Abstract
The abnormal activation of epidermal growth factor receptor (EGFR) is strongly associated with a variety of human cancers but the underlying molecular mechanism is not fully understood. By using direct stochastic optical reconstruction microscopy (dSTORM), we find that EGFR proteins form nanoclusters in the cell membrane of both normal lung epithelial cells and lung cancer cells, but the number and size of clusters significantly increase in lung cancer cells. The formation of EGFR clusters is mediated by the ionic interaction between the anionic lipid phosphatidylinositol-4,5-bisphosphate (PIP2) in the plasma membrane and the juxtamembrane (JM) region of EGFR. Disruption of EGFR clustering by PIP2 depletion or JM region mutation impairs EGFR activation and downstream signaling. Furthermore, JM region mutation in constitutively active EGFR mutant attenuates its capability of cell transformation. Collectively, our findings highlight the key roles of anionic phospholipids in EGFR signaling and function, and reveal a novel mechanism to explain the aberrant activation of EGFR in cancers.
Similar content being viewed by others
Introduction
The epidermal growth factor receptor (EGFR) is a receptor tyrosine kinase of the ErbB family that regulates a variety of important biological events including cell growth, proliferation, differentiation and migration1,2. The ligation of EGFR with its ligands such as epidermal growth factor (EGF) induces the dimerization of EGFR extracellular domain, the N-terminal interaction of the transmembrane helice and the dimerization of the juxtamembrane (JM) region, which promotes the formation of an asymmetric active dimer of EGFR kinase core and thus activates downstream signaling3,4,5,6,7. Genetic alterations including gene amplification, kinase domain mutations and in-frame deletions often lead to overexpression or constitutive activation of EGFR protein, which is frequently observed in different types of tumors and effectively drives cancer development8. Given its strong association with malignant progression in different epithelial carcinoma types, EGFR has received much attention and is considered as a valuable therapeutic target. Small-molecule inhibitors gefitinib and erlotinib that specifically target the tyrosine kinase activity of EGFR have been proven effective towards lung cancer patients with EGFR mutations9.
The plasma membrane is an important defense system with asymmetric lipid bilayer and regulates many fundamental physiological processes10,11. As an integral membrane protein, EGFR is surrounded by the lipid environment of the cellular membrane, and is potentially modulated by its niche. However, how the lipid components in the plasma membrane regulate the activity of EGFR remains largely unknown. Recent studies have reported that the anionic phospholipids, which are minor lipid species present in the inner-leaflet of the plasma membrane, can ionically interact with proteins carrying polybasic sequences and regulate their spatial distribution, structure and function12. The protein list is quickly growing, now including membrane receptors, ion channels, small GTPases, kinases and others. For example, our previous study shows that the tyrosine residues of T-cell antigen receptor (TCR) can be sequestered in the membrane bilayer by the dynamic interaction between TCR tyrosine motifs and anionic phospholipids in the plasma membrane13,14. The SNAP (Soluble NSF (N-ethylmaleimide Sensitive Fusion protein) Attachment Protein) receptor protein syntaxin-1A forms microclusters in the plasma membrane via the ionic interaction between its polybasic JM region and the strong anionic lipid phosphatidylinositol-4,5-bisphosphate (PIP2), which is essential for the vesicle docking and membrane fusion during neuronal exocytosis15. As a critical co-factor, PIP2 can also induce big conformational change in inward-rectifier K+ (Kir) 2.2 channel through the ionic interaction with polybasic amino acids at the interface of transmembrane domain and cytoplasmic domain16. Previous studies with Ras superfamily members show that PIP2 and PIP3 can target small GTPases to the plasma membrane via ionic interactions17. All these data highlight the importance of ionic protein-lipid interactions in regulating various cellular events.
Although studies of the different structural domains of EGFR itself provide considerable insight into its activation mechanism3,4,5,6,7, few works have been done in terms of the relationship between EGFR and its lipid niche. Previous study using near-field scanning optical microscopy (NSOM) suggested that EGFR might be organized in aggregates with distinct diameters in the membrane of paraformaldehyde-fixed Hela cells18. However, the detailed information of EGFR distribution on cell surface, especially on living cell surface, still remains largely elusive. Moreover, the mechanisms involved in regulating EGFR spatial distribution have not been brought to light yet. Therefore, further study using improved imaging technology is necessary and timely urgent. The recent development of super-resolution fluorescence microscopy techniques, such as stochastic optical reconstruction microscopy (STORM)19,20, direct STORM (dSTORM)21, photoactivated localization microscopy (PALM)22 and stimulated-emission depletion (STED) microscopy15, has broken the diffraction limit of light and provided super-resolution imaging (tens of nanometers) at a single-molecule level in living cells.
Therefore, we used dSTORM to study the distribution pattern of EGFR on the surface of cells from malignant tumors and paired normal tissues. Our data uncovered the clustering pattern of EGFR in the plasma membrane of lung cancer cells and normal lung epithelial cells. Moreover, we have identified an important contribution of the ionic interaction between anionic phospholipids and EGFR JM domain in regulating the unique membrane spatial distribution of EGFR as well as its activation and function.
Results
EGFR forms clusters in the plasma membrane of both normal lung epithelial cells and lung cancer cells
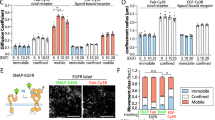
To investigate the spatial distribution pattern of EGFR under both physiological and pathological conditions, we freshly isolated both human lung cancer cells and paired pathologically normal lung epithelial cells, which are indicated by positive staining of epithelial cell marker EpCAM (epithelial cell adhesion molecule; Supplementary information, Figure S1A and S1B)23,24. We then used these epithelial cells for dSTORM analyses via the labeling of Alexa647-conjugated Cetuximab (Supplementary information, Figure S1B), an anti-EGFR antibody known to compete for the binding of EGFR ligands and inhibit EGFR activation25. Interestingly, we found that the majority of EGFR proteins formed clusters with sizes ranging from 70 to 660 nm in the plasma membrane of lung cancer cells, with an average of 293 ± 124 nm (SD) (Figure 1A and 1B). In contrast, fewer EGFR clusters were visible in the plasma membrane of normal lung epithelial cells; these clusters were smaller in size, ranging from 60 to 460 nm, with an average of 207 ± 69 nm (SD) (Figure 1A and 1C). Consistently, statistic analyses showed that the cell surface density of EGFR clusters was much higher in lung cancer cells in comparison with normal lung epithelial cells (Figure 1D).
EGFR forms more clusters with bigger size in the plasma membrane of lung cancer cells compared with normal lung epithelial cells. (A) Reconstructed dSTORM images of Alexa647-Cetuximab-labeled EGFR in the plasma membrane of lung cancer cells (LC) and normal lung epithelial cells (NL). Scale bar, 10 μm. (B, C) Size distribution of EGFR clusters on cell surface of LC (B) and NL (C). (D) Surface density of EGFR clusters for LC and NL. (E) Percentage of EGFR proteins in cluster on cell surface of LC and NL. (F) Average number of clusters containing different fluorescence intensity unit of EGFR proteins on cell surface of LC and NL. All the cells were labeled with Alexa647-Cetuximab for dSTORM analyses. All error bars denote SD. ***P < 0.001, two-tailed unpaired t-test.
We further found that about 65.6% of EGFR on lung cancer cell surface were assembled as the form of clusters whereas this percentage dropped to about 35.4% on normal lung epithelial cell surface (Figure 1E). To analyze the size difference of EGFR clusters, we semiquantified the number of EGFR proteins within each cluster by the number of bound Alexa647-conjugated Cetuximab, and defined it as “fluorescence intensity unit”. This value is calculated through dividing the total Alexa647 number inside a cluster by the number of Alexa647 carried by each Cetuximab molecule. We hereafter refer the fluorescence intensity unit to the relative EGFR protein number. We found that these clusters were composed of different numbers of EGFR proteins, ranging from 3 to over 30 (Figure 1F). Interestingly, most of the EGFR proteins existed as relatively moderate-sized clusters containing 7–14 EGFR proteins on the surface of lung cancer cells, while the clusters of normal lung epithelial cells were much smaller, mainly containing 3–5 proteins (Figure 1F). Lung cancer cells contained significantly more moderate- and big-sized clusters with variant protein amounts from 6 to over 30 than normal lung epithelial cells, but there was no obvious difference for small clusters with 3–5 proteins (Figure 1F). Meanwhile, clusters containing more than 15 EGFR proteins could be readily observed on lung cancer cell surface, but are almost absent on the surface of normal lung epithelial cells (Figure 1F). A high proportion of EGFR proteins tend to stay in clusters rather than to be sporadically distributed on lung cancer cell surface, which might be important for EGFR activation. Indeed, these lung cancer cells displayed a strong activation of EGFR signaling (Supplementary information, Figure S1C). Using Alexa647-conjugated EGF as an additional probe, we obtained similar findings in both normal lung epithelial cells and lung cancer cells (Supplementary information, Figure S1D–S1I). Moreover, similar observations that EGFR clusters mainly contained 3–8 proteins were obtained in COS-7 cells, which are frequently used for the study of EGFR activation4 (Figure 2B and 2E).
Constitutive PIP2 depletion disrupts the EGFR clustering in the plasma membrane. (A) Two-color dSTORM and PALM images of EGFR labeled with Alexa647-Cetuximab (red, left) or Alexa647-EGF (red, right) and PIP2 labeled with PH-PLCδ-mEos3.2 (green) in COS-7 cells. Scale bar, 10 μm. Inset 1 and 2 show the complete and partial overlap of EGFR clusters and PIP2 clusters, respectively. Scale bar, 200 nm. (B) Reconstructed dSTORM images of control COS-7 cells (Ctrl) and SJ2-mCitrine-expressing COS-7 cells (SJ2) labeled with Alexa647-Cetuximab. Scale bar, 10 μm. (C) Surface density of EGFR clusters in Ctrl and SJ2 COS-7 cells. (D) Percentage of EGFR proteins in cluster on the surface of Ctrl and SJ2 COS-7 cells. (E) Average number of clusters containing different amount of EGFR proteins on the surface of Ctrl and SJ2 COS-7 cells. (F) The number of EGFR clusters on the surface of Ctrl and SJ2 COS-7 cells, which is normalized by corresponding total amount of surface EGFR protein. All error bars denote SD. ***P < 0.001, two-tailed unpaired t-test.
PIP2 depletion in the plasma membrane impairs the EGFR cluster formation
We then studied the molecular mechanisms involved in the EGFR cluster formation. Previous studies have shown that the high accumulation of PIP2 is required for receptor protein syntaxin-1A clustering15. To test whether PIP2 was involved in the EGFR clustering, we labeled PIP2 in COS-7 cell membrane with a PIP2 probe consisting of the Pleckstrin homology (PH) domain26 of phospholipase C delta fused to a photoactivatable fluorescent protein mEos3.227 (PH-PLCδ-mEos3.2) and analyzed the spatial distribution of PIP2 and EGFR using PALM and dSTORM, respectively. Our data showed that PIP2, similar to EGFR protein, also formed clusters in the cell membrane (Figure 2A). Interestingly, from magnified images, we could clearly observe the overlap between these two different types of clusters (Figure 2A). Measurement of Mander's colocalization coefficients28 showed that about 60% of EGFR clusters were colocalized with PIP2 clusters, highlighting the potential contribution of PIP2 in EGFR cluster formation. Considering the potential influence of the membrane mobility of living cells, we performed imaging in fixed cells and obtained consistent results showing that about 58.6% of EGFR clusters were colocalized with PIP2 clusters (Supplementary information, Figure S2A). To further test the role of PIP2, we transfected COS-7 cells with an inositol-polyphosphate 5-phosphatase Synaptojanin 2 (SJ2) that can dephosphorylate the D5 position phosphate from PIP2. Our data showed that the total amount as well as the surface density of EGFR clusters dramatically decreased on COS-7 cell surface after ectopic SJ2 expression (Figure 2B and 2C). The percentage of EGFR proteins in clusters dropped from 57.1% to 31.1% after PIP2 depletion (Figure 2D). Furthermore, statistic analyses showed that the clusters containing different number of EGFR proteins were significantly reduced after PIP2 depletion; and those large clusters with more than 11 proteins inside were hardly detectable on the surface of cells expressing SJ2 (Figure 2E). These data suggest that EGFR could not form large-sized clusters without PIP2, further supporting the important role of PIP2 in the EGFR cluster formation.
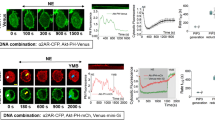
To avoid potential artifacts caused by the constitutive PIP2 depletion by SJ2, we established an inducible PIP2 depletion system to modify PIP2 level in the plasma membrane29. We transfected COS-7 cells with two constructs: Lyn-FKBP12 and mCitrine-FRB-SJ2. FKBP12 fusion protein is targeted to the plasma membrane by its N-terminal myristoylation and palmitoylation sequence of human GAP43 protein (Lyn), whereas the PIP2 phosphatase SJ2 fused with a fluorescent protein mCitrine and a FRB domain localizes in cytosol under normal condition. Rapamycin treatment triggers the heterodimerization of FRB and FKBP12 domains and thus targets SJ2 to the plasma membrane for PIP2 depletion (Figure 3A and 3B). To monitor the PIP2 level in the plasma membrane, we used a PIP2 probe with the PH domain of PLCδ fused to a red fluorescence protein mCherry, and the depletion of PIP2 would result in the dissociation of the PH-PLCδ-mCherry from the plasma membrane (Figure 3A and 3B). Our data showed that PIP2 depletion induced by rapamycin treatment resulted in a significant reduction of EGFR clusters in the plasma membrane (Figure 3C and Supplementary information, Figure S2B). Consistently, the surface density of EGFR clusters, the percentage of EGFR in clusters and the number of clusters containing different amount of EGFR dramatically decreased after rapamycin treatment (Figure 3D–3F and Supplementary information, Figure S2C–S2E), similar to the observation from the cells with constitutive SJ2 expression. In contrast, rapamycin treatment in control cells did not cause significant change of EGFR distribution pattern (Supplementary information, Figure S3A–S3F).
Inducible depletion of PIP2 significantly impairs the EGFR clustering in the plasma membrane. (A) Schematic diagram of the components (left) and principle (right) of inducible PIP2 depletion system. (B) Validation of the system in COS-7 cells visualized by laser scanning confocal microscope. Scale bar, 20 μm. (C) dSTORM images of COS-7 cells transfected with the inducible PIP2 depletion system before (Rapa−) and after (Rapa+) rapamycin treatment. Scale bar, 10 μm. (D) Surface density of EGFR clusters in COS-7 cells before (Rapa−) and after (Rapa+) rapamycin treatment. (E) Percentage of EGFR proteins clustering on the surface of COS-7 cells before (Rapa−) and after (Rapa+) rapamycin treatment. (F) Average number of clusters containing different amount of EGFR proteins on the surface of COS-7 cells before (Rapa−) and after (Rapa+) rapamycin treatment. (G) The number of EGFR clusters on the surface of COS-7 cells before (Rapa−) and after (Rapa+) rapamycin treatment, which is normalized by corresponding total amount of surface EGFR protein. All the cells were labeled with Alexa647-Cetuximab for dSTORM analyses. All error bars denote SD. ***P < 0.001, two-tailed unpaired t-test.
Our data showed that PIP2 depletion did not affect the total amount of surface EGFR (Supplementary information, Table S1). We also did the normalization of the EGFR cluster number by total surface EGFR amount and found that the cluster number was still much less after constitutive or inducible PIP2 depletion (Figures 2F and 3G and Supplementary information, Figure S2F). Together, these data demonstrated that PIP2 played an important role in EGFR clustering in the plasma membrane.
The JM region of EGFR ionically binds to PIP2
Next, we studied how PIP2 regulates the membrane distribution of EGFR. The N-terminus of EGFR JM region contains a highly conserved stretch including nine positively charged residues (645RRRHIVRKRTLRR657). Previous in vitro studies showed that the N-terminus of JM peptide (residues 645–660) binds strongly to the anionic phospholipid-containing vesicles30,31. As two major anionic phospholipids in the plasma membrane, phosphatidylserine (PS) and PIP2 have a net charge of −1 and −4, respectively. To further characterize the interaction between EGFR JM region and anionic phospholipids, we labeled the full-length JM peptide (residues 645–682) with Alexa488 and measured its binding affinity to lipid bicelles containing either PS or PIP2 by fluorescence polarization assay. We found that the full-length JM peptide showed a stronger binding affinity to bicelles containing 10% PIP2 than to bicelles containing 50% PS despite of more negative charges on the PS bicelles (Figure 4A), suggesting a specific binding between PIP2 and the JM peptide. JM peptide did not bind to bicelles composed of zwitterionic phosphatidylcholine (PC). Consistent with previous studies4, the JM peptide adopted α-helix folding after binding to PIP2-containing bicelles, as measured by circular dichroism (CD; Figure 4B).
The JM region of EGFR ionically binds to the anionic phospholipid PIP2 in vitro and in vivo. (A) Analysis of the association of JM peptide with the membrane bilayer by fluorescence polarization (FP) technique. The FP value increased gradually with the increase of lipid concentration. (B) Analysis of the secondary structural change of JM peptide by a far UV CD experiment. Free JM displayed as an unstructured flexible peptide in solution, while the presence of 50% POPS/POPC or 10% DOPIP2/DOPC bicelles induced partial helical folding of JM. (C) Schematic diagram of two EGFR JM region mutations. JM-PM, point mutation; JM-DM, deletion mutation. (D) Cellular localization of N-myristoyl JM peptide or JM-PM peptide fused with mCherry protein in 293T cells visualized by laser scanning confocal microscopy. Scale bar, 10 μm. (E) The location of mCherry-JM-N-CVIM fusion protein and mCitrine-FRB-SJ2 were scanned by confocal microscopy in 293T cells transiently transfected with corresponding constructs before and after loading rapamycin and incubating at 37 °C for 1 h (left). The fluorescence intensity distributions along the white lines were shown on the right before and after rapamycin treatment. Scale bar, 10 μm.
To test whether the JM-PIP2 interaction is physiologically relevant, we first developed a method to directly visualize the binding of the JM region to the plasma membrane in living cells. We designed a construct in which the JM peptide was fused to an N-terminal myristoylation site and a C-terminal mCherry fluorescence protein. This fusion protein mainly localized at the plasma membrane when it was exogenously expressed in 293T cells that do not have endogenous EGFR expression (Figure 4D, left and Supplementary information, Figure S4A). To confirm the specificity of this binding, a JM mutant, named JM-PM, in which the eight conserved basic residues were mutated to serines (Figure 4C), was analyzed in parallel. As expected, the JM-PM mutant lost the specific binding to the plasma membrane (Figure 4D, right). To test whether PIP2 is involved in this ionic binding between the JM region and the plasma membrane, we fused the N-terminus of EGFR JM region (JM-N, residues 645–664) to an N-terminal mCherry protein and a C-terminal K-Ras4b prenylation motif CVIM17. This fusion protein (mCherry-JM-N-CVIM) was exogenously expressed in 293T cells together with the inducible PIP2 depletion system. After the rapamycin-induced PIP2 depletion, we found an obvious dissociation of the JM region from the plasma membrane (Figure 4E).
We further applied a fluorescence resonance energy transfer (FRET)-based assay to confirm the ionic JM-PIP2 interaction14. As previously described13, we generated a KIR2DL3-JM region-mTFP1 chimera to study the FRET signal between the JM region and the plasma membrane. The cytoplasmic tail of an irrelevant monomeric membrane protein KIR2DL3 was replaced by the EGFR JM region (residues 645–682) followed by a C-terminal mTFP1 fluorescence protein. The mTFP1 acted as a FRET donor while an octadecyl rhodamine B (R18) dye that can be incorporated into the plasma membrane was used as a FRET acceptor. The mTFP1 and R18 can generate FRET signal only when mTFP1 is in close proximity to the plasma membrane, e.g., when the EGFR JM region binds to the membrane. To roughly translate the FRET efficiency to distance information, we used three control constructs in which the JM region sequence was replaced by flexible linkers with different lengths (3 a.a., 25 a.a., 50 a.a.). We exogenously expressed these FRET constructs in Jurkat cells and sorted the transfectants with comparable chimera expression levels. A donor dequenching method was used to measure the FRET efficiency between mTFP1 and R18 (Figure 5A–5C). The short linker construct (3 a.a.) showed a high FRET efficiency (38.8%, Figure 5A), while the 25 a.a. and 50 a.a. linker constructs exhibited relatively low values (20.5% and 12.3%, respectively, Figure 5A). A relatively high FRET efficiency was detected from the EGFR-JM construct (29.3%, Figure 5A), which was significantly higher than that of the 25 a.a. construct, suggesting that most of the EGFR JM region (38 a.a.) should largely bind to the plasma membrane. However, we observed a significantly reduced FRET efficiency for the JM-PM mutant (19.0%, Figure 5A), confirming the importance of basic residues in regulating the membrane binding of the JM region. Then we introduced the inducible PIP2 depletion system into the Jurkat transfectants with JM-mTFP1 expression, and found that the FRET efficiency of JM region significantly decreased after rapamycin treatment, indicating that PIP2 is the primary binding partner of the JM region at the plasma membrane (Figure 5B). Collectively, these cellular and biochemical data showed that the JM region of EGFR could ionically bind to PIP2 in the plasma membrane.
The JM region binds to PIP2 in the plasma membrane via ionic interaction. (A, B) FRET efficiencies were measured between mTFP1 and the membrane dye R18 by dequenching approach in living Jurkat cells. 3a.a.-mTFP1, 25a.a.-mTFP1 and 50a.a.-mTFP1 served as control constructs. The measured FRET efficiencies were compared between wild-type JM (JM-mTFP1) and mutated JM (JM-PM-mTFP1) in A, or between the conditions before and after PIP2 depletion in B. (C) R18-labeled cells were imaged in both channels and the acceptor R18 was then photobleached to release the quenched fluorescence. BP, before photobleaching; AP, after photobleaching. Scale bar, 5 μm. The FRET efficiencies were measured for 16–18 cells per condition. Each dot represents the FRET value from one individual cell. Unpaired two tailed Student's t-test was used for the statistical analysis.
Previous studies showed that the ionic interaction between PIP2 and syntaxin-1A could glue the protein molecules together and also remove the charge-charge repulsion between protein molecules, thus mediating syntaxin-1A clustering15. Our finding of the JM-PIP2 interaction suggests that EGFR clustering might be regulated by PIP2 via the same mechanism.
The JM-PIP2 interaction is important for EGFR cluster formation
We further tested whether EGFR clustering is dependent on the ionic JM-PIP2 interaction. We expressed either wild-type full-length EGFR (EGFR-WT) or EGFR mutants (either EGFR-PM with the basic residue mutation or EGFR-DM with the deletion of the polybasic stretch (residue 645–657); Figure 4C) in 293T cells and analyzed their distribution patterns on the cell surface through dSTORM. Consistently, exogenous EGFR-WT proteins formed clusters in the 293T cell membrane (Figure 6A and Supplementary information, Figure S4B). However, we observed a significant decrease of the cluster numbers and sizes of the two mutants (Figure 6A and Supplementary information, Figure S4B), as well as dramatically reduced cluster density, cluster number normalized by total surface EGFR protein and the percentage of EGFR proteins in clusters (Figure 6B–6D and Supplementary information, Figure S4C–S4E). Moreover, mutation or deletion of the JM region resulted in a decrease of the number of clusters containing different amount of EGFR protein, and the clusters with more than eight EGFR proteins were hardly observed in the cell membrane compared to EGFR-WT, especially for EGFR-DM (Figure 6E and Supplementary information, Figure S4F). Collectively, these data further supported the notion that JM-PIP2 interaction is important for EGFR clustering in the plasma membrane.
The JM region regulates the EGFR clustering in the plasma membrane. (A) dSTORM images of 293T cells expressing comparable levels of exogenous full-length EGFR-WT, PM or DM. Scale bar, 10 μm. (B) Surface density of EGFR clusters in 293T cells expressing EGFR-WT, PM or DM. (C) The number of EGFR clusters on the surface of 293T cells expressing EGFR-WT, PM or DM, which is normalized by corresponding total amount of surface EGFR protein. (D) Percentage of EGFR proteins clustering on cell surface of 293T expressing EGFR-WT, PM or DM. (E) Average number of clusters containing different amount of EGFR proteins on cell surface of 293T expressing EGFR-WT, PM or DM. All the cells were labeled with Alexa647-Cetuximab for dSTORM analyses. All error bars denote SD. *P < 0.05, ***P < 0.001, two-tailed unpaired t-test.
The JM-PIP2 interaction regulates EGFR activity and function
We further asked whether the JM-PIP2 interaction-dependent EGFR clustering on the cell surface was important for EGFR activity. Growth factor receptor-bound protein 2 (Grb2) is a classic receptor tyrosine kinase-related adaptor protein well known for its ability to link EGFR tyrosine kinase to the activation of Ras and its downstream kinases, such as Erk1/232,33. Considering the fact that the binding of Grb2 to the tyrosine phosphorylated C-terminus of EGFR through its SH2 domain is an indication of EGFR activation32, we used the SH2 domain of human Grb2 protein with an N-terminal mEos3.2 (mEos3.2-Grb2/SH2) as a probe for the detection of EGFR activation in COS-7 cells34. After EGF stimulation, the mEos3.2-Grb2/SH2 probe translocated to the plasma membrane as expected but started to be internalized after 20 min, which should be caused by the EGFR receptor internalization (Figure 7A). To avoid the influence of the receptor internalization, we fixed the cells after 10 min of EGF stimulation and then visualized the spatial distribution of EGFR and mEos3.2-Grb2/SH2 in the plasma membrane with or without persistent SJ2 expression through dSTORM and PALM, respectively. Our data consistently showed that the mEos3.2-Grb2/SH2 formed clusters on the plasma membrane and significantly colocalized with EGFR clusters after EGF stimulation (Figure 7B), indicative of the activation of EGFR. Conversely, in resting cells labeled with Cetuximab or cells stimulated with EGF at 4 °C, few mEos3.2-Grb2/SH2 clusters were formed on the plasma membrane (Supplementary information, Figure S5A–S5C), and the percentage of the colocalization with EGFR clusters was nearly ten times lower than that in activated cells (Supplementary information, Figure S5D). Importantly, PIP2 depletion resulted in a significant decrease of the colocalization of mEos3.2-Grb2/SH2 and EGFR clusters after EGF stimulation, from 62.7% to 18.8% (Figure 7B and 7C), highlighting the involvement of PIP2 in regulating EGFR activation. Consistently, western blot data showed that either inducible or constitutive PIP2 depletion inhibited EGFR phosphorylation and the activation of downstream PI3K-Akt and MAPK pathways (Figure 7D and 7E).
The JM-PIP2 interaction regulates the EGFR activation, signaling and biological function. (A) Cellular location of mEos3.2-Grb2/SH2 (green) in COS-7 cells after different time of EGF treatment visualized by laser scanning confocal microscopy. Scale bar, 10 μm. (B) Two-color PALM and dSTORM images of exogenous mEos3.2-Grb2/SH2 (green) and endogenous EGFR labeled with Alexa647-EGF (red) in COS-7 cells with (SJ2) or without (Ctrl) SJ2 expression after 10 min of Alexa647-EGF treatment at 37 °C. Scale bar, 10 μm. (C) Percentage of colocalized mEos3.2-Grb2/SH2 clusters and EGFR clusters at the plasma membrane of COS-7-Ctrl or COS-7-SJ2 cells. Error bars denote SD. ***P < 0.001, two-tailed unpaired t-test. (D) Western blot analysis of EGFR activation and its downstream signal transduction in COS-7 cells stably expressing Lyn-FKBP12-T2A-mCitrine-FRB-SJ2 in response to EGF stimulation (50 ng/ml) before (Rapa−) or after (Rapa+) rapamycin treatment. (E) Western blot analysis of the effect of SJ2 constitutive expression on the activation of endogenous EGFR and its downstream signaling in COS-7 cells. (F, G) Western blot analyses of EGFR activation and its downstream signal transduction in 293T cells stably (F) or transiently (G) expressing exogenous wild-type (WT) or mutated full-length EGFR (EGFR-PM, EGFR-DM). (H) Cell proliferation of Ba/F3 cells stably expressing EGFR-L858R or EGFR-L858R with mutated JM region (L858R-PM or L858R-DM) in the absence of IL-3 was analyzed. Error bars denote SD.
We also studied EGFR signaling in 293T cells stably expressing either EGFR-WT, or EGFR-PM or EGFR-DM. We found that EGFR phosphorylation level dramatically decreased after the mutation of basic residues and this effect was even more severe when the whole positively charged stretch (residues 645–657) in JM region was deleted (Figure 7F). Interestingly, the phosphorylation level of Erk1/2 did not show obvious downregulation after 5 min of EGF stimulation, but decreased rapidly after 15 and 30 min of treatment (Figure 7F), indicating a temporal hysteresis of the downstream signaling. PI3K-Akt pathway was not dramatically affected by JM region mutations (Figure 7F). Similar results were observed in 293T cells transiently transfected with EGFR-WT, EGFR-PM or EGFR-DM (Figure 7G). To further test the biological consequence of disrupting EGFR clustering by the JM region mutation or deletion in EGFR, we took advantage of the Ba/F3 cell transformation assay, which is commonly used for testing the function of oncogenes including kinase domain-mutated EGFR gene and RET gene fusions35,36,37,38. The transformation of Ba/F3 cells results in the cell growth independent of interleukin-3 (IL-3)35,36,37,38. Consistently, the constitutively activated mutant of EGFR, named EGFR-L858R, was capable of transforming Ba/F3 cells to IL-3-independent cell growth whereas this growth was attenuated when the JM region was either mutated or deleted (L858R-PM and L858R-DM, Figure 7H). Together, these data demonstrated that the PIP2-dependent EGFR clustering played an essential role in EGFR signaling.
Discussion
In summary, we have uncovered the unique pattern of EGFR spatial distribution on the surface of living cells through super-resolution imaging: EGFR are found to form nanoclusters comprised of a variable number of proteins in the plasma membrane (Figure 8A). More importantly, there are much bigger and more EGFR clusters present in the plasma membrane of cancer cells compared to normal cells (Figure 1), suggesting that more EGFR clustering might lead to higher EGFR activity in cancer cells. Moreover, we find that either the depletion of PIP2 or the mutation of EGFR JM region results in the significant decrease of EGFR clustering. Since both modulations do not affect the total EGFR level on cell surface, these data highlight the importance of the ionic interaction between anionic PIP2 in plasma membrane and the JM region of EGFR in essentially regulating the unique distribution pattern of EGFR (Figure 8A and 8B). The EGFR clustering plays an important role in EGFR activation and its downstream signaling (Figure 8C) and thus might contribute to human tumor malignancy progression.
A schematic illustration of ionic protein-lipid interaction-mediated EGFR membrane clustering model. (A) On the cell surface, EGFR can form clusters comprised of 3–5 proteins, partially overlapping with the PIP2 clusters in the plasma membrane, or distributes sporadically. (B) The N-terminus of the JM region of EGFR binds to the anionic PIP2 clusters in the inner-leaflet of the plasma membrane through ionic interaction, which facilitates EGFR aggregation and cluster formation. (C) This special distribution pattern makes EGFR ready for a quick response to ligand stimulation and a strong activation of downstream signaling.
Our work, for the first time, depicts the EGFR membrane distributions in living cells at single-molecule level with a resolution up to ∼20 nm, and uncovers the underlying mechanism for receptor clustering. Besides its critical functions in many physiological processes such as ion channel activation, second messenger production and cytoskeletal attachment11, the anionic lipid PIP2, in our work, is given prominence for its role in regulating EGFR clustering via the interaction with the basic JM region of EGFR. This ionic protein-lipid interaction between EGFR and its “niche” regulates its distribution, and finally affects its activation and biological function. Currently, we still do not fully understand how the EGFR cluster formation is initiated though PIP2 seems to participate in this process. In consideration of spatially confined PIP2 clusters or pools on the plasma membrane39,40,41,42, it is possible that EGFR proteins might follow the track of PIP2 on membrane to aggregate and form clusters in PIP2-enriched region. Therefore, depletion of PIP2 can disassemble the EGFR clusters into relatively small clusters or even sporadic distribution (Figures 8A and 3C–3F). Nevertheless, it is also possible that EGFR clusters first and subsequently recruits PIP2 to stabilize the cluster structure. A better explanation may be that clustering of PIP2 and EGFR in the cell membrane is mutually supportive. Furthermore, our data show that about 40% of EGFR clusters are not colocalized with PIP2 clusters (Figure 2A). This indicates that other mechanisms could exist besides the JM region-PIP2 interaction, e.g., protein-protein interaction or cholesterol involvement in the regulation of EGFR clustering. Future work will provide more detailed mechanisms.
Large EGFR clusters containing a high amount of receptors should endow the cells with enhanced capability of EGFR signaling activation (Figure 7B and 7D-7G), because those receptors within the cluster are easier to aggregate and get activated after EGF stimulation, which might play an important role in tumor development. Previous studies have indicated that EGFR might exist as preformed dimers in the absence of ligands43,44,45. From our data analyses, we also find that EGFR form clusters without ligand engagement (Figures 1A and 3C) and the clusters contain from a few to a dozen of EGFR proteins (Figures 1F and 3F). Although we are not clear how these EGFR proteins exactly assemble inside the clusters, we reason that they might form dimers or tetramers.
Although non-small cell lung cancer (NSCLC) patients with specific EGFR mutations respond dramatically to gefitinib or erlotinib treatment, the emergence of newly acquired drug resistance greatly limits the drug effects46. Considering the inhibitory effect of JM region mutation and deletion on the constitutively active EGFR-L858R-promoted cell proliferation (Figure 7H), our work provides a hint that disrupting EGFR cluster formation in drug-resistant cancers would be a promising approach to fight with cancers.
Materials and Methods
Antibody or ligand labeling
Cetuximab (Merck) or EGF (Sigma-Aldrich) was labeled with Alexa647 (Invitrogen) in an appropriate concentration. The NHS group of the dyes can react with the amino group of the protein. 100 μg/ml Cetuximab or EGF was stained by 0.2 μl Alexa647 (1 mg/ml, dissolved in DMSO) in dark and vortexed for 2 h at room temperature. After reacting completely, the solution was filtered by gel filtration using Illustra NAP-5 columns (GE Healthcare) to remove excess dye. The Alexa647 labeling efficiency of either Cetuximab or EGF was determined to be 0.7–1 dye/protein by absorption spectroscopy assay.
Cell culture
HEK-293T cells were purchased from ATCC (American Type Culture Collection). COS-7 cells were generously provided by Dr Xinyuan Liu. Ba/F3 cells were generously provided by Dr Liang Chen. HEK-293T and COS-7 cells were cultured in Dulbecco's modified Eagle's medium (Hyclone, Thermo Scientific, USA) supplemented with 8% fetal bovine serum (FBS; Biochrom AG, Germany) and antibiotics (100 U/ml streptomycin and 100 μg/ml penicillin; Invitrogen, USA). Jurkat cells were cultured in RPMI-1640 medium (Hyclone, Thermo Scientific, USA) with 8% FBS and antibiotics. Ba/F3 cells were cultured in RPMI-1640 medium with 8% FBS, 10% WEHI conditional medium and antibiotics. The cells were maintained at 37 °C in a humidified atmosphere with 5% CO2 and passaged every 2 days.
Plasmids and reagents
Constructs (with the backbone of pHAGE-fEF1ap) expressing mCitrine-SJ2, PH-PLCδ-mEos3.2 (a gift from Prof Pingyong Xu and Prof Tao Xu), myristoyl JM-mCherry, myristoyl JM-PM-mCherry, Lyn-FKBP12-T2A-mCitrine-FRB-SJ2, KIR2DL3-3aa/25aa/50aa/JM/JM-PM-mTFP1 and constructs (with the backbone of pCDH-CMV-EF1-Puro) expressing EGFR-WT-mCherry, EGFR-PM-mCherry, EGFR-DM-mCherry, mEos3.2-Grb2/SH2, were used to package lentiviruses for establishing stable cell lines.
Western blot analyses were done with the following primary antibodies: anti-EGFR (Santa Cruz, sc-03), anti-p-EGFR (Y1173) (Cell Signaling, #4407), anti-Akt (Cell Signaling, #9272), anti-p-Akt (S473) (Cell Signaling, #4060), anti-Erk1/2 (Cell Signaling, #9102), anti-p-Erk1/2 (T202/Y204) (Cell Signaling, #9106), anti-actin (Sigma, A2228). Peroxidase-conjugated donkey anti-goat IgG, goat anti-rabbit IgG or goat anti-mouse IgG (Santa Cruz) was used as the secondary antibodies.
Lentiviral packaging and establishment of stable cell lines
Lentiviruses were produced by transfecting HEK293T cells with plasmids and the helper plasmids (pMD2.G and psPAX for pHAGE-fEF1ap; pCMV-Δ8.2 and pCMV-VSVG for pCDH-CMV-EF1-Puro). The cell supernatants were harvested 48 h after transfection and used for infection during 1 week. COS-7 cells, 293T cells and Jurkat cells were infected by corresponding lentiviruses for ∼24 h and then cultured with normal medium. The cells were sorted by FACS to acquire stable cell lines with persistent fluorescent protein expression.
Human specimen, primary cell isolation and pretreatment
This study was approved by the ethics review board at the Second Hospital of Jilin University and the clinical specimens including lung cancer and paired pathologically normal lung specimens were collected with written informed consents from all patients. Lung cancer cells and normal lung epithelial cells were freshly isolated from the lung cancer specimens and paired pathologically normal lungs and subjected to dSTORM analyses. Briefly, the human clinical samples were rinsed with 1× PBS for 3–5 times and then divided into several pieces with one for dSTORM imaging and one fixed in 4% PFA for immunohistochemistry and H&E analyses. For imaging, the lung cancer specimens were fully minced and homogenized in 1 ml ACL4 medium with 5% FBS47; the normal lung tissues were incubated with Collagenase III (Invitrogen, final concentration of 1 mg/ml) at 37 °C for 1-h shaking. These tissues in solution were then passed through a 60–80 μm2 filter, and the cell suspension was centrifuged at 1 500 rpm for 4 min and resuspended in 1× PBS with 3% bovine serum albumin (BSA) for labeling with FITC anti-human CD326 (EpCAM; Biolegend; diluted at 1:40 ratio by PBS) by shaking at 4 °C in dark for 20 min. The cells were then washed with PBS twice and incubated with Alexa647-conjugated Cetuximab or EGF at 4 °C in dark for 20 min. After washing with PBS, the pretreated cells were dropped onto precleaned glass slides and kept at 4 °C for adhesion. Before imaging, oxygen scavenging solution was replaced with an imaging buffer containing 50 mM Tris-HCl (pH 7.5), 10 mM NaCl, 10% (w/v) glucose, 0.5 mg/ml glucose oxidase (Sigma-Aldrich), 40 μg/ml catalase (Sigma-Aldrich) and 1% (v/v) β-mercaptoethanol48. For the fixed samples, H&E staining was performed for detection of histology; Immunostaining of phosphorylated EGFR (pY1173, diluted at 1:70 ratio by 3% BSA) was done as previously described49.
The stable cell lines were seeded onto the glass slides at an appropriate density and cultured overnight for adhesion. After the cell density reached ∼80%, the slides were picked up and blocked by 0.1% BSA, and then incubated with Alexa647-conjugated Cetuximab or EGF at 4 °C in dark for 10 min for imaging analyses. Three constructs expressing Lyn-FKBP12, mCitrine-FRB-SJ2 and PH-PLCδ-mCherry were transfected together into COS-7 cells by X-tremeGENE HP (Roche) reagent for establishing inducible PIP2 hydrolysis system. After labeling with Cetuximab or EGF, COS-7 cells were treated with 1 μM rapamycin and those with location change of mCitrine and mCherry were subjected to dSTORM analyses. 293T cells expressing comparable level of myristoyl JM/JM-PM-mCherry protein were imaged by laser scanning confocal microscopy (Zeiss confocal LSM710, Zeiss, Germany).
dSTORM, PALM imaging and data analyses
dSTORM or PALM imaging was performed on a Ti-E microscope with a 100 × 1.49 NA TIRF lens (Nikon, Japan) using a 405 nm laser (20 mw), 561 nm laser (200 mw) and 640 nm laser (100 mw). Fluorescence images were recorded at rates of 25 Hz in laser-excitation widefield. For single-color dSTORM imaging of EGFR, the sample stained with Alexa647-Cetuximab or Alexa647-EGF was excited by 640 nm laser power of 30 mW (at the sample) only. For two-color dSTORM and PALM imaging, a stable cell line expressing photoactivatable protein mEos3.2 was stained with Alexa647-Cetuximab or Alexa647-EGF. The sample was first excited by 640 nm laser power of 30 mW (at the sample) to obtain EGFR imaging. To record fluorescence from mEos3.2, periodical short pulses of 405 nm illumination were controlled by an acousto-optic tunable filter (AOTFnC-400.650, A-A Opto-Electronic, Orsay Cedex, France) and used for photoactivation. The 405 nm laser intensity was adjusted between 10–20 μW at the sample according to the protein expression levels. The 561 nm laser was adjusted to 20 mw and continuously applied to photoswitch and localization of mEos3.2 molecules. The system was equipped with one dichroic filter which reflects 405 nm, 561 nm and 647 nm, and widened by a telescope consisting of the achromatic doublet lenses before injection into the microscope. The fluorescence was filtered with emission filters and imaged on an EMCCD camera (Photometrics, Cascade II). We initially captured 20 000 frames of EGFR clusters in the COS-7 cell membrane, and reconstructed two super-resolution images from the first 5 000 frames and all the frames, respectively (Supplementary information, Figure S6). These two images had no significant differences in cluster morphology, and the number of localization spots was 43 627 from 5 000 frames and 44 329 from 20 000 frames. In the following imaging process, we recorded a time series of 5 000 frames per cell for the reconstruction of the super-resolution imaging, and the time is long enough to exhaust a great majority of the fluorescent molecules. Obtaining a single dSTORM or dual color super-resolution image usually took less than 8 min. During this short acquisition time, the z-drift was eliminated by the means of a focus lock, and 150-nm diameter fluorescent beads were embedded as a fiducial marker to correct the x-y drift of the sample. The optical registration for dual color imaging was characterized via TetraSpeck microspheres (Invitrogen), and the average deviation between Alexa647 and mEOS3.2 channels was 8–12 nm. To avoid any possible EGFR endocytosis induced by Alexa647-EGF labeling, all imaging processes were executed at low temperature (0–4 °C), whereas Cetuximab-labeled EGFR was imaged at room temperature.
For imaging process and data analyses, we used a public plug-in for Image J named QuickPALM and Matlab to analyze raw images. Image TIFF stacks were first preprocessed via background subtraction. For each frame, particles corresponding to single photoemission events were identified with a minimum SNR of 2–4. Then fluorescence peaks were identified in each frame and fitted a least-squares fit with an elliptical Gaussian function. Individual least-squares fit estimates were performed by a threshold of the peak height and the peak widths in the two lateral dimensions. After rejecting the poor fit and asymmetric PSFs, the centroid positions of peaks were determined by selected fit. STORM images were reconstructed using the precise localization data of single fluorescent molecules obtained from the fits.
To detect the resolution in our set-up instrument, single-molecule localization precision was measured50. Adequately diluted Alexa647-Cetuximab/EGF molecules (≈10 nM) were added onto an APTES-coverslip, incubated for 40 min and imaged. Repetitively switching fluorophores allowed us to determine the localization precision to be 28 nm for Alexa647-conjugated Cetuximab and 24 nm for Alexa647-conjugated EGF by measuring the full-width at half-maximum (FWHM) of the localization distribution of individual fluorophores (Supplementary information, Figure S7A–S7B). Similar localization precision (30 nm for Alexa647-conjugated Cetuximab and 27 nm for Alexa647-conjugated EGF) were observed when we performed the labeling of living COS-7 cells by adequately diluted Alexa647-Cetuximab/EGF molecules (≈10 nM) (Supplementary information, Figure S7C–S7D).
To analyze spatial EGFR clusters on the plasma membrane, we used Ripley's K function to analyze the characteristic parameters of the clusters. Supplementary information, Figure S8A showed a reconstructed dSTORM image of EGFR on the surface of a lung cancer cell. The examined region with an area of 4 × 4 μm2 in the reconstructed image was selected, which was boxed by a white rectangle. Then Ripley's K function was calculated to measure the deviation of the observed distribution from a random distribution as previously stated51,52,53. The average diameter of clusters in the chosen region was the value of rmax corresponding to the maximum of L(r) – r (Supplementary information, Figure S8B). The values of L(r) generated by each particle were used to produce a cluster map by interpolating a surface plot with L(r) as the z-axis (Supplementary information, Figure S8C). After that, a binary cluster map was generated through an appropriate L(r) threshold (Supplementary information, Figure S8D). Finally the information of clustering was extracted from the binary map, such as the number and the size of the clusters. All calculations and image processing were performed with Matlab.
The size (diameter) of EGFR clusters was extracted from the binary cluster map obtained through Ripley's K function analysis. Two-dimension diameters (the longest and the shortest diameter) of each cluster were measured, and the size of each cluster was the average of the two values. Eight regions from each cell were selected for analysis. The surface density of clusters was calculated via dividing the total number of clusters from every cell by the individual surface area of those cells. The percentage of EGFRs in clusters was calculated via dividing the number of EGFR proteins in clusters by total surface number of EGFR proteins. The number of EGFR in clusters was normalized by the number of bound single Cetuximab or EGF. The average number of localized spots in a single Alexa647-labeled Cetuximab and EGF was 28.2 ± 5.1 (SD) and 23.5 ± 4.8 (SD) (100 dyes analyzed), respectively. Using the statistical method and keeping the experimental conditions as same as possible, we could semiquantitatively estimate the fluorescence intensity unit (relative protein amount) of EGFR in clusters via dividing the total number of localized spots in the cluster by the average localized spot number of a single Cetuximab or EGF molecule on glass.
Mander's colocalization coefficients28 M1 and M2 were used to measure the degrees of spatial association between EGFR and mEos3.2-Grb2/SH2 clusters. The colocalization coefficients represent the amount of fluorescence of the colocalized area relative to the fluorescence of total area covered by each component. Since the areas of EGFR and mEos3.2-Grb2/SH2 clusters were usually different, M1 and M2 were not equal. The two kinds of clusters were defined as colocalization if they exhibited an M1 or M2 value > 0.33. Detail information could be found in23.
Preparation of bicelles
Bicelles (q = 0.8) were prepared by mixing appropriate amount of long-chain phospholipids POPS, POPC, DOPIP2 or DOPC (Avanti) powder with the short-chain phospholipid DHPC (Dihexanoyl-phosphatidylcholine) in 20 mM Tris-HCl, pH 7.4 at a mole ratio of 0.8:1. The mixture was subjected to five freeze-thaw cycles with pipetting and vortexing until the lipids were dissolved. In 50% POPS/POPC bicelles, the molar ratio between POPS and POPC is 1:1, while in 10% DOPIP2/DOPC bicelles, the molar ratio between DOPIP2 and DOPC is 1:9.
JM peptide expression, purification and labeling
JM peptide was expressed as a GST fusion protein in Escherichia coli, separated from GST tag by tobacco etch virus protease cleavage and purified by reverse-phase HPLC. For fluorescence polarization experiment, peptides were expressed with an additional N-terminal cysteine and labeled by an Alexa488-maleimide derivative (Invitrogen). For CD spectroscopy, the JM peptide was directly used for detection of its secondary structure.
Fluorescence polarization assay
To measure the binding kinetics of JM peptide to phospholipid-containing bicelles, an Alexa488-labeled JM peptide was incubated with 50% POPS/POPC bicelles (q = 0.8) or 10% DOPIP2/DOPC bicelles (q = 0.8) at a wide range of concentration (0, 1, 2.5, 5, 10, 25, 50, 100, 250 μM) in a 384-well plate for 20 min at 25 °C. The final concentration of peptide in reaction solution is 200 nM. The sample buffer was 20 mM Tris-HCl, pH 7.4, 150 mM NaCl. Fluorescence polarization values were measured using a Tecan Infinite M1000Pro Microplate Reader.
CD spectroscopy
Far ultraviolet (UV) CD spectra of JM peptide alone, JM peptide with POPC bicelles (q = 0.8), DOPC bicelles (q = 0.8), 50% POPS/POPC bicelles (q = 0.8) or 10% DOPIP2/DOPC bicelles (q = 0.8) in solution of 20 mM Tris-HCl, pH 7.4, 150 mM NaF were recorded on a JASCO J-715 spectropolarimeter at 25 °C. 20 M peptide and 0 or 1 mM mixture bicelles were present in the solution.
FRET measurements
Constructs were delivered into human Jurkat cells by lentiviral infection and pools of stable cells with similar mTFP1 fluorescence were isolated by FACS. Lyn-FKBP12-T2A-BFP-FRB-SJ2 plasmid was used for inducible PIP2 depletion. To induce SJ2 translocation and PIP2 depletion, 1 μM rapamycin was added into the dish, which was then incubated at room temperature for additional 5 min. After labeling the plasma membrane with 2.5 μg/ml octadecyl rhodamine B (R18) (Invitrogen, USA), the dish was then mounted onto a Leica TCS SP5 Confocal microscope (Leica, Germany) for imaging. FRET efficiency between the C-terminal mTFP1 domain and the membrane dye R18 was measured using a donor dequenching approach in which mTFP1 fluorescence was measured before and after photobleaching of the R18 acceptor. mTFP1 was excited with the Argon 458 nm laser line and visualized using detection window at 470–560 nm. R18 was excited with the Helium/Neon 561 nm laser line, visualized using detection window at 590–650 nm. The dequenching FRET efficiency was calculated as E = ((DQ-Q)/DQ) × 100%, where DQ and Q are dequenched and quenched donor fluorescence. Unpaired two-tailed Student's t-test was used for the statistical analysis of all FRET data.
Cell-based signaling analysis
Cells underwent serum starvation after reaching proper confluence. COS-7 cells were starved for 24 h, while 293T cells were starved for 12 h. Ligand stimulation was performed by using 50 ng/ml EGF at 37 °C afterwards. Besides stable cells, for 293T cells, 1.5 μg constructs expressing full-length EGFR-WT, PM or DM were transfected per 35-mm dish by calcium phosphate transfection assay when the cell confluence reaches 50%–60%. The cells were starved and stimulated with EGF as above. For inducible PIP2 hydrolysis system, after being starved for 24 h, COS-7-Lyn-FKBP12-T2A-mCitrine-FRB-SJ2 stable cells underwent 1 μM rapamycin treatment at 37 °C for 15 min and then 50 ng/ml EGF stimulation at 37 °C. The cells were washed with ice-cold 1× PBS after EGF stimulation and then scraped and lysed in sample buffer (0.125 M Tris-HCl, 3.1% SDS, 10% glycerol, 5% β-mercaptoethanol). The concentration of the proteins was measured by BCA Protein Assay Reagent (Beyotime). Appropriate micrograms of protein were separated on 7.5% or 10% SDS-polyacrylamide gels and electrophoretically transferred to PVDF membranes (PALL). After being blocked in 5% skim milk for 3 h, the membranes were incubated in sequence with primary antibodies diluted in 5% BSA and secondary antibodies diluted in TBS-T with 5% skim milk. The blot signals were developed by enhanced chemiluminescence.
Ba/F3 cell transformation assay
pCDH-CMV-EF1-Puro vector containing EGFR-L858R, L858R-PM or L858R-DM and the empty pCDH were used to package lentiviruses. Ba/F3 cells were infected with corresponding viruses for 48 h and then selected by 2 μg/ml puromycin for 2 days to establish stable cells. IL-3 was removed from Ba/F3 cells 48 h before seeding. IL-3-independent cell proliferation was analyzed by MTS assay (G3580, Promega, Wisconsin, USA) for 5 days according to the manufacturer's recommendation. A total of 1 500 cells were plated in each well of a 96-well plate with 200 μl of medium.
References
Hynes NE, Lane HA . ERBB receptors and cancer: the complexity of targeted inhibitors. Nat Rev Cancer 2005; 5:341–354.
Citri A, Yarden Y . EGF-ERBB signalling: towards the systems level. Nat Rev Mol Cell Biol 2006; 7:505–516.
Schlessinger J . Ligand-induced, receptor-mediated dimerization and activation of EGF receptor. Cell 2002; 110:669–672.
Jura N, Endres NF, Engel K, et al. Mechanism for activation of the EGF receptor catalytic domain by the juxtamembrane segment. Cell 2009; 137:1293–1307.
Endres NF, Das R, Smith AW, et al. Conformational coupling across the plasma membrane in activation of the EGF receptor. Cell 2013; 152:543–556.
Arkhipov A, Shan Y, Das R, et al. Architecture and membrane interactions of the EGF receptor. Cell 2013; 152:557–569.
Zhang X, Gureasko J, Shen K, Cole PA, Kuriyan J . An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell 2006; 125:1137–1149.
Hynes NE, MacDonald G . ErbB receptors and signaling pathways in cancer. Curr Opin Cell Biol 2009; 21:177–184.
Sharma SV, Bell DW, Settleman J, Haber DA . Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer 2007; 7:169–181.
Fadeel B, Xue D . The ins and outs of phospholipid asymmetry in the plasma membrane: roles in health and disease. Crit Rev Biochem Mol Biol 2009; 44:264–277.
McLaughlin S, Murray D . Plasma membrane phosphoinositide organization by protein electrostatics. Nature 2005; 438:605–611.
Li L, Shi X, Guo X, Li H, Xu C . Ionic protein–lipid interaction at the plasma membrane: what can the charge do? Trends Biochem Sci 2014; 39:130–140.
Xu C, Gagnon E, Call ME, et al. Regulation of T cell receptor activation by dynamic membrane binding of the CD3epsilon cytoplasmic tyrosine-based motif. Cell 2008; 135:702–713.
Shi X, Bi Y, Yang W, et al. Ca2+ regulates T-cell receptor activation by modulating the charge property of lipids. Nature 2013; 493:111–115.
van den Bogaart G, Meyenberg K, Risselada HJ, et al. Membrane protein sequestering by ionic protein-lipid interactions. Nature 2011; 479:552–555.
Hansen SB, Tao X, MacKinnon R . Structural basis of PIP2 activation of the classical inward rectifier K+ channel Kir2.2. Nature 2011; 477:495–498.
Heo WD, Inoue T, Park WS, et al. PI(3,4,5)P3 and PI(4,5)P2 lipids target proteins with polybasic clusters to the plasma membrane. Science 2006; 314:1458–1461.
Abulrob A, Lu Z, Baumann E, et al. Nanoscale imaging of epidermal growth factor receptor clustering: effects of inhibitors. J Biol Chem 2010; 285:3145–3156.
Rust MJ, Bates M, Zhuang X . Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat Methods 2006; 3:793–795.
Dani A, Huang B, Bergan J, Dulac C, Zhuang X . Superresolution imaging of chemical synapses in the brain. Neuron 2010; 68:843–856.
Heilemann M, van de Linde S, Schuttpelz M, et al. Subdiffraction-resolution fluorescence imaging with conventional fluorescent probes. Angew Chem Int Ed Engl 2008; 47:6172–6176.
Betzig E, Patterson GH, Sougrat R, et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science 2006; 313:1642–1645.
Balzar M, Winter MJ, de Boer CJ, Litvinov SV . The biology of the 17–1A antigen (Ep-CAM). J Mol Med (Berl) 1999; 77:699–712.
Winter MJ, Nagtegaal ID, van Krieken JH, Litvinov SV . The epithelial cell adhesion molecule (Ep-CAM) as a morphoregulatory molecule is a tool in surgical pathology. Am J Pathol 2003; 163:2139–2148.
Messersmith WA, Ahnen DJ . Targeting EGFR in colorectal cancer. N Engl J Med 2008; 359:1834–1836.
Hurley JH, Meyer T . Subcellular targeting by membrane lipids. Curr Opin Cell Biol 2001; 13:146–152.
Zhang M, Chang H, Zhang Y, et al. Rational design of true monomeric and bright photoactivatable fluorescent proteins. Nat Methods 2012; 9:727–729.
Manders EMM, Verbeek FJ, Aten JA . Measurement of colocalization of objects in dual-color confocal images. J Microsc (Oxf) 1993; 169:375–382.
Varnai P, Thyagarajan B, Rohacs T, Balla T . Rapidly inducible changes in phosphatidylinositol 4,5-bisphosphate levels influence multiple regulatory functions of the lipid in intact living cells. J Cell Biol 2006; 175:377–382.
McLaughlin S, Smith SO, Hayman MJ, Murray D . An electrostatic engine model for autoinhibition and activation of the epidermal growth factor receptor (EGFR/ErbB) family. J Gen Physiol 2005; 126:41–53.
Sengupta P, Bosis E, Nachliel E, et al. EGFR juxtamembrane domain, membranes, and calmodulin: kinetics of their interaction. Biophys J 2009; 96:4887–4895.
Lowenstein EJ, Daly RJ, Batzer AG, et al. The SH2 and SH3 domain-containing protein GRB2 links receptor tyrosine kinases to ras signaling. Cell 1992; 70:431–442.
Skolnik EY, Lee CH, Batzer A, et al. The SH2/SH3 domain-containing protein GRB2 interacts with tyrosine-phosphorylated IRS1 and Shc: implications for insulin control of ras signalling. EMBO J 1993; 12:1929–1936.
Saito Y, Furukawa T, Arano Y, Fujibayashi Y, Saga T . Basic study on SH2 domain of Grb2 as a molecular probe for detection of RTK activation. Int J Oncol 2010; 37:281–287.
Jiang J, Greulich H, Janne PA, Sellers WR, Meyerson M, Griffin JD . Epidermal growth factor-independent transformation of Ba/F3 cells with cancer-derived epidermal growth factor receptor mutants induces gefitinib-sensitive cell cycle progression. Cancer Res 2005; 65:8968–8974.
Yuza Y, Glatt KA, Jiang J, et al. Allele-dependent variation in the relative cellular potency of distinct EGFR inhibitors. Cancer Biol Ther 2007; 6:661–667.
Lipson D, Capelletti M, Yelensky R, et al. Identification of new ALK and RET gene fusions from colorectal and lung cancer biopsies. Nat Med 2012; 18:382–384.
Vaishnavi A, Capelletti M, Le AT, et al. Oncogenic and drug-sensitive NTRK1 rearrangements in lung cancer. Nat Med 2013; 19:1469–1472.
van Rheenen J, Achame EM, Janssen H, Calafat J, Jalink K . PIP2 signaling in lipid domains: a critical re-evaluation. EMBO J 2005; 24:1664–1673.
Ellenbroek WG, Wang YH, Christian DA, Discher DE, Janmey PA, Liu AJ . Divalent cation-dependent formation of electrostatic PIP2 clusters in lipid monolayers. Biophys J 2011; 101:2178–2184.
Janmey PA, Lindberg U . Cytoskeletal regulation: rich in lipids. Nat Rev Mol Cell Biol 2004; 5:658–666.
Golub T, Caroni P . PI(4,5)P2-dependent microdomain assemblies capture microtubules to promote and control leading edge motility. J Cell Biol 2005; 169:151–165.
Low-Nam ST, Lidke KA, Cutler PJ, et al. ErbB1 dimerization is promoted by domain co-confinement and stabilized by ligand binding. Nat Struct Mol Biol 2011; 18:1244–1249.
Chung I, Akita R, Vandlen R, Toomre D, Schlessinger J, Mellman I . Spatial control of EGF receptor activation by reversible dimerization on living cells. Nature 2010; 464:783–787.
Sako Y, Minoguchi S, Yanagida T . Single-molecule imaging of EGFR signalling on the surface of living cells. Nat Cell Biol 2000; 2:168–172.
Kobayashi S, Boggon TJ, Dayaram T, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med 2005; 352:786–792.
Zheng C, Sun YH, Ye XL, Chen HQ, Ji HB . Establishment and characterization of primary lung cancer cell lines from Chinese population. Acta Pharmacol Sin 2011; 32:385–392.
Zhuang X . Nano-imaging with Storm. Nat Photonics 2009; 3:365–367.
Ji H, Li D, Chen L, et al. The impact of human EGFR kinase domain mutations on lung tumorigenesis and in vivo sensitivity to EGFR-targeted therapies. Cancer Cell 2006; 9:485–495.
Wu J, Gao J, Qi M, et al. High-efficiency localization of Na(+)-K(+) ATPases on the cytoplasmic side by direct stochastic optical reconstruction microscopy. Nanoscale 2013; 5:11582–11586.
Ripley BD . Tests of randomness for spatial point patterns. J R Stat Soc Ser B Stat Methodol 1979; 41:368–374.
Owen DM, Rentero C, Rossy J, et al. PALM imaging and cluster analysis of protein heterogeneity at the cell surface. J Biophotonics 2010; 3:446–454.
Aaron JS, Carson BD, Timlin JA . Characterization of differential Toll-like receptor responses below the optical diffraction limit. Small 2012; 8:3041–3049.
Acknowledgements
We thank Drs Pingyong Xu, Tao Xu, Xinyuan Liu and Liang Chen for material contribution and Drs Bo Huang, Zhaocai Zhou, Yan Feng, Tian Xiao and Yijun Gao for helpful discussion. We thank Yujuan Jin, Xiangkun Han, Yuetong Wang, Qibiao Wu, Fei Li, Shun Yao, Rong Liu, Haijiao Xu, Yibing Bai, Yan Jiang, Wei Yang, Jun Guo and Huizhen Wang for their technical support. This work was supported by MOST (2012CB910800 to HJ and CX, 2011CB933600 to HW, 2010CB912102 to HJ, and 2011CB910901 to CX), NSFC (31370747 and 81325015 to HJ, 21373200 to HW, 31070738 to CX, and 31330082 to JJ), CAS (the Strategic Priority Research Program XDB08020100 to CX, the 100 Talent Program to HW and CX, the “Cross and cooperation in science and technology innovation team” program to HJ, HW, CX and XD), and Science and Technology Commission of Shanghai Municipality (12JC1409800 to HJ). We also gratefully acknowledge the support of SA-SIBS scholarship.
Author information
Authors and Affiliations
Corresponding authors
Additional information
( Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary information
Supplementary information, Figure S1
EGFR form clusters in the plasma membrane of lung cancer cells and normal lung epithelial cells. (PDF 991 kb)
Supplementary information, Figure S2
PIP2 regulates the EGFR clustering in the plasma membrane. (PDF 218 kb)
Supplementary information, Figure S3
Rapamycin treatment of COS-7 cells does not affect the surface EGFR clustering. (PDF 248 kb)
Supplementary information, Figure S4
The JM region of EGFR is important for EGFR clustering in the plasma membrane. (PDF 232 kb)
Supplementary information, Figure S5
PIP2 is important for binding of downstream adaptor to EGFR during EGFR signaling transduction. (PDF 262 kb)
Supplementary information, Figure S6
No significant difference is observed between the dSTORM images of EGFR clusters from two different imaging time. (PDF 48 kb)
Supplementary information, Figure S7
Quantitative analysis of the localization precision of single Alexa647 conjugated Cetuximab or EGF in vitro and on living COS-7 cell surface. (PDF 164 kb)
Supplementary information, Figure S8
Ripley's K function analysis of EGFR membrane clusters. (PDF 117 kb)
Supplementary information, Table S1
The relative expression level of EGFR on the cell surface of different groups (PDF 34 kb)
Rights and permissions
About this article
Cite this article
Wang, Y., Gao, J., Guo, X. et al. Regulation of EGFR nanocluster formation by ionic protein-lipid interaction. Cell Res 24, 959–976 (2014). https://doi.org/10.1038/cr.2014.89
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cr.2014.89
Keywords
This article is cited by
-
Mutant APC reshapes Wnt signaling plasma membrane nanodomains by altering cholesterol levels via oncogenic β-catenin
Nature Communications (2023)
-
Electron transfer in protein modifications: from detection to imaging
Science China Chemistry (2023)
-
Mechanistic insights into HuR inhibitor MS-444 arresting embryonic development revealed by low-input RNA-seq and STORM
Cell Biology and Toxicology (2022)
-
Lattice complex assembled by noncompetitive anti-EGFR antibodies regulates actin cytoskeletal reorganization
Cancer Cell International (2020)
-
Cholesterol transport through the peroxisome-ER membrane contacts tethered by PI(4,5)P2 and extended synaptotagmins
Science China Life Sciences (2019)