Abstract
Although particular chromosomal syndromes are phenotypically and clinically distinct, the majority of individuals with autosomal imbalance, such as aneuploidy, manifest mental retardation. A common abnormal phenotype of Down syndrome (DS), the most prevalent autosomal aneuploidy, shows a reduction in both the number and the density of neurons in the brain. As a DS model, we have recently created chimeric mice from ES cells containing a single human chromosome 21. The mice mimicked the characteristic phenotypic features of DS, and ES cells showed a higher incidence of apoptosis during early neuronal differentiation in vitro. In this study, we examined the induction of anomalous early neural development by aneuploidy in mouse ES cells by transferring various human chromosomes or additional mouse chromosomes. Results showed an elevated incidence of apoptosis in all autosome-aneuploid clones examined during early neuronal differentiation in vitro. Further, cDNA microarray analysis revealed a common cluster of down-regulated genes, of which eight known genes are related to cell proliferation, neurite outgrowth and differentiation. Importantly, targeting of these genes by siRNA knockdown in normal mouse ES cells led to enhanced apoptosis during early neuronal differentiation. These findings strongly suggest that autosomal imbalance is associated with general neuronal loss through a common molecular mechanism for apoptosis.
Similar content being viewed by others
Introduction
Aneuploidy is a condition in which an organism or cell possesses a higher or lower number of chromosomes than normal. Extra copies of large autosomal segments, as well as autosomal trisomies, are generally incompatible with human and mouse embryonic development. However, postnatal survival is possible in trisomies of human chromosome 21, 18 and 13 as semilethal conditions accompanied by severe developmental anomalies, known as Down (DS) and Edwards and Patau syndromes 1. The most frequent and thoroughly studied condition, trisomy of chromosome 21 (DS), is associated with approximately 80 clinical phenotypes, of which a variable subset is present in all DS individuals. Although high phenotypic variability is a characteristic feature of aneuploid syndromes, the underlying mechanism remains unclear. To analyze DS, we have previously generated mouse embryonic stem (ES) cell lines that contain a single human chromosome 21 2, 3. Chimeric mice derived from these cell lines showed a wide variety of phenotypic traits of DS, including behavioral impairment and cardiac anomalies 3. In addition, these ES cell lines showed an increased susceptibility to apoptosis at the neuronal stem cell differentiation stage in the presence of PA6 stromal cell-derived inducing activity (SDIA) 4.
In this study, hypothesizing that imbalance in gene dosage caused by aneuploidy plays an important role in enhanced apoptosis during early neural differentiation in vitro, we first constructed mouse ES cell clones with aneuploidy using two methods: one set of cell clones were constructed to include a single human chromosome 21, 11, or 6p using microcell-mediated chromosome transfer (MMCT) 5, 6, 7, 8, whereas the other contained mouse chromosome aneuploidies isolated by sub-cloning parental ES cell clones. We then investigated apoptosis in the two cell systems during neurogenesis in vitro. Differentiation of ES cells on the feeder layer of PA6 mouse stromal cells results in extremely efficient neuronal differentiation in vitro by SDIA 9.
Here, we investigated apoptosis during neuronal differentiation in two different chromosome-aberrant ES cell systems, which either contain an introduced human chromosome or show mouse chromosome aneuploidy. We showed that these chromosomal aberrations induce enhanced apoptosis in early neuronal differentiation, which are possibly associated with mental retardation. We then performed transcript analysis to reveal the underlying molecular mechanism that might be responsible for the enhanced apoptosis during early neurogenesis.
Results
Construction of mouse ES cells with chromosome abnormalities
We constructed various mouse ES cell clones with aneuploidy by two means. First, mouse ES cell clones with a single transferred human chromosome 21, 11 or 6p were constructed using the MMCT technique. A library of mouse A9 cells containing single human chromosomes was screened for cells carrying an independent and intact human chromosome 21, 11 or 6p. Microcells prepared from donor A9 cells were then fused with mouse ES cells (E14: 40, XY or TT2F: 39, XO), and positive hybrids were identified by antibiotic selection. The presence of a transferred human chromosome in each line was confirmed by polymerase chain reaction (PCR) and fluorescence in situ hybridization (FISH) analyses (Figure 1A). FISH analysis revealed the retention of a single and independent transferred human chromosome in each clone. Second, aneuploid mouse ES clones carrying extra mouse chromosomes 1, 8 and 17 were isolated by sub-cloning normal mouse TT2F (39, XO) and TT2 (40, XY), followed by confirmation by SKY analysis of ES cells (+mChr 1), (+mChr 8), (+mChr 8, 17), (+mChr 1,8) and (−mChr Y) (Figure 1B). All clones showed extra chromosome load in ES cells, but with different modal karyotypes (Table 1).
Chromosomal analysis of modal karyotypes of ES cells with various aneuploidies. (A) Transferred human chromosomes were detected by digoxigenin-labeled human Cot-1 DNA (red): TT2F (hChr 21)-10, TT2F (hChr 11) and TT2F (hChr 6p) contain human chromosome 21, 11 and 6p in parental TT2F, respectively, whereas E14 (hChr 11) contains human chromosome 11 in parental E14. The extra human chromosome was detected within the otherwise normal mouse karyotype of each ES cell. (B) SKY analysis revealed karyotypes of sub-cloned TT2F cell clones: TT2Fs (+mChr 1) shows a modal karyotype 40, XO +1, and TT2Fs (+mChr 8,17) a modal karyotype 41, XO +8, +17.
Neuronal differentiation of parental and chromosome-aberrant ES cell lines
We then assessed these ES cell lines for the induction of neuronal differentiation by SDIA. Expression of nestin, a neural marker, after neural induction results in neurogenesis, which closely resembles the in vivo development of neurons in the central nervous system 9, and the production of a full dorsal-ventral range of neuro-ectodermal derivatives by ES cell-derived neural precursors upon SDIA treatment 10. Differentiation proceeding from the default ectodermal to neuronal pathway was observed. Immunocytochemical analysis revealed successful differentiation of neural stem cells in all ES cell clones (Figure 2). Further, the frequency of neuronal differentiation in aneuploid ES clones was comparable to that of parental ES clones (data not shown).
Neural differentiation of ES cells by SDIA treatment (left panel). PA6-induced ES colonies stained with nestin, a neural precursor marker (right panel). Parental TT2F, TT2F (hChr 6p) and TT2F (+mChr 1) cultured for three days on PA6 are shown. In normal cultures, nestin was negative. Aneuploid and parental ES clones progressed to the neural stem cell differentiation stage. Each colony was grown from a single ES cell for three days on PA6 cells. Scale bar: 50 μm.
Neuronal differentiation-associated apoptosis
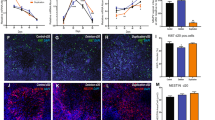
We also investigated apoptosis during neurogenesis in parental and aneuploid clones by TUNEL staining. On the third day (D3) of differentiation on PA6, higher proportions of TUNEL-positive cells were observed in aneuploid than in normal ES cell clones (Figure 3A). Flow cytometry analysis revealed 11.1%, 4.3% and 10.5% apoptotic cells in parental E14-1, TT2F and TT2 cells, respectively. Further, the number of developing neural stem cells undergoing apoptosis significantly increased (by one- to three-fold) in all autosomal aneuploid ES cell clones compared with controls (Figure 3B). Single and double trisomy resulted in a similar increase in neuronal apoptosis. In contrast, no enhancement of apoptosis was observed after deletion of the Y chromosome (39, XO in TT2s (−mChr Y)) (Figure 3B), suggesting that sex chromosome aneuploidy does not cause deterioration in neuronal apoptosis.
Detection of apoptotic cells in parental and chromosome-aberrant ES cells. (A) TUNEL assay in parental E14, E14 (hChr 11)-1, TT2F and TT2F (hChr 6p). Each clone was cultured for three days on either MEF or PA6. As part of the normal development in the vertebrate nervous system 36, ES cells with a normal karyotype showed a low level of apoptosis on PA6. (B) Quantitative analysis of apoptotic cells was performed by flow cytometry. Cells were subjected to staining with Annexin-V. On day 3 (D3), neural apoptosis in normal karyotypic and chromosome-aberrant clones during neurogenesis on PA6 was compared. Comparisons of normal and hChr.11 E14 clones, normal and hChr. 6P, 11, or 21 TT2F clones, as well as sub-cloned TT2F and TT2 clones are shown. The apoptotic rate of aneuploid clones was calculated on the basis of the apoptotic percentage of parental clones. Results show 11.1%, 4.3% and 10.5% apoptotic cells in parental E14-1, TT2F and TT2 cells, respectively. Data are represented as the mean ± SD; n=three independent experiments, *P < 0.05 compared with control using Student's t-test.
Next, we evaluated the enhanced apoptosis observed in aneuploid ES cell lines for its specific occurrence during neuronal differentiation. Because a highly efficient system of ES cell differentiation into a single lineage is not available except for neuronal differentiation, we performed in vivo analysis by subcutaneous injection of ES cells into nude mice, which would lead to teratoma formation with random differentiation. Transplanted aneuploid and euploid ES cells gave rise to typical teratomas after 5 weeks. Histological analyses showed that the tumors contained derivatives of all three embryonic layers including squamous epithelium, cartilage, skeletal muscle, osteoid tissue, glandular epithelium and ciliated epithelium (Figure 4A). Few apoptotic cells were noted in these differentiated areas or tissues. On the other hand, a good number of apoptotic cells were observed in the non-differentiated areas, consisting of cells with higher nuclear/cytoplasmic ratio. Importantly, no significant difference was noted in the rate of apoptotic cells between aneuploid and euploid ES cell lines (Figure 4A). Further, we evaluated apoptosis during embryoid body formation with random differentiation. Again, no significant difference was noted in the rate of apoptotic cells between aneuploid and euploid ES cell lines (Figure 4B). These results suggest that the enhanced apoptosis observed in aneuploid ES cells is specific to neuronal differentiation. Nevertheless, one cannot exclude the possibility that enhanced apoptosis may also occur in other cell lineages that were not reflected by the present assays.
Detection of apoptotic cells in parental and chromosome-aberrant ES cells. (A) Histological analysis of teratomas derived from TT2Fs (+mChr 1, 8) and TT2F. Apoptotic cells were indicated by arrows. (B) TUNEL assay in a random differentiation system during embryoid body formation derived from TT2Fs (+mChr 1), TT2Fs (+mChr 1, 8) and TT2F. The apoptotic rate of aneuploid clones was calculated on the basis of the apoptotic percentage of parental clone at D3, in which the rate of apoptotic cell was 3.3%. A similar result was obtained in TT2 cell clones (data not shown).
Common down-regulated genes in aneuploid ES clones at the neural stem cell stage
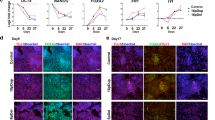
To determine the underlying molecular mechanism involved in defective neuronal differentiation and enhanced apoptosis in autosome-imbalanced neural stem cells, we surveyed the expression patterns of more than 15 012 genes (Supplementary information, Figure S1). Although no cluster specific to aneuploid ES cells was observed in undifferentiated state (Supplementary information, Figure S1), a differentially expressed cluster of 14 genes, which are commonly and significantly down-regulated from D2 to D6 of neuronal differentiation (Figure 5 and Table 2), was identified in both 40, XO (single trisomy of either mChr 1 or mChr 11) and 41, XO (double trisomy of either mChr 1+8 or mChr 8+17) clones (data available from CIBEX at http://cibex.nig.ac.jp/index.jsp). These genes locate to different chromosomes but not the trisomic chromosomes, suggesting that down-regulation of these genes is caused by the aneuploidy status. Of the 14 identified genes, 8 were already known, and quantitative real-time PCR analyses confirmed their down-regulation in aneuploid ES clones after induction of neuronal differentiation (Figure 6A), which was not observed under normal conditions nor after induction of random differentiation in the embryoid body (Figure 6B and Supplementary information, Figure S2).
Identification of a gene cluster in autosome-imbalanced TT2F clones during early neuronal differentiation. Results from clustering analysis (See Supplementary information, Figure S1) are shown. The dynamic changes in relative gene expression are plotted against the different days of neuronal differentiation. Significant expression suppression of the listed genes in 40, XO (lower left panel) and 41, XO (upper right panel) clones was observed compared with minor changes in expression profiles of 39, XO clones (upper panels).
Quantitative real-time PCR results for the listed genes. (A) After neural differentiation induction, expression levels of target genes are significantly down-regulated in chromosome-aberrant compared with normal mouse ES cells. Error bars indicate standard deviation obtained from three independent experiments. The opened and closed bars represent normal and chromosome-aberrant ES cells, respectively. (B) In a random differentiation system during embryoid body formation, no alteration in expression levels of target genes was observed between chromosome-aberrant and normal mouse ES cells.
Effect of RNAi of candidate genes on neurogenesis
We investigated the functional relationship between the eight identified genes and enhanced apoptosis during neuronal differentiation of ES cells, by performing RNAi experiments on parental TT2F clones. Transfection of each siRNA against these genes resulted in a dramatic and reproducible reduction in the levels of respective mRNAs compared with negative control (Figure 7A). Further, similar to aneuploid ES cell clones, defective neurogenesis and enhanced apoptosis were observed in six of the knockdowns (Figure 7B), suggesting that down-regulation of these genes is associated with enhanced apoptosis during neuronal differentiation. The precise mechanism by which the knockdown of these genes contributes to apoptosis, however, remains to be determined.
Dysregulated genes in early neuronal apoptosis. (A) siRNA knockdown of target genes. The white, black and gray bars represent non-treated, siRNA-transfected, and negative control siRNA-transfected TT2F, respectively. An approximately 0.3- to 0.6-fold knockdown in gene expression levels was observed in siRNA-transfected compared with normal TT2F. Error bars indicate standard deviation obtained from three independent experiments. (B) Target gene knockdown mimics defective neurogenesis and apoptosis in aneuploid clones. siRNAs targeting eight genes (Table 2) were transfected into normal TT2F, followed by neuronal differentiation induction by SDIA treatment. The graph compares the apoptosis rate between normal and siRNA-transfected TT2F during neurogenesis on D3. The white, closed and shaded bars represent non-treated, siRNA-transfected and negative control siRNA-transfected TT2F, respectively. Data are represented as mean ± SD; n=three independent experiments, *P<0.05 compared with control using Student's t-test.
Discussion
In this study, we present evidence suggesting that specific ES cell aneuploidy is not directly related to enhanced apoptosis at the early stage of neural stem cell differentiation in vitro. Owing to the involvement of a large number of genes in neuronal differentiation, an additional or missing chromosome in any chromosome pair carrying these genes may affect neuronal differentiation and result in apoptosis. Alternatively, an abnormal total number of chromosomes results in non-specific genetic imbalance, which may also induce enhanced apoptosis at the early stage of neural stem cell differentiation. This study suggests that autosomal imbalance triggers apoptosis during early neuronal differentiation by altering the expression of a set of genes, which were identified as a common cluster and reported to be involved in cell proliferation, as well as neurite outgrowth and differentiation 11, 12, 13, 14, 15, 16, 17, 18. We suggest that dysregulation of these genes leads to excess apoptosis during early neurogenesis by altering the cell cycle progression and differentiation program. Alternatively, these genes could be components of a pathway unidentified to date, the disturbance of which leads to apoptosis in early neuronal differentiation. In particular, genes identified as down-regulated in aneuploid ES clones are located at different chromosomal positions in the genome, showing that trisomy triggers the dysregulation of euploid genes 19, 20, 21, 22. Aneuploidy-associated down-regulation of the 14 common genes is possibly due to the effect of putative genes that monitor genomic imbalance 23, 24, 25. In addition, aneuploidy for various chromosomes should also modify the expression of different groups of target genes, thus producing unique phenotypes.
Apoptosis occurs naturally during normal development, and a number of studies have reported the importance of cell death during neural development in mammals 26, 27. Recently, several groups have shown the important role of the caspase family in apoptosis associated with neural development. In particular, studies showed that caspase 3 is crucial during this period and under pathological conditions such as cerebral ischemia 28, 29, 30. A few studies reported the involvement of Sonic hedgehog (Shh) in the ongoing neurogenesis in postnatal rodent brains 31, 32, 33. Further, a study in Ts65Dn mice showed a substantially reduced mitogenic response to Hedgehog signaling 34. These molecular mechanisms are possibly linked to the enhanced apoptosis induced by aneuploidy during early neurogenesis. It would also be important to understand the functional roles of the identified cluster of down-regulated genes, for example, by further in-depth analysis in knockout mice. Further studies are also required to better understand whether the enhanced apoptosis we have observed is caused by an increase in naturally occurring apoptosis or by an additional pathway not observed during normal neural development.
In summary, our results show that autosomal imbalance commonly enhances apoptosis during early neuronal differentiation of mouse ES cells, and alters the expression of a common cluster of genes. In addition, this effect is possibly limited to autosomes as no enhancement of apoptosis was observed in TT2s cells with a missing Y chromosome. These findings are consistent with the suggested relationship between dysregulated developmental apoptosis in early neurogenesis and mental retardation in patients with autosomal aneuploidy.
Materials and Methods
ES cell culture and construction of chromosome-aberrant ES cell clones
Three mouse ES cell lines were used: E14, TT2F and TT2. Parental and chromosome-aberrant ES cell clones were maintained in an undifferentiated state on a mitomycin C-treated mouse embryonic fibroblast (MEF) feeder. The ES culture medium was composed of high glucose Dulbecco's modified Eagle's medium (DMEM) with 15% FBS, 0.1 mM β-mercaptoethanol, 1 mM sodium pyruvate, 1× nonessential amino acids, 2 mM glutamine, and 1 000 U/ml recombinant human leukemia inhibitory factor (LIF; Gibco).
Chromosome-aberrant ES cell clones contained human chromosome 21, 11 or 6p and were transferred using MMCT, as described previously 3, 7. Human chromosomes 21, 11 and 6p were transferred into TT2F, whereas human chromosome 11 was transferred into E14. Chromosome-aberrant ES cell clones with mouse chromosome aneuploidy were constructed by sub-cloning normal TT2F cells (39, XO) derived from TT2 cells (40, XY) 35. Sub-cloned TT2F and TT2 cells were named “TT2Fs” and “TT2s”, respectively. The passage number of ES clones used was unchanged throughout the experiment.
Fluorescence in situ hybridization
ES cells were treated with 0.075 M KCl and fixed in a 3:1 solution of methanol and acetic acid. Human chromosome painting was performed on fixed metaphase spreads using digoxigenin-labeled human specific Cot-1 DNA (Gibco) as a probe. The probe was detected with anti-digoxigenin-rhodamine (Boehringer) and chromosomes were counterstained with DAPI (Sigma).
Spectral karyotyping (SKY)
For SKY analysis, slides were processed using the ASI spectral mouse karyotyping kit (Applied Spectral Imaging (ASI), Carlsbad, CA) according to the manufacturer's instructions. The slides were mounted in antifade reagent (ASI) and imaged using the SKY system with SkyView v. 1.6 and Spectral Imaging v. 3.0 (ASI).
Induction of ES cell differentiation to neuron, teratoma formation and embryoid body formation
Differentiation of ES cells was induced using the SDIA method as reported previously 4. Briefly, PA6 cells were cultured to confluence in α-MEM (SIGMA) supplemented with 10% FBS and 2 mM L-glutamine, and then used as feeder layers. To remove the MEF feeder cells, ES cells were dissociated and plated on a gelatin-coated dish in ES medium for more than 30 min and 5 × 104 cells were plated on PA6 cells in 10-cm dishes. The differentiation medium was composed of G-MEM (Gibco) supplemented with 10% KSR (Gibco), 1 mM sodium pyruvate, 0.1 mM non-essential amino acids, 0.1 mM 2-mercaptoethanol and 2 mM L-glutamine. ES cells were cultured on PA6 feeder cells for three days, washed five times in PBS, dissociated using cell dissociation solution (SIGMA) and passed through a cell strainer (Falcon) to remove any cell clumps. The resulting cell suspensions were used for flow cytometry. Formation of teratoma and embryoid body was performed as reported previously 4, 37.
Fluorescence immunocytochemical analysis for nestin
Anti-nestin (Pharmingen) antibodies were used for immunocytochemical analysis. ES cells differentiated in chamber slides (NUNC) for three days were fixed in 4% paraformamide, permeabilized in 0.2% Triton X-100 (Sigma), and treated in 1× Block-ace solution (Snow Brand Milk Products). Cy-3-conjugated secondary antibodies (Chemicon) were used and detected using a laser confocal microscope (Bio-Rad).
TUNEL staining
To detect DNA fragmentation using in vitro nick end labeling (TUNEL), differentiated ES cells grown in chamber slides for three days were stained using the APOPtag red in situ apoptosis detection kit (Intergene) and DNA was counter-stained with DAPI.
Flow cytometry analysis
Annexin-V was used as marker for apoptosis. Following dissociation of cells, parental and aneuploid ES cell clones were subjected to flow cytometry analysis using FITC-conjugated Annexin-V kit (MBL) according to the manufacturer's protocol. All samples were analyzed using an EPICS ALTRA flow cytometer (Beckman Coulter).
Microarray
Total RNA was isolated from ES cells after D0, D2, D3, D4 and D6 of neuronal differentiation by SDIA treatment, reversed transcribed into cDNA using a T7 promoter-dT(24) primer, which was then linearly amplified using the MEGA-script™ Kit (Ambion) according to the manufacturer's protocol. A total of 2 μg target amplified RNA was subjected to a reverse transcription reaction with random hexamer primers and labeled with either Cy3 (D0 as reference) or Cy5 (D2, D3, D4 or D6) dCTP/dUTP (Amersham, Buskinghamshire, UK). Labeled cDNA was hybridized to a microarray (NIG-NIA 15K Mouse Array Sets) adapted from the NIA 15K mouse cDNA clone set containing 15 012 unique validated mouse probes and 976 control spots. Hybridized arrays were scanned using a ScanArray BS2000 (GSi Lumonica, Wilmington, MA) and images analyzed using the QuantArray software (GSi Lumonica) as reported previously 36.
Quantitative real-time RT-PCR
Total RNA was isolated from ES cells after D3 of neuronal differentiation by SDIA treatment using an RNeasy Plus Mini Kit (Qiagen) and converted to cDNA using an oligo(dT)15 primer (Roche) and SuperScript III RT (Invitrogen). Real-time PCR was carried out using the ABI Prism 7900 Sequence Detection System and 2× converted SYBR Green PCR Master Mix reagent (PE Applied Biosystems) in 10-μl volume. b-actin was used as internal standard. The following primers were used: b-actin, 5′-GGG GTG TTG AAG GTC TCA AA-3′ and 5′-TGT TAC CAA CTG GGA CGA CA-3′; Pctk1, 5′-TTG TCT GAG ATT GGC TTT GG-3′ and 5′-TTC AAA GAC AAG TGT GAG GG-3′; Atg7, 5′-TAG CCC AGA TTG TCC TAA AG-3′ and 5′-CAA ATT CAT ACA GAG GCT TGC-3′; Lcp1, 5′-CAG GAA AAC TTG AAC TTG GC-3′ and 5′-GCT GAA GTT GGT GAT TTT GG-3′; Pdhx, 5′-CAA GTT GGA GGT GGT TTC ACA GT-3′ and 5′-TAG GAG ACA GTG ATG GCA TTA GGA-3′; Mdm2, 5′-ATG GCA AAA AGC TGA CAG AG-3′ and 5′-TCA TTT GGA TTG GCT GTC TG-3′; Zfp91, 5′-GTG GCA CAC AAA GCA AAA AG-3′ and 5′-ATA TCC GAG TTC ATG ACC AG-3′; Pax6, 5′-TTA CCC AAG AGC AGA TTG AG-3′ and 5′-TAG GAA TGT GAC TAG GAG TG-3′; Dnmt1, 5′-GCA AAA CAA TCC TGA TGC TG-3′ and 5′-ACT TCT CAA TCT GCT CTG AG-3′.
siRNA transfection
The Stealth Select RNAi oligonucleotides for the targeted genes were synthesized by Invitrogen. BLOCK-iT RNAi Designer (Invitrogen), a web-based program for designing siRNA targets, was used to select siRNA for Pctk1, Atg7, Lcp1, Pdhx, Mdm2, Zfp91, Pax6 and Dnmt1. The Stealth Select RNAi negative control Duplex was used as a control oligonucleotide. On D0 of neuronal differentiation, transfections were performed on PA6 stromal cells using Lipofectamine 2000 (Invitrogen) following the manufacturer's protocol.
( Supplementary information is linked to the online version of the paper on the Cell Research website.)
References
Pearson PL . In: Brenner S, Miller JH, eds. Encyclopedia of Genetics. Vol. 4. London: Academic Press, 2001:2056–2058.
Kazuki Y, Shinohara T, Tomizuka K, et al. Germline transmission of a transferred human chromosome 21 fragment in transchromosomal mice. J Hum Genet 2001; 46:600–603.
Shinohara T, Tomizuka K, Miyabara S, et al. Mice containing a human chromosome 21 model behavioral impairment and cardiac anomalies of Down's syndrome. Hum Mol Genet 2001; 10:1163–1175.
Kadota M, Shirayoshi Y, Oshimura M . Elevated apoptosis in pre-mature neurons differentiated from mouse ES cells containing a single human chromosome 21. Biochem Biophys Res Commun 2002; 299:599–605.
Fournier RE, Ruddle FH . Microcell-mediated transfer of murine chromosomes into mouse, Chinese hamster, and human somatic cells. Proc Natl Acad Sci USA 1977; 74:319–323.
Koi M, Shimizu M, Morita H, Yamada H, Oshimura M . Construction of mouse A9 clones containing a single human chromosome tagged with neomycin-resistance gene via microcell fusion. Jpn J Cancer Res 1989; 80:413–418.
Tomizuka K, Yoshida, H, Uejima H, et al. Functional expression and germline transmission of a human chromosome fragment in chimaeric mice. Nat Genet 1997; 16:133–143.
Kugoh H, Mitsuya K, Meguro M, Shigenami K, Schulz TC, Oshimura M . Mouse A9 cells containing single human chromosomes for analysis of genomic imprinting. DNA Res 1999; 6:165–172.
Kawasaki H, Mizuseki K, Nishikawa S, et al. Induction of midbrain dopaminergic neurons from ES cells by stromal cell-derived inducing activity. Neuron 2000; 28:31–40.
Mizuseki K, Sakamoto T, Watanabe K, et al. Generation of neural crest-derived peripheral neurons and floor plate cells from mouse and primate embryonic stem cells. Proc Natl Acad Sci USA 2003; 100:5828–5833.
Unoki M, Okutsu J, Nakamura Y . Identification of a novel human gene, ZFP91, involved in acute myelogenous leukemia. Int J Oncol 2003; 22:1217–1223.
Robinson BH, MacKay N, Petrova-Benedict R, Ozalp I, Coskun T, Stacpoole PW . Defects in the E2 lipoyl transacetylase and the X-lipoyl containing component of the pyruvate dehydrogenase complex in patients with lactic acidemia. J Clin Invest 1990; 85:1821–1824.
Matsudaira P . Modular organization of actin crosslinking proteins. Trends Biochem Sci 1991; 16:87–92.
Tucker KL, Talbot D, Lee MA, Leonhardt H, Jaenisch R . Complementation of methylation deficiency in embryonic stem cells by a DNA methyltransferase minigene. Proc Natl Acad Sci USA 1996; 93:12920–12925.
Mizushima N, Noda T, Yoshimori T, et al. A protein conjugation system essential for autophagy. Nature 1998; 395:395–398.
Besset V, Rhee K, Wolgemuth DJ . The cellular distribution and kinase activity of the Cdk family member Pctaire1 in the adult mouse brain and testis suggest functions in differentiation. Cell Growth Differ 1999; 10:173–181.
Hack MA, Sugimori M, Lundberg C, Nakafuku M, Gotz M . Regionalization and fate specification in neurospheres: the role of Olig2 and Pax6. Mol Cell Neurosci 2004; 25:664–678.
Hipfner DR, Cohen SM . Connecting proliferation and apoptosis in development and disease. Nat Rev Mol Cell Biol 2004; 5:805–815.
Vacik T, Ort M, Gregorova S, et al. Segmental trisomy of chromosome 17: a mouse model of human aneuploidy syndromes. Proc Natl Acad Sci USA 2005; 102:4500–4505.
Mao R, Zielke CL, Zielke HR, Pevsner J . Global up-regulation of chromosome 21 gene expression in the developing Down syndrome brain. Genomics 2003; 81:457–467.
Lyle R, Gehrig C, Neergaard-Henrichsen C, Deutsch S, Antonarakis SE . Gene expression from the aneuploid chromosome in a trisomy mouse model of down syndrome. Genome Res 2004; 14:1268–1274.
FitzPatrick DR . Transcriptional consequences of autosomal trisomy: primary gene dosage with complex downstream effects. Trends Genet 2005; 21:249–253.
Devlin RH, Holm DG, Grigliatti TA . The influence of whole-arm trisomy on gene expression in Drosophila. Genetics 1988; 118:87–101.
Guo M, Birchler JA . Trans-acting dosage effects on the expression of model gene systems in maize aneuploids. Science 1994; 266:1999–2002.
Niehrs C, Pollet N . Synexpression groups in eukaryotes. Nature 1999; 402:483–487.
Oppenheim RW . Cell death during development of the nervous system. Annu Rev Neurosci 1991; 14:453–501.
Thomaidou D, Mione MC, Cavanagh JF, Parnavelas JG . Apoptosis and its relation to the cell cycle in the developing cerebral cortex. J Neurosci 1997; 17:1075–1085.
Nicholson DW . Caspase structure, proteolytic substrates, and function during apoptotic cell death. Cell Death Differ 1999; 6:1028–1042.
Porter AG, Janicke RU . Emerging roles of caspase-3 in apoptosis. Cell Death Differ 1999; 6:99–104.
Zheng TS, Hunot S, Kuida K, Flavell RA . Caspase knockouts: matters of life and death. Cell Death Differ 1999; 6:1043–1053.
Lai K, Kaspar BK, Gage FH, Schaffer DV . Sonic hedgehog regulates adult neural progenitor proliferation in vitro and in vivo. Nat Neurosci 2003; 6:21–27.
Machold R, Hayashi S, Rutlin M, et al. Sonic hedgehog is required for progenitor cell maintenance in telencephalic stem cell niches. Neuron 2003; 39:937–950.
Ahn S, Joyner AL . In vivo analysis of quiescent adult neural stem cells responding to Sonic hedgehog. Nature 2005; 437:894–897.
Roper RJ, Baxter LL, Saran NG, Klinedinst DK, Beachy PA, Reeves RH . Defective cerebellar response to mitogenic Hedgehog signaling in Down [corrected] syndrome mice. Proc Natl Acad Sci USA 2006; 103:1452–1456.
Uchida M, Tokunaga T, Aizawa S, Niwa K, Imai H . Isolation and nature of mouse embryonic stem cell line efficiently producing female germ-line chimeras. Anim Sci Technol (Jpn) 1995; 66:361–367.
Wang CC, Kadota M, Nishigaki R, et al. Molecular hierarchy in neurons differentiated from mouse ES cells containing a single human chromosome 21. Biochem Biophys Res Commun 2004; 314:335–350.
Kazuki Y, Hoshiya H, Kai Y, et al. Correction of a genetic defect in multipotent germline stem cells using a human artificial chromosome. Gene Ther 2008; 15:617–624.
Acknowledgements
We thank Naoyo Kajitani, Toko Yoshino, Mitsutaka Kadota, Hidetoshi Yamada and Mitsuhiko Osaki (Tottori University) for their technical assistance. This study was supported in part by the 21st Century COE program: The Research Core for Chromosome Engineering Technology from the Ministry of Education, Culture, Sports, Science and Technology of Japan, a Grant-in-Aid for Scientific Research in Priority Areas from the Ministry of Education, Culture, Sports, Science and Technology of Japan (No. 18012033), as well as the Li Ka Shing Institute of Health Sciences and Direct Grants, Faculty of Medicine, The Chinese University of Hong Kong, Hong Kong SAR, China. CCW, YK and MT are Research Fellows of the Japan Society for the Promotion of Science. TW was supported by a Grant for Scientific Research in Priority Areas (15080211) and a project for the realization of regenerative medicine (research towards technical developments in stem cell manipulation) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary information, Figure S1
High-stringency self-organizing maps (SOM) hierarchical clustering of biological replicated gene expression profiles. (PDF 234 kb)
Supplementary information, Figure S2
Relative expression levels of the listed genes in normal cultures. (PDF 103 kb)
Rights and permissions
About this article
Cite this article
Kai, Y., Wang, C., Kishigami, S. et al. Enhanced apoptosis during early neuronal differentiation in mouse ES cells with autosomal imbalance. Cell Res 19, 247–258 (2009). https://doi.org/10.1038/cr.2008.305
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cr.2008.305