish. We found that in 12-week-old gpr183+/− adult fish (gpr183−/− adult fish were not viable), both the cell number in the head kidney and size of the organ were significantly reduced (Figure 2F), suggesting a defect in definitive hematopoiesis upon gpr183 haploinsufficiency. Furthermore, time course experiments in gpr183 mutant and clotrimazole treatment revealed that gpr183 was not required before HE specification (Supplementary information, Figure S4A-S4D). Taken together, these results indicate that Gpr183 is required for HSPC development in zebrafish.
Gpr183 plays an indispensable role during definitive hematopoiesis in zebrafish. (A) Generation of gpr183 mutant using the CRISPR/Cas9 technique. PAM sequence is 'TGG' (in blue); alignment of WT and mutated sequences is listed below. (B, C) Reduced expression of cmyb (or runx1) and rag1 in gpr183 mutant embryos (B) and clotrimazole-treated embryos (C). Red arrowheads indicate cmyb or runx1 expression in AGM, and white circles denote rag1 expression in the thymus. (D) The number of HE in Tg(gpr183−/−;flk1:mCherry/runx1:en-GFP) embryos is attenuated significantly compared with that in control embryos. White arrowheads denote HE in the AGM region. (E) Dot-plot quantification analysis of HE in sibling control and gpr183 mutant. Error bar represents SD. ****P < 0.0001. (F) Hematoxylin-eosin staining of head kidney section showing hematopoietic defect in 12-week-old gpr183 heterozygous mutant fish.
Gpr183 specifically appears to regulate HSPC emergence. We did not observe any general developmental defects, or abnormality in HSPC niche, or aberration in cell proliferation and apoptosis in gpr183 morphants or mutant (Supplementary information, Figure S5A-S5D).
Notch signaling is downstream of Gpr183
We noticed that expression level of arterial marker ephrinB2a was increased in embryos treated with clotrimazole (Supplementary information, Figure S6A). Because ephrinB2a is a Notch target, we wondered whether Notch signaling is involved in the regulation of HSPC emergence by Gpr183. We thus examined another Notch target gene hey2 and found that its expression was significantly increased upon gpr183 signaling inhibition from 33 hpf (Figure 3A and Supplementary information, Figure S6B). DBZ, a Notch signaling inhibitor, reversed cmyb and hey2 phenotype in gpr183−/− embryos (Figure 3A-3C). Moreover, the upregulation of Notch target genes including ephrinB2, hey2, her6, and her9 and downregulation of cmyb in clotrimazole-treated embryos were also reversed by DBZ treatment (Supplementary information, Figure S6A-S6D). These data together suggest that Notch signaling acts downstream of Gpr183. To define whether Gpr183 modulates Notch signaling in a cell-autonomous manner, we treated Tg(flk1:mCherry/runx1:en-GFP) embryos with DMSO or clotrimazole and sorted the flk1+runx1− and flk1+runx1+ cells at 28 hpf for qPCR analysis. Expression levels of Notch target genes her6 and her9 increased remarkably in the flk1+runx1+ cells (HECs) but not in the flk1+runx1− cells (non-hemogenic ECs) after clotrimazole treatment compared with the DMSO control (Figure 3D), supporting that Gpr183 modulates Notch signaling in HE in a cell-autonomous manner.
Downregulation of Notch signaling is required for definitive hematopoiesis. (A) The increase in hey2 expression in gpr183 mutant is reversed by DBZ treatment. Right panel shows the qPCR quantification. (B) The decrease in cmyb expression in gpr183 mutant is reversed by DBZ treatment. In A and B, red arrowheads indicate hey2 or cmyb expression in the AGM region at 33 hpf. (C) Dot-plot analysis of cmyb+ cell numbers in AGM. (D) Clotrimazole treatment increases expression levels of her6 and her9 in flk1+runx1+ cells at 28 hpf. (E) Notch-active endothelial cells increase in gpr183 morphant (n = 15 for control and morpholino treatment). Right panel shows the quantification analysis. (F) EHT from 37 (shown as 00:00:00) to 43 hpf in Tg(runx1:en-GFP/tp1:mCherry) embryo. Asterisks label individual cells. (G, H) cmyb expression in the AGM region of control embryos and embryos in which Notch signaling is inhibited. Tg(hsp70:dn-MAML) embryos expressing a dominant negative (dn) isoform of the murine mastermind-like (MAML) protein fused to GFP under a hsp70 promoter were heat shocked at 20 or 26 hpf; cmyb expression was examined at 36 hpf. GFP− embryo serves as a control. Representative images are shown in G and the dot-plot quantification analysis is shown in H. (I, J) cmyb expression in the AGM region of control and Notch signaling-activated embryos. Tg(UAS:NICD) (NICD, control) and Tg(hsp70:GAL4/UAS:NICD) (hs:NICD) embryos were heat shocked at 20 or 26 hpf and cmyb expression was examined at 36 hpf. Representative images are shown in I, and the dot-plot quantification analysis is shown in J. In G and I, red arrowheads mark the staining signals in AGM. Error bar represents SD. *P < 0.05, **P< 0.01, ***P < 0.001.
Notch signaling downregulation is required for HSPC development
Previous findings have demonstrated that Notch signaling is indispensable for definitive hematopoiesis, whereas some recent studies have also suggested that activated Notch signaling impedes HSPC development15,28. Our study provides several lines of evidence that suggest that Notch signaling is not continuously required during HSPC development, and downregulation of Notch signaling is a prerequisite for HSPC emergence. First, fli1+tp1+ cells in Tg(fli1a:eGFP/tp1:mCherry) embryos (tp1:mCherry is a Notch reporter) were significantly increased upon gpr183 knockdown (Figure 3E). Second, we monitored EHT in Tg(cmyb:GFP/tp1:mCherry) or Tg(runx1:en-GFP/tp1:mCherry) embryos and found that there were no tp1+cmyb+ or tp1+runx1+ cells undergoing EHT (even though some emerging cells were initially runx1+tp1+, they eventually developed into runx1+tp1− cells; Figure 3F, Supplementary information, Movies S3 and S4).
Moreover, in the transgenic line Tg(hsp70:dn-MAML), which expresses a dominant-negative form of MAML (a transcriptional co-activator for Notch signaling) upon heat shock treatment29, cmyb expression at 36 hpf was significantly lower than the level observed in control embryos when the heat shock treatment was applied at 20 hpf, but was significantly higher than the control when embryos were heat shocked at 26 hpf (Figure 3G and 3H). Finally, in the Tg(hsp70:GAL4/UAS:NICD)30 embryos, which express Notch intracellular domain (NICD) upon heat shock treatment, cmyb expression was significantly higher than the level observed in control embryos (Tg(UAS:NICD)) when the heat shock treatment was applied at 20 hpf, but was significantly lower than the control when embryos were heat shocked at 26 hpf (Figure 3I and 3J).
Gpr183 cooperates with Arrb1 to suppress Notch signaling
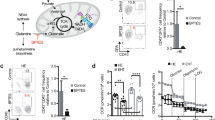
We next explored the mechanism by which Gpr183 represses Notch signaling in HE. Previous results have revealed that Arrb1 is responsible for GPCR desensitization and internalization31, and Arrb1 can be recruited by Gpr18324. To test this interaction in zebrafish, we injected gpr183-eGFP and arrb1-mCherry mRNA into one-cell stage embryos and observed that the GFP signal at cell membrane was internalized upon 7α-25-OHC stimulation (Figure 4A). Next, we transfected gpr183-Flag and arrb1-Myc constructs into 293T cells and found that Flag-tagged Gpr183 was precipitated by anti-Myc in the presence of 7α-25-OHC (Figure 4B), supporting that Arrb1 is recruited by Gpr183 after Gpr183 activation. To determine the function of Arrb1, we knocked down arrb1 using antisense morpholino and found that runx1 expression was normal at 24 hpf, but cmyb expression level was decreased at 30 hpf (Figure 4C). In addition, hey2 expression level was increased in arrb1 morphants at 30 hpf (Figure 4C), consistent with its increase in gpr183 mutants (Figure 3A). Taken together, we conclude that Gpr183 cooperates with Arrb1 to downregulate Notch signaling.
Gpr183 cooperates with Arrb1 and Nedd4 to downregulate Notch signaling. (A) Internalization of Gpr183-GFP in 90% epiboly embryos (n = 6 for each treatment). White arrowheads denote the internalized Gpr183-GFP in cytoplasm. (B) Gpr183 interacts with Arrb1 in 7α-25-OHC-stimulated cells. 293T cells were transfected with Gpr183-Flag and/or Arrb1-Myc and stimulated with 7α-25-OHC for 0.5 h. Cell lysates were subjected to immunoprecipitation using anti-Myc followed by western blot analysis. (C) Expression of runx1, cmyb and hey2 in arrb1 morphants. Red arrowheads label the staining signals in AGM. (D) NICD expression in zebrafish embryos treated with 7α-25-OHC (from 1-cell stage to 36 hpf) or MG132 (from 26 to 36 hpf). Right panel shows the quantification analysis. (E) MS analysis of Notch1-interacting proteins in control or 7α-25-OHC-treated AGM cells. Mouse E10.5 AGM cells were cultured in the presence of hematopoietic cytokines for three days, and then treated with DMSO or 7α-25-OHC for 1 h. (F) Polyubiquitination of NICD-Flag (zebrafish) induced by zebrafish Nedd4a-GST (Nedd4a(Z)) or mouse Nedd4-GST (Nedd4(M)). (G) GST pull-down assay showing the interaction between Nedd4-GST and NICD-Flag. (H) runx1 expression is reduced in nedd4a morphants and increased in nedd4a-overexpressing embryos. (I) fli1a+tp1+ cells (white arrowheads) are increased in nedd4a morphants and are reduced in nedd4a-overexpressing embryos. Right panel shows the quantification result. (J) NICD expression in nedd4a morphants or nedd4a-overexpressing embryos. Right panel shows the quantification result. (K) HE in Tg(flk1:mCherry/runx1:en-GFP) embryos after nedd4a knockdown or 7α-25-OHC treatment. White arrowheads indicate HE in the AGM region at 36 hpf. Error bar represents SD. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Gpr183-Arrb1 complex promotes the degradation of Notch1 through Nedd4-mediated ubiquitination
Because the expression of Notch ligands including deltC, deltD, dll4, and jagged1 was not altered upon Gpr183 deficiency (data not shown), we asked whether Notch receptors were affected. There are four Notch receptors in vertebrates. Among them, Notch1 is known to regulate artery specification and HE specification11,14, while Notch3 is required within the somites to specify HSPCs in a cell non-autonomous manner32. Considering that Gpr183 cell-autonomously modulates Notch signaling in HECs, we reasoned that Notch1, but not Notch3, is the target of Gpr183 signaling cascade. To examine if the Gpr183-Arrb1 complex represses Notch1 signaling, we first sorted zebrafish flk1+runx1+ cells at 28 hpf and found that notch1a and notch1b mRNA levels did not increase after clotrimazole treatment (data not shown). We next asked whether the Gpr183-Arrb1 complex regulates Notch1 protein stability. To this end, we treated zebrafish embryos with 7α-25-OHC and found that expression of NICD at 36 hpf was significantly reduced compared with untreated control embryos, and this reduction was partially reversed by a proteasome inhibitor, MG132 (Figure 4D). Treatment with a lysosome inhibitor chloroquine did not reverse the attenuation of NICD levels after 7α-25-OHC stimulation (data not shown), suggesting that Notch1 is degraded by a Gpr183 signaling-induced proteasome pathway.
To identify the E3 ligase responsible for Notch1 degradation downstream of Gpr183-Arrb1, we performed mass spectrometry (MS) to identify Notch1-binding proteins using E10.5 mouse AGM cells treated with 7α-25-OHC (Figure 4E, Supplementary information, Tables S2 and S3). Among hits were two ubiquitin ligases: the HECT domain type E3 ligase Nedd4 and the RING finger domain type E3 ligase Pias3. An in vitro ubiquitination assay revealed that Nedd4 facilitated poly-ubiquitination of NICD (Figure 4F). GST pull-down assay further demonstrated a direct interaction between Nedd4 and NICD (Figure 4G). We next examined if Nedd4 modulates Notch1 expression in vivo. Indeed, expression of HSPC marker runx1 in nedd4a morphants was significantly reduced, whereas in nedd4a-overexpressing embryos, runx1 expression was markedly increased (Figure 4H). In addition, knockdown of nedd4a elevated the tp1+ signal in endothelial cells (Figure 4I). Moreover, western blot analysis revealed that NICD expression was augmented in nedd4a morphants but was reduced in nedd4a-overexpressing embryos (Figure 4J), further confirming that nedd4a negatively regulates Notch signaling in vivo. We also observed that the increase in HEC numbers induced by 7-25-OHC treatment could be disrupted by nedd4a knockdown (Figure 4K). All these results together indicate that Nedd4 functions downstream of Gpr183 to attenuate Notch activity.
To address the cellular mechanism by which Notch signaling represses HSPC development, we examined the expression of tight junction gene ve-cadherin and found an upregulation of ve-cadherin upon inhibition of gpr183 signaling (Figure 5A). Moreover, a large proportion of cmyb-GFP positive cells (potential HSPCs) were converted to non-hemogenic ECs after clotrimazole treatment beginning at 30 hpf (Figure 5B, 5C and Supplementary information, Figure S7A). Based on these results and the observation that expression levels of arterial markers are elevated in the absence of gpr183 signaling (Figure 3A, Supplementary information, Figure S6A and S6B), we speculate that when Notch signaling is activated, the specified HE lose their hematopoietic potential and instead, maintain their arterial fate33.
7α-25-OHC stimulation facilitates HSPC development in zebrafish. (A) Expression of ve-cadherin increases in clotrimazole-treated embryos. (B) Transition of HE to HSPCs or non-hemogenic ECs after DMSO or clotrimazole treatment beginning at 30 hpf. White asterisk indicates the same cell in different frames. (C) Quantification of HE (flk1+cmyb+, yellow cells in B) differentiating to HSPCs (flk1−cmyb+, green cells in B) or non-hemogenic ECs (flk1+cmyb−, red cells in B) (n = 5). Error bar represents SD. (D, E) runx1 expression in embryos treated with 7α-25-OHC or clotrimazole and examined at 36 hpf by WISH (D) and western blotting (E). Red arrowheads in A and D label the staining signals in AGM.
7α-25-OHC treatment facilitates HSPC emergence
To gain further support for a role of Gpr183-Arrb1-Notch1 cascade during the differentiation of HSPCs from HECs, we asked if 7α-25-OHC treatment would facilitate the generation of HSPCs. Both runx1 mRNA level and protein level were significantly upregulated in 7α-25-OHC-treated embryos (Figure 5D and 5E). Moreover, the frequency of cells undergoing EHT in Tg(flk1:mCherry/cmyb:GFP) embryos was markedly increased after 7α-25-OHC treatment (Supplementary information, Movie S2). These results support the notion that the activated Gpr183-Arrb1-Notch1 cascade facilitates HSPC emergence in vivo.
Regulation of HSPC emergence by Gpr183 is conserved in mammals
We next asked whether the regulation of HSPC emergence by gpr183 is conserved in mammals. Among three populations sorted from the AGM region of E11 mice: non-hemogenic EC (c-Kit−CD31+CD41−CD45−Ter119−), HEC (c-Kit+CD31+CD41−CD45−Ter119−) and HSPC (c-Kit+CD34+), HE had the greatest level of gpr183 expression, consistent with the results in zebrafish (Figure 6A). Expression levels of ch25h and cyp7b1 in the mouse AGM region increased markedly from E10.5 to E11.5 (Figure 6B), and these enzymes were highly enriched in non-hemogenic ECs (c-Kit−CD31+CD41−CD45−Ter119−) (Supplementary information, Figure S7B). Moreover, the colony forming ability of cells isolated from AGM was significantly reduced after clotrimazole treatment in a colony forming unit in spleen (CFU-S) assay (Figure 6C-6E), confirming the role of Gpr183 in hematopoiesis. To examine directly whether gpr183 regulates EHT, pan-ECs (CD31+CD41−CD45−Ter119−) were sorted from the E10 caudal half and cultured with clotrimazole (Figure 6F)34. Remarkably, clotrimazole treatment reduced HSPC population (CD45+c-Kit+)34 in a dose-dependent manner in OP9 co-cultures (Figure 6G and 6H), indicating that ablation of gpr183 signaling suppresses HSPC development. To determine whether gpr183 signaling specifically affects HE, we sorted non-hemogenic ECs, HECs or HSPCs from the E11 AGM region and treated them with clotrimazole. As shown in Supplementary information, Figure S7C, only HECs displayed a significant decreased hematopoietic capability, supporting that gpr183 signaling specifically targets HECs but not non-hemogenic ECs and HSPCs. To further determine whether gpr183 signaling contributes to the emergence of HSPCs, we conducted a long-term transplantation assay, in which cells dissected from AGM region of E11 embryo (CD45.1/2) were treated with DMSO or clotrimazole and then transplanted into irradiated mice (CD45.2; Figure 6F). Four months after the transplantation, all 6 recipients were successfully repopulated with DMSO-treated cells, with a donor chimerism in bone barrow ranging from 30% to 80% (Figure 6I). Multiple hematopoietic lineages derived from the donor cells were detected in peripheral blood of recipients (Figure 6J). The donor cells also displayed a potent self-renewal capacity upon transplantation into secondary recipients (Figure 6K-6L). In contrast, the AGM cells treated with clotrimazole failed to reconstitute any recipients (Figure 6I; 0/6, 2 independent experiments), suggesting that HSPC development was disrupted upon Gpr183 signaling deficiency. Taken together, we conclude that Gpr183 signaling is a conserved regulatory mechanism during HSPC emergence in vertebrates.
Gpr183 function during HSPC development is conserved in mammals. (A) gpr183 expression by RT-PCR in non-hemogenic ECs, HECs and HSPCs sorted from the AGM region of E11 mouse embryos. (B) Expression of ch25h and cyp7b1 in mouse AGM region (E10.5 and E11.5; n = 3 for each time point). (C) Flowchart of CFU-S assay. (D, E) CFU-S colony number is significantly reduced in mice transplanted with clotrimazole-treated cells. (F) Schematic illustration of flow cytometry analysis and long-term transplantation in mouse. E10 mouse caudal half and E11 mouse AGM region were dissected and dissociated by collagenase. Pan-ECs (CD31+CD41−CD45−Ter119−) from E10 caudal half were sorted and cultured with DMSO or clotrimazole before flow cytometry analysis. AGM cells from E11 mouse (CD45.1/2) embryos (but not sorted pan-ECs as they often resulted in poor reconstitution) were treated with DMSO or clotrimazole for 4 days and transplanted into irradiated C57BL/6J (CD45.2) mice. HC, hematopoietic cells; EC, pan-ECs; OP9, feeder cells used in co-culture. (G, H) Representative FACS plots (G) and quantification (H) of CD45+c-Kit+ HSPCs from OP9 co-cultures. (I) Donor-derived chimerism in recipients. Each symbol represents the percentage of donor chimerism in bone marrow of a recipient 4 months post-transplantation. (J) Multiple hematopoietic lineages including myeloid (Gr1+/Mac-1+), B lymphoid (B220+), and T lymphoid (CD3+) populations in peripheral blood of the recipients (CD45.2) transplanted with E11 AGM cells (CD45.1/2) pre-treated with DMSO. (K, L) Donor-derived chimerism (K) and multi-lineage potential (L) in peripheral blood of secondary recipients (IIo) transplanted with bone marrow cells of repopulated primary recipients (Io). Symbols in K represent the donor chimerism in peripheral blood of individual secondary recipients 12 weeks post-transplantation. Symbols of same color indicate the donor and the related secondary recipients. Data are presented as mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001.
The Gpr183-Arrb1-Notch1 signaling cascade is conserved in mammals
To determine whether Gpr183-Arrb1 represses Notch1 expression in mammals, we dissected cells from E10.5 mouse AGM region and treated them with either 7α-25-OHC or clotrimazole. Both Notch1 and NICD expression levels were decreased after 7α-25-OHC stimulation, but increased after clotrimazole treatment (Figure 7A and 7B). We next performed immuneprecipitation (IP) assay using anti-Arrb1 with cells dissected from E10.5 mouse AGM region and found that after 7α-25-OHC stimulation, both Gpr183 and Notch1, as well as the E3 ligase Nedd4, were precipitated by anti-Arrb1 (Figure 7C). Interestingly, Gpr183 was precipitated by an antibody against Notch1 even in the absence of 7α-25-OHC stimulation (Figure 7C), indicating that Gpr183 is localized adjacent to Notch1 and serves as an important switch to downregulate the Notch1 level before the onset of HSPC emergence. Moreover, consistent with results shown in Figure 4D, MG132 treatment rescued the reduction of NICD protein levels induced by 7α-25-OHC stimulation in mouse AGM cells (Figure 7D). Taken together, we conclude that activated Gpr183 recruits Arrb1 and E3 ligase Nedd4 to degrade Notch1 through the ubiquitin-proteasome pathway in mouse.
Gpr183-Arrb1-Notch1 signaling cascade is conserved in mammals. (A, B) Notch1 and NICD expression in E10.5 mouse AGM cells treated with clotrimazole or 7α-25-OHC for 1 h. (C) Western blot detection of Gpr183 or Notch1 in anti-Arrb1 or anti-Notch1 IP product from E10.5 AGM cells. Equal amounts of Notch1 were used in the co-IP assay as 7a-25-OHC treatment decreases Notch1 expression and thus it is difficult to detect Notch1-precipitated proteins in the IP assay. (D) Decrease in NICD expression in 7α-25-OHC-treated E10.5 AGM cells is rescued by MG132. Right panel shows the quantification of western blot data. (E) Flowchart for isolation, culture and transplantation of mouse AGM cells. For flow cytometry, pan-ECs (CD31+CD41−CD45−Ter119−) were sorted from cell suspension obtained from E11 mouse AGMs and treated with DMSO or 7α-25-OHC for 4 days. For CFU-S or long-term transplantation assay, total E11 AGM cells were treated with DMSO or 7α-25-OHC for 4 days. (F) Flow cytometry showing an increased amount of c-Kit+CD45+ cells from 7α-25-OHC-treated E11 mouse AGM pan-ECs. (G) Quantification of results from the CFU-S assay (n = 5). (H) Donor-derived chimerism. AGM cells from actin-GFP transgenic mice were treated with DMSO or 7α-25-OHC (1 μM) for 4 days, mixed with bone marrow carrier cells, and were injected intravenously into irradiated female mice (GFP−). Three months later, bone marrow cells were collected for the analysis of GFP+ chimerism. Error bar represents SD. *P < 0.05.
In Tie2-positive endothelial cells, Notch1 expression level was decreased after 7α-25-OHC treatment (Supplementary information, Figure S7D and S7E). Pan-ECs (CD31+CD41−CD45−Ter119−) from the E11 mouse AGM generated markedly more HSPCs in the presence of 7α-25-OHC (Figure 7E and 7F). Furthermore, 7α-25-OHC treatment promoted the production of functional HSPCs in vivo in a CFU-S assay (Figure 7E and 7G). In addition, 7α-25-OHC treatment promoted the rate of chimerism in recipients in a three-month transplantation assay (Figure 7H, Supplementary information, Figure S7F and S7G). These results, together with the data obtained in zebrafish, demonstrate the conservation of Gpr183-Arrb1-Notch1 signaling cascade as an essential regulatory mechanism for the development of HSPCs in vertebrates (Supplementary information, Figure S8).
Discussion
It is well known that HSPCs are generated from specified HE in AGM, but the mechanism governing this process remains poorly understood. The EHT involves cell shape changes, cell migration and cell-cell communication5,6,7, and is regulated by a complex network of cell-intrinsic and cell-extrinsic signals. Exploring the signals that induce EHT is of great importance for our understanding of HSPC development and may provide insights into clinical applications in regenerative medicine. Our genome-wide transcriptome analyses of non-hemogenic ECs, HECs, and HSPCs reveal that gpr183 begins to be highly expressed in HECs, and is downregulated in HSPCs. Functional analysis demonstrates that Gpr183, upon activation by its endogenous ligand 7α-25-OHC, specifically modulates HSPC development through repressing Notch1 signaling. An unexpected finding is that Notch1 signaling is not continuously required for HSPC development, and persistent activation of Notch1 signaling after HE specification abrogates EHT, suggesting a temporally restricted role of Notch1 during HSPC development. Moreover, our results show that 7α-25-OHC-activated Gpr183 signaling is a conserved signaling mechanism regulating HSPC development.
RNA-Seq using a Tg(runx1:en-GFP) line identifies an important regulator of HSPC emergence
Our newly generated runx1:en-GFP transgenic line, with robust and specific fluorescence signals, has been proved recently to be capable of labeling functional HSPCs22. High-throughput RNA-Seq using this transgenic line in current study uncovers significant transcriptome variations among non-hemogenic ECs, specified HECs and HSPCs during HSPC development in zebrafish. Moreover, GO analysis identifies various GPCRs that are enriched in ECs, HECs or HSPCs. Based on our results and several previous findings19,20,35, we postulate that multiple GPCRs influence HSPC development through complex mechanisms that are still poorly understood. Gpr183 was previously reported to guide immune cell migration23,24. Our current work assigns a new role for Gpr183 during embryonic HSPC development based on the observations that blocking Gpr183 signaling significantly attenuates HSPC generation, while activating Gpr183 leads to more functional HSPCs. Similar results obtained from two animal models, zebrafish and mouse, indicate Gpr183 is a conserved and essential regulator of HSPC emergence in vertebrates.
After HE specification, continuous Notch signaling inhibits HSPC development
Recent studies using pluripotent stem cells (PSCs) in in vitro culture systems have indicated that Notch signaling is essential for the generation of functional HSPCs33,36. However, a previous study using mouse AGM-derived cell lines as feeder cells to support the differentiation of hematopoietic cells from human embryonic stem cells has also reported that downregulation of Notch signaling facilitates the production of hematopoietic cells37. Based on previous studies on Notch signaling during HSPC development and our results here, we speculate that HSPC development is intricately regulated and in vitro culture systems might not properly recapitulate the endogenous signaling processes. Our work supports that Notch signaling is not continuously required for HSPC development. Blocking Notch signaling before HE specification nearly completely disrupts HSPC generation, consistent with previous findings11; however, after HE specification, persistent Notch signaling can convert HECs into non-hemogenic ECs, thus abrogating subsequent HSPC development. The temporal regulation of HSPC development by a specific signaling pathway is not rare. For instance, Wnt signaling is essential for HE specification, but has to be downregulated to allow HSPC development to proceed further38,39. These findings together imply the complexity of mechanisms governing HSPC development, and each signaling pathway essential for hematopoiesis must be tightly regulated at different developmental stages.
Gpr183 downregulates Notch1 through Nedd4-mediated ubiquitination
Our work reveals that activated Gpr183 recruits Arrb1 and Nedd4 to degrade Notch1 through the ubiquitination-proteasome process, and this Gpr183-Arrb1-Nedd4-Notch1 signaling cascade is evolutionarily conserved in both zebrafish and mouse. The contribution of Arrb1 and Nedd4 to HSPC development has not been reported before, although Arrb1 is known to regulate primitive hematopoiesis by relieving PcG-mediated repression of cdx4-hox signaling40. Here we show that disruption of each component in the Gpr183-Arrb1-Nedd4-Notch1 cascade leads to a HSPC differentiation defect, highlighting the importance of this signaling pathway during HSPC development. A recent study has indicated that the HECT family E3 ligase Itch inhibits homeostasis of the bone marrow HSPCs by degrading Notch141. However, we did not detect an enriched expression of Itch in our MS dataset. In vitro ubiquitination assay further confirms that Itch is not involved in Notch1 ubiquitination in embryonic HSPCs (data not shown).
What remains to be clarified is how Notch1 in HE inhibits EHT. Based on our results we speculate that the specified HE possess both hemogenic and endothelial properties held in a fine balance, and an elevation in the Notch1 protein level, similar to the enhanced ERK signaling as we reported before42, is able to strengthen the arterial cells identity16, resulting in a loss of hematopoietic marker expression accompanied by an increase of adhesion molecule expression, and an aborted EHT.
7α-25-OHC serves as a potent agonist of HSPC emergence
Expression of Ch25h and Cyp7b1 is enriched in endothelial cells in the AGM region of both zebrafish (data now shown) and mouse, suggesting that 7α-25-OHC derived from the HSPC niche activates Gpr183 through a paracrine mechanism. Here, we find that Gpr183 serves as a switch to abrogate Notch1 activity after 7α-25-OHC stimulation, indicating that specified HE receive signals from the surrounding niche to trigger EHT. 7α-25-OHC-stimulated Gpr183 signaling facilitates HSPC development, which is reminiscent of recent findings that small molecules, such as PGE2 and epoxyeicosatrienoic acids (EETs), can serve as GPCR ligands to facilitate HSPC differentiation or expansion19,43. The mechanisms of their action are emerging. EETs promote HSPC development in zebrafish embryos through an unknown GPCR that activates AP-1, which in turn upregulates runx1 expression in both AGM and CHT in zebrafish embryos. In addition, EETs also efficiently enhance HSPC homing and engraftment in mammals43. These findings and ours demonstrate that GPCR signaling pathways are evolutionarily conserved important regulators of HSPC development and function, and their ligands may have clinical applications in blood transplantation and regenerative medicine.
Materials and Methods
Zebrafish
Zebrafish strains including Tubingen, Tg(runx1:en-GFP), Tg(flk1:mCherry/runx1:en-GFP), Tg(flk1:mCherry/cmyb:GFP), Tg(tp1:mCherry)44, Tg(fli1a:eGFP/tp1:mCherry), Tg(sclα:dsRed)45, Tg(rag2:dsRed) (provided by Zilong Wen), Tg(hsp70:dn-MAML)29, Tg(hsp70:Gal4)30, Tg(UAS:NICD)30 and gpr183 heterozygous mutant, were raised in the experimental animal center in the Institute of Zoology, Chinese Academy of Science. Embryos were staged according to the number of somites. For imaging, embryos were raised in the presence of 0.003% 1-phenyl-2-thiourea to avoid pigment formation. gpr183−/− was generated in AB/Tuebingen background. This study was approved by the Ethical Review Committee of the Institute of Zoology, Chinese Academy of Sciences, China.
Morpholinos, mRNA synthesis, qPCR, western blotting, BrdU/TUNEL assay, and double fluorescence in situ hybridization (dFISH)
All morpholinos were purchased from GeneTools and are listed in Supplementary information, Table S4 along with the dosages used in this study. mRNA synthesis, qPCR, western blotting, BrdU/TUNEL assay, and dFISH were performed as previously described46,47. qPCR primers are listed in Supplementary information, Table S5. For the Gpr183-GFP internalization experiment, gpr183-eGFP and arrb1-mCherry mRNA were co-injected into one-cell stage embryos. At about 90% epiboly, embryos were dechorionated and incubated with Hoechst for 30 min, and treated with DMSO or 7α-25-OHC (1 μM, Sigma, SML0541) for 10 min. Embryos were crushed by cover glass for observation.
Cell sorting and RNA-deep sequencing
The trunk region of 28 hpf Tg(flk1:mCherry/runx1:en-GFP) embryos was dissected and dissociated into single cells as previously described48. mCherry+GFP−, mCherry+GFP+, mCherry−GFP+ cells were sorted using BD FACS Aria I. 1 ng of total RNA extracted using QIAGEN RNeasy Micro kit was reversely transcribed and amplified as previously described49. cDNA samples were sequenced using Illumina HiSeq2500 to produce 101 bp paired-end sequence reads. Quality of raw RNA-seq reads for each sample was controlled by FastQC software and bases with quality scores less than 20 were removed by Trimmomatic. The remaining reads were mapped to the Zebrafish reference cDNA sequences (zv9, from Ensembl) by BWA.
Differentially expressed genes between two groups were determined using the R-package DEseq2, with criterion P value cutoff of 0.001 and fold change cutoff of 2. Heatmap was generated by R-package Heatmap2 according to RPKMs (reads per kilobase per million reads) of each gene in multiple samples. The RNA-Seq dataset is shown in Supplementary information, Table S1.
Cell transfection and co-IP assay
Zebrafish gpr183 and arrb1 coding sequences were ligated into the pCMV-Flag and pcDNA4ToMycHisB plasmids, respectively. According to the Lipo2000 (Invitrogen) transfection protocol, the plasmids were transfected into 293T cells for the co-IP assay. 24 h later, cells were treated with 1 μM 7α-25-OHC for 0.5 h before harvesting. Proteins were purified in RIPA buffer (50 mM Tris, 150 mM NaCl, 10% Glycerol, 5 mM MgCl2, 0.5% NP40, and Roche cocktail protease inhibitor) and mixed with anti-c-Myc agarose affinity gel antibody (Sigma, A7470). 3-4 h later, beads were collected for western blotting. Antibodies used were anti-c-Myc (Sigma, M4439, 1:2 000) and anti-Flag (Sigma, F7425, 1:1 000).
Zebrafish confocal imaging
Embryos for microscopic observation and photography were prepared as previously described50. Supplementary information, Movie S1 was captured using ZEISS LSM 510 confocal microscopy. Supplementary information, Movie S2 and fluorescent images were taken using Nikon A1 confocal microscopy. Supplementary information, Movies S3 and S4 were captured using Perkin Elmer spinning disk confocal microscopy.
Chemical treatment
Zebrafish embryos at 10 somite stage (14 hpf) or 26 hpf were treated with clotrimazole (10 μM; Sigma, C6019) or DBZ (8 μM; Sigma, SML0649). 7α-25-OHC (4.2 pg) was injected into one-cell stage embryos. Embryos were treated with MG132 (10 μM, Sigma C2211) from 26 to 36 hpf.
Mice
Mice were kept at the Animal Center of the Academy of Military Medical Sciences according to institutional guidelines. Embryos were staged by somite number: E10, 31-40 sp, and E11, 41-50 sp. This study was approved by the Ethical Review Committee of the Academy of Military Medical Sciences, China.
Mouse co-IP assay and western blotting
E10.5 mouse AGM regions were dissected and dissociated into single cell suspension using 0.1% collagenase. After a 3-day culture in the presence of hematopoietic cytokines (50 ng/ml SCF, 50 ng/ml IL-3, and 20 ng/ml FLT3), cells were starved for 4 h and pretreated with MG132 (10 μM, Sigma C2211) for 2 h, and then treated with 7α-25-OHC for 0.5 h (or 1 h in Figure 4F) before harvesting. Cells were lysed in RIPA buffer (50 mM Tris, 150 mM NaCl, 10% Glycerol, 5 mM MgCl2, 0.5% NP40, and Roche cocktail protease inhibitor), and incubated with Arrb1 (Abcam, ab32097, 1:100) or Notch1 (Abcam, ab65297, 1:100) antibody for 4 h at 4 °C. Protein A agarose (Invitrogen, 15918014) was added next to the mixture which was incubated for another 3-4 h. Beads were collected for western blotting using following antibodies: β-actin (cell signaling, 4967, 1:2000), Notch1 (Abcam, ab128076, 1:1 000), Notch1 (Abcam, ab65297, 1:1 000), NICD (Abcam, ab83232, 1:500), Gpr183 (Abcam, ab168744, 1:1 000), Arrb1 (Abcam, ab32097, 1:5 000), and ubiquitin (Santa Cruz, sc8017, 1:1 000).
Mass spectrometry
E10.5 mouse AGM was dissected and dissociated into single cells, which were cultured for three days in the presence of hematopoietic cytokine, before being treated with either DMSO or 7α-25-OHC for 1 h. Anti-Notch1 (Abcam, ab65297, 1:100) was used for the IP. LTQ Orbitrap Elite was used to perform the MS. The entire dataset is shown in Supplementary information, Tables S2 and S3.
Protein purification
His-Ubc4, His-Ub, RGSH-Flag-NICD, GST-Ub E1, GST-Nedd4 and other Ub E3 ligases were first affinity purified, then fractionated by size exclusion chromatography on Superdex75 and Superdex200 column in a reaction buffer containing 50 mM Tris-HCl, pH 8.0, 150 mM KCl, 5% glycerol and 2 mM MgCl.
In vitro ubiquitination assay
Ubiquitination experiment was carried out as follows: 60 nM E1, 300 nM Ubc4 (E2), 300 nM Nedd4 (E3) and 500 nM NICD were incubated with 1 μM ubiquitin at 30°C in a reaction buffer containing 25 mM Tris-HCl, pH 7.4, 2 mM MgCl/ATP, and 0.2 mM DTT. Samples were collected 2 h later, and were mixed with the Laemmli buffer in the presence of a reducing reagent for western blotting.
GST Pull-down
Nedd4-GST was incubated with GST beads at 4 °C for 2 h (GST protein was used as a negative control). The beads were centrifuged and washed in NP40 buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl and 0.5% NP40) three times. RGSH-FLAG-NICD was added to the mixture, which was incubated for additional 2 h at 4 °C. Following the incubation, the beads were centrifuged and washed 3 times in NP40 buffer, before a loading buffer was added. The samples were heated at 65 °C for 5 min and analyzed by western blotting.
AGM explant and CFU-S assay
Mouse AGMs were dissected and cultured as explants for 3 days as previously described51. In brief, AGMs were deposited on nylon filters (Millipore) placed on metallic supports and cultured in a medium for myeloid long-term culture (Stem Cell Technologies) supplemented with 10 μM hydrocortisone (Stem Cell Technologies). To this air-liquid interphase culture we added either DMSO, or 1 μM 7α-25-OHC, or 10 μM clotrimazole. The AGMs after treatment were dissociated by 0.04% collagenase for 40 min at 37 °C. Cells were injected intravenously into female C57BL/6J mice that had been exposed to a split dose of 9 Gy γ-irradiation (60Co, 4.5 + 4.5 Gy with 2 h interval). After 11 days, animals were sacrificed and their spleens were removed and fixed in Bouin's fixative. Spleen colonies were counted under a dissecting microscope.
OP9 co-cultures
FACS-sorted CD31+CD41−CD45−Ter119− pan-ECs from E10 caudal half were cultured on OP9 stromal cells in the presence of cytokines (100 ng/ml SCF, 100 ng/ml IL-3, and 100 ng/ml FLT3), and were treated with DMSO, 1 μM 7α-25-OHC or 10 μM clotrimazole for 2 days. After additional 5 days of co-culture, cells were harvested by trituration using a pipet for subsequent flow cytometry analysis.
Long-term transplantation assay
Male C57BL/6J (CD45.2) and female C57BL/6J (CD45.1) mice were mated to obtain CD45.1/2 embryos. The AGMs of E11 (43-45 sp) embryos were digested in 0.1% collagenase for 30 min at 37 °C. Individual cells were cultured in the presence of DMSO or 10 μM clotrimazole plus hematopoietic cytokines (100 ng/ml SCF, 100 ng/ml IL-3, and 100 ng/ml FLT3). After 4 days of culture, cells were harvested by pipetting and enzymatic digestion. These cells, together with 2 × 104 bone marrow carrier cells (CD45.2), were injected intravenously into irradiated female C57BL/6J mice (CD45.2). Chimerism in peripheral blood and bone marrow was measured 16 weeks after the transplantation. To examine the self-renewal potential, secondary transplantation was performed, in which a total of 3 × 106 cells from the bone marrow of a primary recipient were intravenously injected into a secondary recipient. Donor-derived chimerism was assessed by flow cytometry using anti-CD45.2-PE and anti-CD45.1-APC antibodies (eBioscience) 12 weeks after transplantation. Mice demonstrating over 10% donor-derived chimerism (contributing to both myeloid and lymphoid lineages) after a minimum of 12 weeks were considered to be reconstituted. The contribution of transplanted cells to the different lineages was measured by flow cytometry with specific antibodies against CD3, B220, Mac1, Gr1, CD4 and CD8 (eBioscience). For transplantation of 7α-25-OHC-treated cells, 0.3 embryo-equivalent donor cells from actin-GFP transgenic C57BL/6J mice were treated with DMSO or 1 μM 7α-25-OHC, mixed with 2 × 104 bone marrow cells (GFP−), and injected intravenously into irradiated female mice (GFP−). Three months later, bone marrow cells were analyzed for GFP+ chimerism.
Statistical analysis
All experiments were repeated at least three times, except for the long-term transplantation after clotrimazole treatment. Student's two-tailed t-test was used for statistical analysis.
References
Dieterlen-Lievre F . Hematopoiesis: progenitors and their genetic program. Curr Biol 1998; 8:R727–R730.
Jaffredo T, Nottingham W, Liddiard K, Bollerot K, Pouget C, de Bruijn M . From hemangioblast to hematopoietic stem cell: an endothelial connection? Exp Hematol 2005; 33:1029–1040.
Medvinsky A, Rybtsov S, Taoudi S . Embryonic origin of the adult hematopoietic system: advances and questions. Development, 2011; 138:1017–1031.
Kaimakis P, Crisan M, Dzierzak E . The biochemistry of hematopoietic stem cell development. Biochim Biophys Acta 2013; 1830:2395–2403.
Kissa K, Herbomel P . Blood stem cells emerge from aortic endothelium by a novel type of cell transition. Nature 2010; 464:112–115.
Bertrand JY, Chi NC, Santoso B, Teng S, Stainier DY, Traver D . Haematopoietic stem cells derive directly from aortic endothelium during development. Nature 2010; 464:108–111.
Boisset JC, van Cappellen W, Andrieu-Soler C, Galjart N, Dzierzak E, Robin C . In vivo imaging of haematopoietic cells emerging from the mouse aortic endothelium. Nature 2010; 464:116–120.
Zhang CR, Patient R, Liu F . Hematopoietic stem cell development and regulatory signaling in zebrafish. Biochim Biophys Acta 2013; 1830:2370–2374.
Clements WK, Traver D . Signalling pathways that control vertebrate haematopoietic stem cell specification. Nat Rev Immunol 2013; 13:336–348.
Orkin SH, Zon LI . Hematopoiesis: an evolving paradigm for stem cell biology. Cell 2008; 132:631–644.
Burns CE, Traver D, Mayhall E, Shepard JL, Zon LI . Hematopoietic stem cell fate is established by the Notch-Runx pathway. Genes Dev 2005; 19:2331–2342.
Zhong TP, Childs S, Leu JP, Fishman MC . Gridlock signalling pathway fashions the first embryonic artery. Nature 2001; 414:216–220.
Lawson ND, Vogel AM, Weinstein BM . Sonic hedgehog and vascular endothelial growth factor act upstream of the Notch pathway during arterial endothelial differentiation. Dev Cell 2002; 3:127–136.
Robert-Moreno A, Guiu J, Ruiz-Herguido C, et al. Impaired embryonic haematopoiesis yet normal arterial development in the absence of the Notch ligand Jagged1. EMBO J 2008; 27:1886–1895.
Richard C, Drevon C, Canto PY, et al. Endothelio-mesenchymal interaction controls runx1 expression and modulates the notch pathway to initiate aortic hematopoiesis. Dev Cell 2013; 24:600–611.
Lizama CO, Hawkins JS, Schmitt CE, et al. Repression of arterial genes in hemogenic endothelium is sufficient for haematopoietic fate acquisition. Nat Commun 2015; 6:7739.
Wettschureck N, Offermanns S . Mammalian G proteins and their cell type specific functions. Physiol Rev 2005; 85:1159–1204.
Yue R, Li H, Liu H, et al. Thrombin receptor regulates hematopoiesis and endothelial-to-hematopoietic transition. Dev Cell 2012; 22:1092–2100.
North TE, Goessling W, Walkley CR, et al. Prostaglandin E2 regulates vertebrate haematopoietic stem cell homeostasis. Nature 2007; 447:1007–1011.
Solaimani Kartalaei P, Yamada-Inagawa T, Vink CS, et al. Whole-transcriptome analysis of endothelial to hematopoietic stem cell transition reveals a requirement for Gpr56 in HSC generation. J Exp Med 2015; 212:93–106.
Ng CE, Yokomizo T, Yamashita N, et al. A Runx1 intronic enhancer marks hemogenic endothelial cells and hematopoietic stem cells. Stem Cell 2010; 28:1869–1881.
Tamplin OJ, Durand EM, Carr LA, et al. Hematopoietic stem cell arrival triggers dynamic remodeling of the perivascular niche. Cell 2015; 160:241–252.
Hannedouche S, Zhang J, Yi T, et al. Oxysterols direct immune cell migration via EBI2. Nature 2011; 475:524–527.
Liu C, Yang XV, Wu J, et al. Oxysterols direct B-cell migration through EBI2. Nature 2011; 475:519–523.
Heinig M, Petretto E, Wallace C, et al. A trans-acting locus regulates an anti-viral expression network and type 1 diabetes risk. Nature 2010; 467:460–464.
Rose KA, Stapleton G, Dott K, et al. Cyp7b, a novel brain cytochrome P450, catalyzes the synthesis of neurosteroids 7alpha-hydroxy dehydroepiandrosterone and 7alpha-hydroxy pregnenolone. Proc Natl Acad Sci USA 1997; 94:4925–4930.
Lund EG, Kerr TA, Sakai J, Li WP, Russell DW . cDNA Cloning of mouse and human cholesterol 25-hydroxylases, polytopic membrane proteins that synthesize a potent oxysterol regulator of lipid metabolism. J Biol Chem 1998; 273:34316–34327.
Tang Y, Bai H, Urs S, Wang Z, Liaw L . Notch1 activation in embryonic VE-cadherin populations selectively blocks hematopoietic stem cell generation and fetal liver hematopoiesis. Transgenic Res 2013; 22:403–410.
Zhao L, Borikova AL, Ben-Yair R, et al. Notch signaling regulates cardiomyocyte proliferation during zebrafish heart regeneration. Proc Natl Acad Sci USA 2014; 111:1403–1408.
Scheer N, Campos-Ortega JA . Use of the Gal4-UAS technique for targeted gene expression in the zebrafish. Mech Dev 1999; 80:153–158.
Lefkowitz RJ, Whalen EJ . beta-arrestins: traffic cops of cell signaling. Curr Opin Cell Biol 2004; 16:162–168.
Kim AD, Melick CH, Clements WK, et al. Discrete Notch signaling requirements in the specification of hematopoietic stem cells. EMBO J 2014; 33:2363–2373.
Ditadi A, Sturgeon CM, Tober J, et al. Human definitive haemogenic endothelium and arterial vascular endothelium represent distinct lineages. Nat Cell Biol 2015; 17:580–591.
Li Z, Lan Y, He W, et al. Mouse embryonic head as a site for hematopoietic stem cell development. Cell Stem Cell 2012; 11:663–675.
Yue R, Li H, Liu H, et al. Thrombin receptor regulates hematopoiesis and endothelial-to-hematopoietic transition. Dev Cell 2012; 22:1092–1100.
Gori JL, Butler JM, Chan YY, et al. Vascular niche promotes hematopoietic multipotent progenitor formation from pluripotent stem cells. J Clin Invest 2015; 125:1243–1254.
Huang C, Jackson M, Samuel K, Taylor AH, Lowell S, Forrester LM . Haematopoietic differentiation is inhibited when Notch activity is enhanced in FLK1(+) mesoderm progenitors. Stem Cell Res 2013; 11:1273–1287.
Chanda B, Ditadi A, Iscove NN, Keller G . Retinoic acid signaling is essential for embryonic hematopoietic stem cell development. Cell 2013; 155:215–227.
Ruiz-Herguido C, Guiu J, D'Altri T, et al. Hematopoietic stem cell development requires transient Wnt/beta-catenin activity. J Exp Med 2012; 209:1457–1468.
Yue R, Kang J, Zhao C, et al. Beta-arrestin1 regulates zebrafish hematopoiesis through binding to YY1 and relieving polycomb group repression. Cell 2009; 139:535–546.
Rathinam C, Matesic LE, Flavell RA . The E3 ligase Itch is a negative regulator of the homeostasis and function of hematopoietic stem cells. Nat Immunol 2011; 12:399–407.
Zhang C, Lv J, He Q, et al. Inhibition of endothelial ERK signalling by Smad1/5 is essential for haematopoietic stem cell emergence. Nat Commun 2014; 5:3431.
Li P, Lahvic JL, Binder V, et al. Epoxyeicosatrienoic acids enhance embryonic haematopoiesis and adult marrow engraftment. Nature 2015; 523:468–471.
Parsons MJ, Pisharath H, Yusuff S, et al. Notch-responsive cells initiate the secondary transition in larval zebrafish pancreas. Mech Dev 2009; 126:898–912.
Zhen F, Lan Y, Yan B, et al. Hemogenic endothelium specification and hematopoietic stem cell maintenance employ distinct Scl isoforms. Development 2013; 140:3977–3985.
Wang L, Liu T, Xu L, et al. Fev regulates hematopoietic stem cell development via ERK signaling. Blood 2013; 122:367–375.
Wei Y, Ma D, Gao Y, Zhang C, Wang L, Liu F . Ncor2 is required for hematopoietic stem cell emergence by inhibiting Fos signaling in zebrafish. Blood 2014; 124:1578–1585.
Covassin L, Amigo JD, Suzuki K, Teplyuk V, Straubhaar J, Lawson ND . Global analysis of hematopoietic and vascular endothelial gene expression by tissue specific microarray profiling in zebrafish. Dev Biol 2006; 299:551–562.
Tang F, Barbacioru C, Nordman E, et al. RNA-Seq analysis to capture the transcriptome landscape of a single cell. Nat Protoc 2010; 5:516–535.
Renaud O, Herbomel P, Kissa K . Studying cell behavior in whole zebrafish embryos by confocal live imaging: application to hematopoietic stem cells. Nat Protoc 2011; 6:1897–1904.
Medvinsky A, Dzierzak E . Definitive hematopoiesis is autonomously initiated by the AGM region. Cell 1996; 86:897–906.
Acknowledgements
We thank Elaine Dzierzak and David Traver for helpful discussions and critical reading of the paper. We thank Baofa Sun and Yungui Yang for bioinformatics analysis, Anming Meng, Zilong Wen and Jing-Wei Xiong for providing reagents used in this work. This work was supported by the National Basic Research Program of China (2010CB945300 and 2011CB943900), the National Natural Science Foundation of China (31271570 and 31425016), and the Strategic Priority Research Program of the CAS (XDA01010110).
Author information
Authors and Affiliations
Corresponding author
Additional information
( Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary information
Supplementary information, Figure S1
Tg(runx1:en-GFP) can recapitulate runx1 endogenous expression in vivo. (PDF 327 kb)
Supplementary information, Figure S2
Expression profile and loss-of-function assay identify genes essential for definitive hematopoiesis. (PDF 395 kb)
Supplementary information, Figure S3
gpr183 is expressed in the hemogenic endothelium (HE) after 24 hpf, and is essential for adult hematopoiesis in zebrafish. (PDF 324 kb)
Supplementary information, Figure S4
Gpr183 influences HE development after 24 hpf. (PDF 390 kb)
Supplementary information, Figure S5
Gpr183 regulates HE/HSC development specifically. (PDF 441 kb)
Supplementary information, Figure S6
Notch signaling functions downstream of Gpr183. (PDF 334 kb)
Supplementary information, Figure S7
Notch signaling downregulation facilitates HSC emergence. (PDF 453 kb)
Supplementary information, Figure S8
Working model of Gpr183 repressing Notch signaling to facilitate HSPCs emergence. (PDF 216 kb)
Supplementary information, Table S1
RNA-Seq data (XLSX 1278 kb)
Supplementary information, Table S2
Mass spectra-DMSO (XLSX 58 kb)
Supplementary information, Table S3
Mass spectra-7α-25-OHC (XLSX 66 kb)
Supplementary information, Table S4
Morpholinos used in this work. (PDF 79 kb)
Supplementary information, Table S5
The qPCR primers used in this work. (PDF 79 kb)
Supplementary information, Movie S1
The AGM region of the Tg(runx1:en-GFP) embryo was observed from 33 hpf to 39 hpf by 40×oil objective, in Zeiss LSM 510 confocal microscopy. The movie was captured at 6 min/frame, and rendered at 5 frames/s. (AVI 1643 kb)
Supplementary information, Movie S2
The AGM regions of the control, gpr183 mutant and embryo treated with 7α-25-OHC in Tg(flk1:mCherry/cmyb:GFP) background were observed from 36 to 48 hpf by Nikon A1 confocal microscopy. The EHT process is abrogated in gpr183 mutant embryo, and the EHT frequency increases after 7α-25-OHC treatment. The movie was captured at 8 min/frame, and rendered at 5 frames/s. (WMV 1610 kb)
Supplementary information, Movie S3
The AGM region of the Tg(cmyb:GFP/tp1:mCherry) embryo was observed from 33 to 48 hpf by Perkin Elmer spinning disk microscopy. The cells undergoing EHT process are all cmyb+tp1−. The movie was captured at 8 min/frame, and rendered at 5 frames/s. (AVI 1873 kb)
Supplementary information, Movie S4
The AGM region of the Tg(runx1:en-GFP/tp1:mCherry) embryo was observed from 33 to 48 hpf by Perkin Elmer spinning disk microscopy. The cells undergoing EHT process are all runx1+tp1−. The movie was captured at 8 min/frame, and rendered at 5 frames/s. (AVI 2191 kb)
Rights and permissions
This license allows readers to copy, distribute and transmit the Contributionas long as it attributed back to the author. Readers are permitted to alter, transform or build upon the Contribution as long as the resulting work is then distributed under this is a similar license. Readers are notpermitted touse theContributionfor commercial purposes. Please read the full license for further details at - http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Zhang, P., He, Q., Chen, D. et al. G protein-coupled receptor 183 facilitates endothelial-to-hematopoietic transition via Notch1 inhibition. Cell Res 25, 1093–1107 (2015). https://doi.org/10.1038/cr.2015.109
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cr.2015.109
Keywords
This article is cited by
-
Serum from COVID-19 patients promotes endothelial cell dysfunction through protease-activated receptor 2
Inflammation Research (2024)
-
Activation of lineage competence in hemogenic endothelium precedes the formation of hematopoietic stem cell heterogeneity
Cell Research (2023)
-
Engineering a niche supporting hematopoietic stem cell development using integrated single-cell transcriptomics
Nature Communications (2022)
-
Temporal-spatial low shear stress induces heterogenous distribution of hematopoietic stem cell budding in zebrafish
Cellular and Molecular Life Sciences (2022)
-
Beraprost ameliorates postmenopausal osteoporosis by regulating Nedd4-induced Runx2 ubiquitination
Cell Death & Disease (2021)