Abstract
Homozygous human embryonic stem cells (hESCs) are thought to be better cell sources for hESC banking because their human leukocyte antigen (HLA) haplotype would strongly increase the degree of matching for certain populations with relatively smaller cohorts of cell lines. Homozygous hESCs can be generated from parthenogenetic embryos, but only heterozygous hESCs have been established using the current strategy to artificially activate the oocyte without second polar body extrusion. Here we report the first successful derivation of a human homozygous ESC line (chHES-32) from a one-pronuclear oocyte following routine in vitro fertilization treatment. chHES-32 cells express common markers and genes with normal hESCs. They have been propagated in an undifferentiated state for more than a year (>P50) and have maintained a stable karyotype of 46, XX. When differentiated in vivo and in vitro, chHES-32 cells can form derivatives from all three embryonic germ layers. The almost undetectable expression of five paternally expressed imprinted genes and their HLA genotype identical to the oocyte donor indicated their parthenogenetic origin. Using genome-wide single-nucleotide polymorphism analysis and DNA fingerprinting, the homozygosity of chHES-32 cells was further confirmed. The results indicated that 'unwanted' one-pronuclear oocytes might be a potential source for human homozygous and parthenogenetic ESCs, and suggested an alternative strategy for obtaining homozygous hESC lines from parthenogenetic haploid oocytes.
Similar content being viewed by others
Introduction
Human embryonic stem cells (hESCs) are derived from the inner cell mass (ICM) of blastocysts and have the ability to differentiate into the three germ layers of our body 1, 2, which makes them one of the most promising seed cells for future regenerative medicine. One of the main hurdles to their clinical application is that transplantation of their derivatives might lead to allograft rejection 3, 4. Several strategies have been proposed to overcome this obstacle 5, including one relatively practical strategy to establish a hESC bank that contains sufficient cell lines with diverse human leukocyte antigen (HLA) genotypes 6, 7 to serve most of the patients in a region. But this strategy needs hundreds of human embryos and might evoke ethical concerns.
Collecting homozygous hESC lines that have common HLA genotypes significantly reduces the number of cell lines required for the repository and maintains a similar matched rate 6, 7. In this case, tissues differentiated from homozygous hESCs express only one set of histocompatibility antigens and thus are more readily matched to patients with less risk of immunological rejection. In addition, the method for deriving homozygous hESCs is thought to be less ethically controversial. Homozygous hESCs are commonly derived from parthenogenesis, by which an oocyte is activated to develop without fusing with sperm. In mammals, most parthenogenetic embryos die shortly after implantation owing to paternal imprinting defects 8; therefore, it is believed that no 'viable' embryos are created or destroyed when deriving hESC lines by this means.
In many species, including mouse, bovine, rabbit and nonhuman primates, ESCs have been successfully established from parthenogenetically activated oocytes [reviewed in 9]. Several previous studies have reported the successful activation of human oocytes and the formation of parthenogenetic blastocysts thereafter 10, 11, 12, but the successful derivation of parthenogenetic hESCs (pESCs) was reported only recently 13, owing to the inaccessibility of human oocytes and the existing ethical concerns about using viable fresh human oocytes in the process 14.
In humans, spontaneous parthenoactivation of oocytes has been observed in vivo and is related to the formation of benign ovararian teratomas 15, 16, 17. With the widespread introduction of assisted reproductive technology (ART), human oocytes can be manipulated and fertilized in vitro. It is suggested that suboptimal conditions 18, intracytoplasmic sperm injections 19 and cryopreservation 20 might parthenoactivate human oocytes. In most cases, spontaneous parthenogenesis is designated by one-pronucleus formation, and the resulting embryos should never be transferred back into the patients. One-pronuclear oocytes are commonly seen following ART treatment and previous studies have suggested that a small part of these oocytes are of uniparental origin 19, 21, 22, 23; therefore, these 'unwanted' oocytes could be used for the isolation of homozygous human pESCs rather than wasting fresh human oocytes for deliberate parthenogenetic activation.
In this report, one homozygous human pESC line (chHES-32) was successfully isolated from a one-pronuclear oocyte following in vitro fertilization (IVF) treatment. Traditional ESC features and homozygous state were further characterized.
Materials and Methods
Derivation and growth of chHES-32 cells
The experiment was approved by the ethical committee of the CITIC-Xiangya Reproductive and Genetic Hospital. A one-pronuclear oocyte was used for stem cell derivation, with informed consent from a patient undergoing IVF treatment. The oocyte was cultured in a G-series sequential medium (Vitrolife, Gothenburg, Sweden) to the blastocyst stage at 37 °C in 5% CO2, 5% O2 and 90% N2 with 95% humidity. The blastocyst was directly plated on mitotically inactivated human embryonic fibroblasts (HEFs), which were cultured in our laboratory from an aborted fetus of 2-3 months of age, as previously described 24. The cells in the first several passages were cultured in DFSR medium containing DMEM/F12 supplemented with 15% knockout serum replacement, 2 mM nonessential amino acids, 2 mM L-glutamine, 0.1 mM β-mercaptoethanol and 50 ng/ml of basic fibroblast growth factor (bFGF) (all from Invitrogen, Carlsbad, CA). Cells were routinely passaged every 7 days by mechanical cutting. Once established, cells were transferred back to DFSR medium containing 4 ng/ml bFGF for long-term propagation.
In vivo and in vitro differentiation
To carry out an in vivo differentiation assay, 0.5-1 × 106 chHES-32 cells were injected intramuscularly into the rear leg of 6-8-week-old mice with severe combined immunodeficiency disease (SCID). After 8-10 weeks, mice were killed and the xenografts were removed and processed for histological analysis by hematoxylin and eosin (H&E) staining.
To carry out an in vitro differentiation assay, chHES-32 colonies cultured on a feeder layer were mechanically cut into small clumps and detached to grow as aggregates in suspension for 4 days in order to form embryoid bodies in DFSR medium without bFGF. The embryoid bodies were then transferred to a gelatin-coated six-well plate for adherent culture for 2 weeks in the same medium, followed by immunocytochemical analysis, or cells were collected to detect imprinted gene expression as described below.
Karyotype analysis
chHES-32 cells were treated with 100 ng/ml demecolcine (Sigma, St Louis, MO) for 4 h and dissociated with 0.05% trypsin/EDTA (Invitrogen) into single cells for standard G-banding analysis. Usually more than 20 metaphases were analyzed for every sample.
Telomerase activity assay
The telomerase activity of chHES-32 cells was measured using the TRAPeze Telomerase Detection Kit (Chemicon, Temecula, USA), according to the manufacturer's instructions.
DNA fingerprinting analysis
Total genomic DNA was extracted from differentiated chHES-32 cells using the DNeasy Tissue Kit (Qiagen, Hilden, Germany), according to the protocol. Fifteen short tandem repeat (STR) loci and Amelogenin were then amplified using the AmpFlSTR® Identifiler® PCR Amplification Kit (Applied Biosystems, CA, USA), according to the manufacturer's instructions, and detected using the 3100 Genetic Analyzer (Applied Biosystems).
HLA typing analysis
To carry out HLA typing analysis, chHES-32 cells were transferred to gelatin-plated dishes for further culture to exclude the possibility of contamination with HEFs. Approximately 1 × 106 cells were collected for HLA typing. The HLA typing was carried out using the PEL-FREEZ SSP Kit designed for Chinese populations (Dynal Biotech, WI, USA).
Single-nucleotide polymorphism (SNP) analysis
Total genomic DNA was extracted from chHES-32 cells using the DNeasy Tissue Kit (Qiagen) and SNP analysis was performed by the National Engineering Research Center for Beijing Biochip Technology using the GeneChip Human Mapping Nsp Sty Array kit (Affymetrix, Santa Clara, USA). The SNP allelotypes were calculated from scanned data.
Immunocytochemistry
To detect specific hESC markers, chHES-32 cells were fixed in 4% paraformaldehyde in PBS and permeabilized with 0.1% TritonX-100 for 10 min at room temperature. After blocking in 4% goat serum in PBS, the cells were incubated overnight at 4 °C with the following primary antibodies: mouse anti-stage-specific embryonic antigen 1 (SSEA-1) and mouse anti-stage-specific embryonic antigen 4 (SSEA-4) (R&D systems, Minneapolis, USA; 1:50); mouse anti-TRA-1-60 and mouse anti-TRA-1-81 (Chemicon; 1:50); and rat anti-stage-specific embryonic antigen 3 (SSEA-3) and mouse anti-octamer-binding transcription factor (OCT)-3/4 (Santa Cruz Biotechnology, Santa Cruz, USA; 1:50). Localization of antigens was visualized using FITC-conjugated second antibodies (Sigma; 1:400).
Alkaline phosphatase activity was detected using the Fast Red Substrate Pack (Zymed Laboratories, South San Francisco, USA), according to the manufacturer's protocol.
RT-PCR
Total RNA was isolated with TRIzol (Invitrogen) and cDNA was synthesized using 1 mg of total RNA in a 20-μl reaction using the RevertAidTM First Strand cDNA Synthesis Kit (Fermentas Life Sciences, Ontario, USA), according to the manufacturer's instructions. The RT-PCR reaction was carried out under the following conditions: 1.5 min at 95 °C; 30 cycles of 30 s at 95 °C, 30 s at 54–64 °C, 30 s at 72 °C and a 7 min extension at 72 °C. PCR products were separated onto a 1.5% agarose gel, stained with ethidium bromide, visualized, and photographed on a UV transluminator. The product sizes, annealing temperatures and primers that were used are listed in Supplementary information Table S1.
Real-time PCR analysis of imprinted gene expression
Undifferentiated and differentiated chHES-32 cells from p35-45 were collected for imprinted gene analysis, and cells from a normal hESC line (chHES-35) were used as a control. Total RNA was extracted and reverse-transcribed as described above. Each sample was analyzed in triplicate with 28S as the inner control. The final PCR reaction volume of 10 μl contained 5 μl iQTM SYBR Green Supermix (Bio-Rad, Hercules, USA), 2 μl 1:4 diluted cDNA template, 3 μl primer mixture and 250 nM of each primer. Thermal cycling was carried out with a 10-min denaturation step at 95 °C, followed by 40 two-step cycles: 15 s at 95 °C, and 60 s at 58 °C. Amplification data were collected by the ABI PRISM 7900 and analyzed using Sequence Detection System 2.0 software (Applied Biosystems). The primers that were used are listed in Supplementary information Table S1.
Results
Isolation and characterization of chHES-32 cells
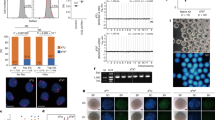
The fertilization status was evaluated 16-18 h after insemination and one oocyte exhibited one pronucleus in the center of the ooplasm with a second body extrusion (Figure 1A). After culture in sequential medium for 5 days, the oocyte developed to the blastocyst stage with poor morphology (Figure 1B). When the blastocyst was directly plated onto mitotic inactivated HEFs, ICM outgrowth occupied most of the colony during the primary culturing. The ICM was mechanically picked up for further serial passaging and a stable hESC line (chHES-32) was successfully derived (Figure 1C). The chHES-32 cells exhibited vigorous growth rate during long-term culture and were routinely passaged every 7 days. In culture, the chHES-32 cells displayed typical features of hESCs, including flat colonies, a high nuclear/cytoplasmic ratio and clearly distinguished nucleoli (Figure 1C inset). These cells are alkaline phosphatase positive and are immunoreactive for SSEA-3, SSEA-4, tumor rejection antigen 1-60 (TRA1-60), tumor rejection antigen 1-81 (TRA1-81) and OCT-4 (Figure 1D–1I), but negative for SSEA-1 (data not shown). In addition, RT-PCR showed that these cells express 10 pluripotency-related transcripts, including OCT-4, NANOG, REX-1, SOX2, LEFTY A, TDGF, FGF4, TERF1, THY1 and DPPA2 (Figure 2A), which are specifically expressed in undifferentiated hESCs as described by others 25, 26.
Isolation and characterization of chHES-32 cells with commonly used markers. (A) The one-pronuclear oocyte used for deriving the chHES-32 cells. (B) The original collapsed blastocyst on d5. (C) The undifferentiated chHES-32 cell colonies growing on mitotically inactivated human feeder layers; the inset is a higher magnified image of one colony. (D–I) Expression of molecular markers in undifferentiated chHES-32 cells. Undifferentiated cells stain positive for AKP (D), SSEA-3 (E), SSEA-4 (F), TRA1-60 (G), TRA1-81 (H) and OCT-4 (I).
(A) RT-PCR analysis of the expression of pluripotency-related genes in chHES-32 cells. RT (-) indicates that the reverse transcriptase was omitted in the cDNA synthesis step, and was subjected to PCR in the same manner with primer sets for β-actin. (B) A telomerase activity assay. Lane 1: negative control, lane 2: positive control, lane 3: heated positive control, lane 4: feeder layer, lane 5: chHES-32 cell sample and lane 6: heated inactivated chHES-32 cell sample. (C) Karyotype analysis of chESC-32 cells: 46, XX.
These cells have been continuously propagated for more than a year (more than 50 passages) and exhibit high proliferation ability. When analyzed at passage 30, chHES-32 cells displayed high telomerase activity comparable with Hela cells (Figure 2B), which reflects their immortal replication ability. More importantly, a stable karyotype of 46, XX could be well maintained (Figure 2C) during long-term culture (analyzed at passages 6 and 49).
Differentiation ability of chHES-32
In vitro, spontaneous differentiation of chHES-32 cells was initiated by suspension culture to induce embryoid body formation, a process that mimics the in vivo gastrulation process and triggers multilineage differentiation of hESCs. On replating day-7 embryoid bodies onto dishes, various types of cells appeared in the outgrowth. Immunocytochemical analysis using antibodies against β-tubulin (ectoderm), smooth muscle antigen (mesoderm) and α-fetoprotein (endoderm) confirmed differentiation into three germ layers (Figure 3A–3C).
In vitro and in vivo differentiation of chHES-32 cells. (A–C) After differentiation in vitro for 3 weeks, the derivatives are positive for β-tubulin (A), SMA (B) and AFP (C). (D–I) Histological analysis of a teratoma derived from chESC-32 cells; cartilage (D), bone (E), adipose tissue (F), neural epithelium (G), sebaceous gland (H) and glandular epithelium (I) were identified.
When chHES-32 cells were injected into the rear legs of SCID mice, teratomas formed later. After 8-10 weeks, teratomas were isolated and histologically examined. Derivatives representing the three germ layers could be found in the tumors, such as cartilage, bone and adipose tissue (mesoderm), neuroepithelium and sebaceous gland (ectoderm) and intestinal epithelium (endoderm) (Figure 3D–3I).
Imprinted gene expression
In pESCs, two alleles of imprinted genes are both maternally inherited. Thus, the expression of paternal imprinted genes is absent, while the expression of maternal imprinted genes is doubled. Six well-defined imprinted genes were analyzed in chHES-32 and compared with a normal hESC line, chHES-35. In normal chHES-35 cells, all analyzed imprinted genes were expressed in undifferentiated hESCs except insulin-like growth factor 2 (IGF2), which is only expressed in differentiated hESCs as described by others 27. By contrast, paternally expressed imprinted genes, including small nuclear ribonucleoprotein polypeptide N (SNRPN), KCNQ1 overlapping transcript 1 (KCNQ1OT1), imprinted in Prader–Willi syndrome (IPW), PEG3 and IGF2 were almost not expressed in both differentiated and undifferentiated chHES-32 cells, whereas the expression of the maternally expressed genes H19 and GNAS-associated transcript NESP55 was strong (Figure 4A). Real-time PCR confirmed these results and further showed that the expression of H19 in undifferentiated chHES-32 cells is almost twice that in normal chHES-35 cells (Figure 4B).
(A) RT-PCR analysis of imprinted gene expression in chHES-32 and chHES-35 cells. The paternally expressed genes IPW, PEG3, KCNQ1OT1, SNRPN and IGF2 are not expressed in chHES-32 cells but are expressed in biparental chHES-35 cells. (The expression of IGF2 was examined in differentiated chHES-32 and chHES-35 cells.) The maternally expressed genes H19 and NESP55 are expressed in both cells. (B) Real-time RT-PCR results show that chHES-32 cells have almost double the expression of the maternal gene H19 and have much lower, almost undetectable expression levels of the paternal genes SNRPN and IGF2 compared with normal chHES-35 cells.
HLA typing and DNA fingerprinting results
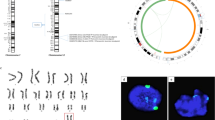
HLA typing showed that the HLA-A, -B and -DRB loci of chHES-32 are all identical to one allele of the oocyte donor but did not match any allele of the sperm donor (Table 1). To confirm this homozygous origin, DNA fingerprinting analysis was performed. All 15 STR loci and the Amelogenin gene analyzed were homologous. Compared with the DNA genotype of the oocyte donor, the alleles were identical in number and size to the alleles for the donor, except D16s539 (Supplementary information Figure S1).
DNA profile
The homozygosity of chHES-32 cells was determined by genome-wide SNP analysis using the Affymetrix 500K microarray. Of a total of 500,447 analyzed SNP sites, 493,314 SNP sites were called by chip hybridization. The homozygous sites comprised 99% of called SNP sites, and the heterozygous sites (listed in Supplementary information Table S2) exhibited a normal distribution among different chromosomes (Table 2).
Discussion
In IVF treatment, the parthenogenetic origin of one-pronuclear oocytes can only be identified when the resulting embryos are haploid. Resultant diploid embryos were previously confirmed to undergo predominantly normal fertilization owing to the asynchronous formation of two pronuclei 19, 21, and a normal diploid hESC line has been derived from these oocytes 28. These results coincide with our unpublished data, which show that, out of 16 hESC lines that are derived from one-pronuclear oocytes, the majority were of biparental origin. Surprisingly, one cell line (chHES-32) was found to be parthenogenetic, as shown in this study. This is the first evidence that human homozygous and parthenogenetic ESC lines can be derived from one-pronuclear oocytes following IVF. The cells were extensively characterized and were found to share common features of self-renewal and pluripotency, features that are suggested to define hESCs 26. For example, they express most of the commonly used hESC markers and specific genes, exhibit a vigorous growth rate and high telomerase activity, have the ability to differentiate into derivatives of all three germ layers both in vivo and in vitro, and maintain a stable diploid karyotype after long-term culture.
The parthenogenetic origin of chHES-32 cells was revealed by imprinted gene expression. Imprinting refers to a phenomenon in which gene expression is regulated in a parentally specific, epigenetic manner. Although several previous studies have reported the epigenetic instability of in vitro cultured hESCs 29, 30, imprinted genes seem less susceptible to in vitro culture conditions 27. Thus, the expression pattern of imprinted genes in hESCs might faithfully reflect the original imprinting state of the embryo from which it originated and might not be an aberrant consequence of the derivation and propagation process. The fact that chHES-32 cells almost did not express five paternal imprinted genes, but expressed the maternal gene H19 at more than double the level compared with normal hESCs, therefore suggests that the original embryo is solely derived from the maternal genome. Such an expression pattern of imprinted genes was also observed in pESCs from other species 31, 32. In addition, 16 STR loci scattered in different regions of 12 different chromosomes, plus three HLA (-A, -B and -DRB) loci showing one allelic expression pattern and matching one allele of the maternal genome add further proof for the uniparental origin of this cell line.
Genome-wide SNP analysis is regarded as a good tool for identifying the homozygous state of pESCs 33, 34. In this study, a high-resolution SNP microarray (500K) was used to identify the homozygosity of chHES-32 cells. More than 99% of the analyzed sites either on the whole genome or on each chromosome showed homozygosity. The heterozygous sites detected made up less than 1% and were distributed normally on each chromosome. They can therefore be regarded as the error of the technology; according to the results of a recent study, the error rate is normally 2-5% when using the same high-resolution SNP microarray to analyze hESCs 34. Thus, we conclude that chHES-32 cells are a homozygous pESC line.
It is particularly interesting to compare the homozygous state of this cell line with the pESCs that are derived from artificially activated oocytes in mice 32, 33 and in humans 13, 34. Artificial parthenogenesis is routinely induced by activating MII oocytes and preventing second polar body extrusion, which is thought to produce diploid embryos that are more likely to develop to the blastocyst stage 9, 13, 31, 32, 33. This method has been proved to produce efficient pESCs, but only predominantly heterozygous pESCs could be obtained owing to early recombination events 13, 32, 33, 34. These cells could be used as autologous stem cells for the oocyte donor, but were of limited use for storage in the hESC bank owing to the heterozygosity of HLA loci. In mice, the incidence of heterozygosity of the major histocompatibility complex (MHC) is 33% in pESCs 33. In recent studies in humans, all established pESCs were heterozygous in HLA loci 13, 34.
Parthenogenesis can also be induced by artificially activating MII oocytes without preventing second polar body extrusion, which results in a one-pronuclear oocyte with a haploid genome. Haploid cells cannot contribute to the post-implantation development of the embryo unless a spontaneous duplication of the haploid genome (diploidization) occurs 35. Pluripotent cell lines have been successfully derived from haploid mouse embryos after diploidization 36, 37. This may provide a putative source for homozygous pESC lines, since no more recombination events will occur in mitosis. The homozygous state of chHES-32 cells when compared with the predominantly heterozygous state in other established human pESC lines 13 suggests that this cell line might originally be haploid with a similar diploidization process to that in mouse occurring thereafter to produce a competent diploid genome for pluripotency. Thus, our results suggest that parthenogenetically activated haploid oocytes may be a source for generating homozygous ESC lines.
Conclusion
Here, we reported the first isolation and further characterization of a homozygous hESC line from a parthenogenetic oocyte following IVF treatment. The results indicate that clinical 'unwanted' one-pronuclear oocytes can be a source for human pESCs, and highlight an alternative strategy of parthenogenetic activation to obtain homozygous hESC lines. This cell line also provides a model to study the function of imprinted genes in cell differentiation.
(Supplementary Information is linked to the online version of the paper on the Cell Research website.)
References
Thomson JA, Itskovitz-Eldor J, Shapiro SS, et al. Embryonic stem cell lines derived from human blastocysts. Science 1998; 282:1145–1147.
Reubinoff BE, Pera MF, Fong CY, Trounson A, Bongso A . Embryonic stem cell lines from human blastocysts: somatic differentiation in vitro. Nat Biotechnol 2000; 18:399–404.
Bradley JA, Bolton EM, Pedersen RA . Stem cell medicine encounters the immune system. Nat Rev Immunol 2002; 2:859–871.
Drukker M, Benvenisty N . The immunogenicity of human embryonic stem-derived cells. Trends Biotechnol 2004; 22:136–141.
Priddle H, Jones DR, Burridge PW, Patient R . Hematopoiesis from human embryonic stem cells: overcoming the immune barrier in stem cell therapies. Stem Cells 2006; 24:815–824.
Taylor CJ, Bolton EM, Pocock S, Sharples LD, Pedersen RA, Bradley JA . Banking on human embryonic stem cells: estimating number of the donor cell lines needed for HLA matching. Lancet 2005; 366:2019–2025.
Fumiaki N, Katsushi T, Norio N . HLA matching estimations in a hypothetical bank of human embryonic stem cell lines in the Japanese population for use in cell transplantation therapy. Stem Cells 2007; 25:983–985.
Kaufman MH, Barton SC, Surani MA . Normal postimplantation development of mouse parthenogenetic embryos to the forelimb bud stage. Nature 1977; 265:53–55.
Cibelli JB, Cunniff K, Vrana KE . Embryonic stem cells from parthenotes. Methods Enzymol 2006; 418:117–135.
Cibelli JB, Kiessling A, Cunniff K, Richards C, Lanza R, West M . Somatic cell nuclear transfer in humans: pronuclear and early embryonic development. J Reprod Med 2001; 2:25–31.
Rogers N, Werb Z . Minireview: parthenogenesis in mammals. Mol Reprod Dev 2004; 59:468–474.
Lin H, Lei J, Wininger D, et al. Multilineage potential of homozygous stem cells derived from metaphase II oocytes. Stem Cells 2003; 21:152–161.
Revazova ES, Turovets NA, Kochetkova OD, et al. Patient-specific stem cell lines derived from human parthenogenetic blastocysts. Cloning Stem Cells 2007; 9:432–449.
Fangerau H . Can artificial parthenogenesis sidestep ethical pitfalls in human therapeutic cloning? A historical perspective. J Med Ethics 2005; 31:733–735.
Linder D, McCaw BK, Hecht F . Parthenogenic origin of benign ovarian teratomas. N Engl J Med 1975; 292:63–66.
Padilla SL, Boldt JP, McDonough PG . Possible parthenogenesis with in vitro fertilization subsequent to ovarian cystic teratomas. Am J Obstet Gynecol 1987; 156:1127–1129.
Oliveira FG, Dozortsev D, Diamond MP, et al. Evidence of parthenogenetic origin of ovarian teratoma: case report. Hum Reprod 2004; 19:1867–1870.
Muechler EK, Graham MC, Huang KE, Partridge AB, Jones K . Parthenogenesis of human oocytes as a function of vacuum pressure. J In Vitro Fertil Embryo Transf 1989; 6:335–337.
Sultan KM, Munné S, Palermo GD, Alikani M, Cohen J . Chromosomal status of uni-pronuclear human zygotes following in-vitro fertilization and intracytoplasmic sperm injection. Hum Reprod 1995; 10:132–136.
Gook DA, Osborn SM, Johnston WI . Parthenogenetic activation of human oocytes following cryopreservation using 1,2-propanediol. Hum Reprod 1995; 10:654–658.
Staessen C, Streirteghem AC . The chromosomal constitution of embryo developing from abnormally fertilized oocytes after intracytoplasmic sperm injection and conventional in-vitro fertilization. Hum Reprod 1997; 12:321–327.
Santos TA, Dias C, Henriques P, et al. Cytogenetic analysis of spontaneously activated noninseminated oocytes and parthenogenetically activated failed fertilized human oocytes—implications for the use of primate parthenotes for stem cell production. J Assist Reprod Genet 2003; 20:122–130.
Van Blerkom J, Davis PW, Merriam J . A retrospective analysis of unfertilized and presumed parthenogenetically activated human oocytes demonstrates a high frequency of sperm penetration. Hum Reprod 1994; 9:2381–2388.
Xie CQ, Lin G, Lu GX . Preliminary study on human fibroblasts as feeder layer for human embryonic stem cells culture in vitro. Chin Sci Bull 2003; 48:354–357.
Ginis I, Luo Y, Miura T, et al. Differences between human and mouse embryonic stem cells. Dev Biol 2004; 269:360–380.
Brivanlou AH, Gage FH, Jaenisch R, Jessell T, Melton D, Rossant J . Stem cells. Setting standards for human embryonic stem cells. Science 2003; 300:913–916.
Sun BW, Yang AC, Feng Y, et al. Temporal and parental-specific expression of imprinted genes in a newly derived Chinese human embryonic stem cell line and embryoid bodies. Hum Mol Genet 2006; 15:65–75.
Suss-Toby E, Gerecht-Nir S, Amit M, Manor D, Itskovitz-Eldor J . Derivation of a diploid human embryonic stem cell line from a mononuclear zygote. Hum Reprod 2004; 19:670–675.
Allegrucci C, Wu YZ, Thurston A, et al. Restriction landmark genome scanning identifies culture-induced DNA methylation instability in the human embryonic stem cell epigenome. Hum Mol Genet 2007; 16:1253–1268.
Maitra A, Arking DE, Shivapurkar N, et al. Genomic alterations in cultured human embryonic stem cells. Nat Genet 2005; 37:1099–1103.
Vrana KE, Hipp JD, Goss AM, et al. Nonhuman primate parthenogenetic stem cells. Proc Natl Acad Sci USA 2003; 100(Suppl 1 ):11911–11916.
Lengerke C, Kim K, Lerou P, Daley GQ . Differentiation potential of histocompatible parthenogenetic embryonic stem cells. Ann NY Acad Sci 2007; 1106:209–218.
Kim K, Lerou P, Yabuuchi A, et al. Histocompatible embryonic stem cells by parthenogenesis. Science 2007; 315:482–486.
Kim K, Ng K, Rugg-Gunn PJ, et al. Recombination signatures distinguish embryonic stem cells derived by parthenogenesis and somatic cell nuclear transfer. Cell Stem Cell 2007; Cell Stem Cell 2007; 1:346–352.
Ito M, Kaneko-Ishino T, Ishino F, Matsuhashi M, Yokoyama M, Katsuki M . Fate of haploid parthenogenetic cells in mouse chimeras during development. J Exp Zool 1991; 257:178–183.
Kaufman MH, Robertson EJ, Handyside AH, Evans MJ . Establishment of pluripotential cell lines from haploid mouse embryos. J Embryol Exp Morphol 1983; 73:249–261.
Robertson EJ, Evans MJ, Kaufman MH . X-chromosome instability in pluripotential stem cell lines derived from parthenogenetic embryos. J Embryol Exp Morphol 1983; 74:297–309.
Acknowledgements
We thank the IVF team at CITIC-Xiangya Reproductive and Genetic Hospital (Changsha, China), Dr Yubing Xie (Changsha Blood Centre, China) for HLA analysis, Qiong Li (Public Security Department of Hunn Province, China) for DNA fingerprinting analysis and Dr Teija Peura (Australian Stem Cell Centre) for editing the English of the manuscript. This work was supported by the Hi-Tech Research and Development Program of China (863 program No. 2006AA02A102), the National Basic Research Program of China (973 program No. 2007CB948103), and the Bureau of Science and Technology Key Project Funds of Hunan Province, China (No. 03SSY2001).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Lin, G., OuYang, Q., Zhou, X. et al. A highly homozygous and parthenogenetic human embryonic stem cell line derived from a one-pronuclear oocyte following in vitro fertilization procedure. Cell Res 17, 999–1007 (2007). https://doi.org/10.1038/cr.2007.97
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cr.2007.97
Keywords
This article is cited by
-
Generation of developmentally competent oocytes and fertile mice from parthenogenetic embryonic stem cells
Protein & Cell (2021)
-
Pronuclear removal of tripronuclear zygotes can establish heteroparental normal karyotypic human embryonic stem cells
Journal of Assisted Reproduction and Genetics (2016)
-
Chromosomal instability in mammalian pre-implantation embryos: potential causes, detection methods, and clinical consequences
Cell and Tissue Research (2016)
-
Ascorbic acid improves pluripotency of human parthenogenetic embryonic stem cells through modifying imprinted gene expression in the Dlk1-Dio3 region
Stem Cell Research & Therapy (2015)
-
Activin A can induce definitive endoderm differentiation from human parthenogenetic embryonic stem cells
Biotechnology Letters (2015)