Abstract
Steroids function as signaling molecules in both animals and plants. While animal steroid hormones are perceived by nuclear receptor family of transcription factors, brassinosteroids (BR) in plants are perceived by a cell surface receptor kinase, BRI1. Recent studies have demonstrated that BR binding to the extracellular domain of BRI1 induces kinase activation and dimerization with another receptor kinase, BAK1. Activated BRI1 or BAK1 then regulate, possibly indirectly, the activities of BIN2 kinase and/or BSU1 phosphatase, which directly regulate the phosphorylation status and nuclear accumulation of two homologous transcription factors, BZR1 and BES1. BZR1 and BES1 directly bind to promoters of BR responsive genes to regulate their expression. The BR signaling pathway has become a paradigm for both receptor kinase signaling in plants and steroid signaling by cell surface receptors in general.
Similar content being viewed by others
Introduction
Since the first discovery of the growth promoting activity in extracts of rape pollen in 1970 1 and the identification of the bioactive compound brassinolide (BL) in 1979 2, many plant steroids with similar structure and functions to BL have been identified throughout the plant kingdom, and they are defined as brassinosteroids (BRs). Physiological studies have demonstrated that BR can induce diverse cellular responses such as stem elongation, pollen tube growth, leaf bending and epinasty, root inhibition, induction of ethylene biosynthesis and fruit ripening, and xylem differentiation 3, 4, 5, 6. The identification of Arabidopsis BR biosynthetic mutants established that BR is a new class of phytohormone with an essential role in plant growth and development 7, 8, 9. Since then, extensive molecular genetic and biochemical studies of BR signaling in Arabidopsis thaliana have illustrated a BR signal transduction pathway from ligand perception on the cell surface to gene expression in the nucleus. This review focuses on recent advances in our understanding of the mechanisms of BR perception, receptor kinase activation and BR regulation of gene expression, with highlights of outstanding questions in the field.
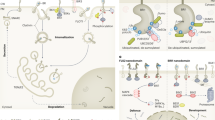
BR is perceived by the BRI1 receptor kinase on cell surface
The brassinosteroid insensitive 1 (bri1) mutant was first identified about 10 years ago 10. Additional alleles of bri1 were isolated in subsequent screens for mutants with similar phenotypes to BR biosynthetic mutants but cannot be rescued by the application of BR 11. Cloning of BRI1 revealed that it encodes a leucine-rich repeat receptor-like kinase (LRR-RLK), containing an extracellular domain, a transmembrane domain and a cytoplasmic serine/threonine kinase domain. The extracellular domain of BRI1 was initially predicted to contain a leucine-zipper motif, 25 tandem LRRs, 2 cysteine pair before and after the LRRs, and a 70 amino acid (70 aa) island inserted between LRR21 and LRR22 11, 12. A recent reannotation of BRI1 predicted no leucine zipper and 24 rather than 25 LRRS, with LRR21 being an unusual methionine-rich repeat 13. The intracellular region was subdivided into a juxtamembrane region (JM), kinase domain, and a short C-terminal extension 13. Mutations in large numbers of bri1 alleles cluster in two regions, the intracellular kinase domain and the 70 aa island domain, suggesting important functions for these two regions 11, 12.
The structure of BRI1 and its essential role in BR response supported a popular hypothesis that BRI1 is the BR receptor. Evidence supporting this hypothesis has been obtained in several studies over the years and the final proof has been achieved recently. First, when the extracellular LRR and transmembrane domains of BRI1 were fused to the serine/threonine kinase domain of Xa21, a rice LRR receptor kinase for disease resistance, the chimeric receptor was able to elicit defense responses in rice cells upon treatment with BL 14. Secondly, BRI1 was co-immunoprecipitated with the BR-binding activity and BR treatment was shown to induce BRI1 autophosphorylation, which indicates that BR interacts with BRI1 and activates its kinase activity 15. However, these studies failed to demonstrate direct binding of BR to BRI1. In fact several lines of evidence supported the possibility that BRI1 might interact with another protein or peptide that binds directly to BR. For example, a secreted serine carboxypeptidase encoded by the BRS1 gene was shown to play an important role in the early stage of the BR signaling and was thus proposed to process an extracellular protein/peptide involved in BR perception 16. Furthermore, there are at least two putative steroid-binding proteins (SBPs) in the Arabidopsis genome, raising the possibility that BR might bind a SBP that interacts directly with BRI1 17. Although the possibility of BRI1 interacting with extracellular proteins cannot be excluded, a recent study from Joanne Chory's lab demonstrated that BR directly binds to the extracellular domain of BRI1 18. Using biotin-tagged photoaffinity castasterone (BPCS), a biosynthetic precursor of brassinolide with partial activity, they demonstrated that BR directly binds to BRI1 in Arabidopsis and to recombinant BRI1. BR binding assays using recombinant BRI1 fragments indicated that the minimal BR binding region in BRI1 consists of the 70-aa island and the carboxy-terminal flanking LRR (ID-LRR22). BR also binds specifically to the ID-LRR22 domain from BRL1 and BRL3, homologs of BRI1 that function as BR receptors in vascular differentiation 19, 20. These studies unambiguously demonstrate that BRI1 is the bona fide BR receptor 18.
BR activation of BRI1 and BAK1 receptor kinases
BR binding activates the BRI1 kinase and causes autophosphorylation of BRI1, as demonstrated by both mobility shift of BRI1 in SDS-PAGE and by increased signal detected by anti-phospho-serine/threonine antibodies in BR treated samples 15, 21. Many phosphorylation sites of BRI1 have recently been identified by mass spectrometry analysis of recombinant BRI1 expressed in E. coli or BRI1 immunoprecipitated from plant extracts 21, 22. Mutation studies of the phosphorylation residues demonstrated that some of the phosphorylation events increase and some reduce the BRI1 activity, as measured by the ability of the mutated BRI1 protein to phosphorylate a synthetic substrate and to rescue a bri1 mutant 21. For example, mutations of Ser or Thr residues in the activation loop and the juxtamembrane and C-terminal domains reduced the kinase activity, whereas mutation of T872A increased the kinase activity 21.
BRI1 has been found to interact with another LRR receptor-like kinase named BAK1 (BRI1 associated receptor kinase 1), which was identified as a component of the BR signaling pathway by two independent groups based on its interaction with BRI1 kinase domain in yeast 23 and suppression of a weak bri1 allele by BAK1 overexpression 24. An obvious structural difference between BRI1 and BAK1 is that BAK1 has only five extracellular LRR repeats and no BR binding domain. Loss-of-function mutation of BAK1 caused a weak dwarf phenotype. However, overexpression of a dominant negative mutant form of BAK1 caused a severe dwarf phenotype similar to strong bri1 mutants, suggesting that BAK1 plays a major role in transducing the BR signal and the weak phenotype of bak1 mutant is likely due to redundant functions of its homologs 24. In vivo interaction between BAK1 and BRI1 has been demonstrated by co-immunoprecipitation assays and fluorescence life time imaging microscopy 23, 24, 25, and it has been shown recently that BR treatment increases the BRI1-BAK1 interaction 21.
BR induced BRI1/BAK1 oligomerization and kinase activation suggests a mechanism of ligand-induced receptor kinase activation similar to those in animals. Studies of receptor kinases in animal systems have shown that activation often involves ligand-induced receptor dimerization or oligomerization, which causes kinase activation by transphosphorylation. For example, response to the transforming growth factor β (TGF-β) is mediated by two receptor serine/threonine kinases Tβ-RI and Tβ-RII. Tβ-RII homodimerizes in the absence of ligand and exhibits constitutive autophosphorylation 26. TGF-β binding to Tβ-RII induces formation of the heterotetramer with Tβ-RI and phosphorylation of Tβ-RI by Tβ-RII 27. Once activated by phosphorylation, Tβ-RI propagates the signal by phosphorylating cytoplasmic substrates that translocate to the nucleus to regulate the expression of TGF-β–responsive genes. In other cases, such as the EGF receptor kinases, monomeric receptors exhibit basal level or no kinase activity, and ligand-induced receptor dimerization leads to transphosphorylation that activates the kinase. Models of BRI1 activation based on either TGF-β or EGF receptors have been proposed 23, 24. In addition, it has been shown that the plant homolog of a mammalian Tβ-RII substrate is a putative BRI1 substrate in Arabidopsis 28, which provides further evidence for the similarity between TGF-β and BR signaling.
Although BR induced BRI1/BAK1 dimerization and kinase activation share similarities with animal receptor kinases, it is not fully understood whether dimerization of BRI1/BAK1 is the cause or result of ligand-induced activation of BRI1 kinase. Analyses of the effects of mutations that abolish kinase activity on the dimerization and transphosphorylation performed in yeast and in vitro yielded slightly conflicting results. Wild type BRI1 and BAK1 co-expressed in yeast interact with each other and show kinase activity. But when wild type BRI1 or BAK1 was co-expressed with a mutant BAK1 or BRI1, respectively, the receptors still interact with each other but no phosphorylation was detected for either wild type or the mutant kinase, suggesting that kinase activation, but not heterodimerization, requires kinase activities of both BRI1 and BAK1 23. In contrast, in vitro assays using the kinase domains expressed in E. coli showed that wild type BRI1 or BAK1 kinases can autophosphorylate as well as transphosphorylate mutant BAK1 or BRI1, respectively. Furthermore, the affinity for the interaction between BRI1 and BAK1 kinases is reduced by mutation of either kinase, with mutation of BRI1 kinase showing stronger effect than mutation of BAK1. The in vitro data is thus consistent with the hypothesis that BR activation of BRI1 kinase activity leads to dimerization with and subsequent activation of BAK1 24.
The second hypothesis is supported by a recent study from Joanne Chory's lab, which provided evidence that receptor activation involves ligand induced conformation changes in the BRI1 kinase 29. In this study, deletion of the C-terminal 40 amino acids (CT) of BRI1 was shown to increase receptor activity, as indicated by increased phosphorylation of BRI1 and growth response in transgenic plants expressing such truncated BRI1 receptor 29. The inhibitory function of the CT region appears to be reduced by phosphorylation, because the kinase activity was increased by mutation of several Ser/Thr residues in this region to Asp, which likely mimics phosphorylation 29, and reduced by mutations of Ser/Thr to Ala, which prevent phosphorylation 21. BRI1 was also shown to interact with itself in vivo and the interaction appears to be increased by BR, suggesting that BR binding stabilizes the BRI1 homodimer. Based on these observations, it was proposed that homodimerization is involved in BRI1 kinase activation 29. In the absence of ligand, BRI1 exists as a preformed inactive homo-oligomer, and its kinase activity is maintained at a basal level via trans- or cis-repression by its CT domain. Steroid binding to the extracellular domain of BRI1 induces a conformational change in the kinase domain and allows trans-phosphorylation of the CT region, which likely enhances BRI1's kinase activity and promotes further phosphorylation of BRI1, thereby providing binding sites for BAK1 29.
Although the model of Wang et al. 29 is consistent with most of the observations, many questions remain to be answered. For example, the possibility that activation of BRI1 kinase requires dimerization with BAK1 has yet to be tested experimentally. BR binding to BRI1 might induce conformational changes in either the extracellular or kinase domain that facilitate interaction with BAK1, and BAK1 might then further activate BRI1 kinase through transphosphorylation. Such a possibility is supported by the ability of BAK1 to phosphorylate mutant BRI1 kinase in vitro 24. It would be interesting to test whether mutants lacking BAK1 and its homologs or the transgenic plants overexpressing the dominant negative bak1 24 show BR-induced BRI1 phosphorylation. Furthermore, the function of the extracellular domain of BAK1 has yet to be explained if interaction with BRI1 is only mediated by the intracellular kinase domains. The extracellular domain of BAK1 is apparently not involved in BR binding, because BR binding activity is not affected in the bak1 null mutant 18 or transgenic plants overexpressing BAK1 29. However, the dominant negative effects of kinase-inactive mutants of BAK1 24 and BRI1 29 suggest that the receptors can interact with their partners independent of the kinase activity. Therefore, it is possible that the extracellular domain of BAK1 binds to the BR-occupied extracellular domain of BRI1 to initiate the heterodimerization. It would be interesting to test whether kinase-inactive mutant BRI1 and BAK1 can interact with each other in a BR-dependent manner in plant cells. The precise sequence of events in BR-induced receptor oligomerization and kinase activation remains to be further defined.
In addition to BAK1, two other putative BRI1 substrates have been identified. Transthyretin-Like (TTL) is an Arabidopsis protein with sequence similarity to vertebrate transthyretin 30. TTL interacts with BRI1 in a yeast two-hybrid assays and the interaction is abolished by mutations that knockout the BRI1 kinase activity. Genetic studies suggested that TTL plays a negative role in BR responses. TTL is localized on the plasma membrane, though it contains no obvious transmembrane domain 30. The function of TTL in BR signal transduction remains unclear 30. AtTRIP1 is an Arabidopsis homolog of the mammalian TGFβ receptor interacting protein 1 (TRIP1). In mammalian, TRIP1 is a cytoplasmic substrate of the TGFβ receptor TβII and functions as a subunit of the eIF3 translation initiation factor. BRI1, but not BAK1, can phosphorylate AtTRIP1 in vitro and interact with AtTRIP1 in vivo. It was proposed that AtTRIP1 might play a role in BR regulation of protein synthesis or in transducing BR signal to downstream components 28.
Tissue specific functions of the homologs of BRI1 and BAK1
BRI1 and BAK1 belong to a large family of plant-specific LRR-RLKs, consisting of more than 200 members in Arabidopsis and over 300 in rice 31. Three close homologs of BRI1 have been characterized 19, 20, 32. BRL1 and BRL3 bind BR with high affinity and VH1 showed very weak BR binding 18. Genetic analysis suggests that they play a role in vascular development 19, 32. BRI1, BRL1, and BRL3 have partially redundant function in BR signaling, and their functional specificities appear to be conferred by their tissue specific promoters rather than specific signaling function of the proteins, because expression of BRL1 and BRL3 using the BRI1 promoter can rescue the bri1 mutant 19 and overexpression of BRL1 can partially suppress bri1-5 20.
BAK1 and four close homologs have been identified as somatic embryogenesis receptor kinases (SERKs) 33 and proposed to play a role in embryo development. A recent study showed that single knockout mutant of either SERK1 or SERK2 shows no obvious phenotypes, but double mutants are male sterile due to defect in tapetum differentiation, a phenotype similar to that caused by mutation of another LRR-RLK, ems1 34. These results suggest that SERK1 and SERK2 are functionally redundant in reproductive development 35. It is unclear whether the function of SERK1 and SERK2 in reproductive development is related to BR signaling. Strong BR-deficient and BR-insensitive mutants are also male sterile, although it remains to be analyzed if they have similar defect in tapetum differentiation. SERK1 was recently shown to be part of a protein complex that includes BRI1 and BAK1 36, suggesting that SERK1 might play a role in BR signaling. How members of BRI1 and BAK1 families pair up in regulating various developmental processes and whether there exist tissue-specific BR signaling mechanisms are interesting questions to be answered in future studies.
Mutations of BRI1 orthologs in rice, pea, and tomato cause BR insensitive dwarf phenotypes 37, 38, 39, suggesting highly conserved function of BRI1. Interestingly, the tomato BRI1 was shown to function as receptor for both BR and systemin, a small peptide involved in plant defense (reviewed by Wang and He, 2004 40). It is currently not known how tomato BRI1 mediates two distinct responses and whether similar peptide ligand of BRI1 exists in Arabidopsis and other plants. Although the function of BRI1 as BR receptor is conserved in Arabidopsis, rice, pea and tomato, the functions of BRI1 homologs in regulation of developmental processes might be different in different species, as suggested by a recent functional study of the rice BRI1 gene family 41.
Downstream components of the BR signal transduction pathway
BR activation of BRI1 and BAK1 kinases initiates a signaling cascade that leads to nuclear gene expression. Downstream components of this cascade include the BIN2 kinase, the BSU1 phosphatase, and the transcription factors BZR1 42, 43 and its homolog BES1/BZR2 44, 45. BZR1 and BES1/BZR2 are substrates of the BIN2 kinase 44, 46 and BSU1 phosphatase 47, and phosphorylated BZR1 is degraded by the proteasome 46. In the absence of BR, BRI1 and BAK1 are inactive, and BIN2 phosphorylates BZR1 and BES1/BZR2 and targets them for degradation by the proteasome. BR induces dephosphorylation and accumulation of BZR1 and BES1/BZR2, possibly by inhibiting the BIN2 kinase or activating the BSU1 phosphatase. BZR1 and BES1 bind to the promoters of BR regulated genes to activate or repress gene expression 45, 46. With direct interactions demonstrated for BR, BRI1 and BAK1, and among BIN2, BZR1/BES1 and promoter DNA, the only gap in the BR signal transduction pathway is between BRI1 or BAK1 and BIN2 or BSU1.
The BIN2 kinase phosphorylates BZR1 and BES1 to target them for degradation by the proteasome
BIN2 shares about 70% sequence similarity with the Drosophila shaggy kinase and mammalian glycogen synthase kinase-3 (GSK-3) in the catalytic domain 48. In animals, GSK3/SHAGGY-like kinases play a key role in the regulation of many cell functions, including signaling by insulin, growth factors and nutrients, and the control of cell division and differentiation, apoptosis, and microtubule dynamics 49. In these pathways, GSK3 functions as a negative regulator that reduces the activity of downstream substrate through inhibiting their biochemical activities, changing their subcellular localization, or promoting their degradation. Phosphorylation by GSK3 often requires pre-phosphorylation of the substrate at a priming site by other kinases. Signaling by cell surface receptors inhibits GSK3 through several mechanisms, including phosphorylation at the N terminal region of GSK3, preventing priming phosphorylation of its substrate that is required for phosphorylation by GSK3 , and disruption of protein complex of GSK3 and its substrate 50.
Like animal GSK3 kinases that inhibit the activity of their substrates, BIN2 functions as a negative regulator in BR signal transduction. Dominant mutations of bin2 that increase the kinase activity or overexpression of the wild type BIN2 gene causes bri1-like BR insensitive phenotypes 52, 53, 54, which is associated with increased phosphorylation and reduced accumulation of BZR1 and BES1 44, 46. BIN2 interacts with and phosphorylates BZR1 and BES1 in yeast and in vitro. Phosphorylated BZR1 is stabilized by MG132, an inhibitor of the proteasome, suggesting that BIN2 phosphorylation leads to degradation of BZR1 by the proteasome. The large number of putative GSK3 phosphorylation sites in BZR1 and BZR2 sequence and the severe mobility shift caused by BIN2 phosphorylation suggests that BIN2 phosphorylates the substrates at multiple amino acid residues 42, 46. The positions of these phosphorylation sites and their effects on BZR1/BES1 accumulation and activity are yet to be determined experimentally.
Unlike animal GSK3 kinase, which requires priming phosphorylation of its substrate, BIN2 relies on a direct kinase-substrate docking interaction to phosphorylate BES1 55. Plant GSK3 kinases also lack the N-terminal Ser residue that is conserved in animal GSK3 kinases and that inhibits kinase activity when phosphorylated 49. Further studies will reveal whether BR signaling inhibits BIN2 kinase through a mechanism similar to those found in animal GSK3 kinases.
The BSU1 phosphatase dephosphorylates BES1
BR treatment induces rapid disappearance of the phosphorylated BZR1 and BES1, possibly due to either degradation by the proteasome or dephosphorylation by a phosphatase. Because BR-induced disappearance of phosphorylated BZR1 is not affected by MG132, a proteasome inhibitor that stabilizes phosphorylated BZR1 in the absence of BR treatment, it was proposed that a phosphatase is involved in BZR1 dephosphorylation 46. Indeed a phosphatase with Kelch-repeat, BSU1, was shown to promote dephosphorylation of BES1. BSU1 was identified as an activation tagged suppressor of bri1-5, and overexpression of BSU1 increased cell elongation and the level of unphosphorylated BES1. Thus BSU1 appears to counteract the activity of BIN2 in regulating phosphorylation status of BES1 and likely also BZR1. In vitro, recombinant BSU1 was able to dephosphorylate BES1 that had been pre-phosphorylated by BIN2, but with very low efficiency. BSU1 did not directly interact with BES1 in vitro, and it was proposed that an adaptor protein might be required for BSU1 to effectively dephosphorylate BES1. BSU1 is constitutively localized in the nucleus, consistent with its action on BES1. It is unclear whether BSU1 activity is regulated by BR signaling 47.
BZR1 and BES1 directly regulate BR responsive gene expression
A genetic screen for mutants resistant to brassinazole lead to the identification of the brassinazole resistant 1-1D (bzr1-1D) mutant, which turned out to suppress the bri1 and bin2 mutants 42. BZR1 encodes a nuclear protein and a P234L mutation in bzr1-1D stabilizes the protein. Same mutation in BZR1's closest homolog, BZR2/BES1, was later identified in a bri1 suppressor screen 44. BZR1 and BES1/BZR2 show 88% sequence identity and their phosphorylation and accumulation are similarly regulated by BR and BIN2 42, 44, 46. While both bzr1-1D and bes1-D suppress bri1 and bin2 and are resistant to brassinazole, they show opposite cell elongation phenotypes when grown in the light. The bzr1-1D plants grown in the light show weak dwarfism with reduced hypocotyl and petiole lengths, while bes1-D has elongated organs 42, 44. The bzr1-1D plants contain reduced level of BR due to increased feedback inhibition of BR synthesis, and its weak dwarf phenotype can be rescued by BR treatment. It has been proposed that BZR1 plays dual roles in mediating feedback inhibition of BR biosynthesis and BR promoted growth responses 43.
The biochemical functions of BZR1 and BES1 have been illustrated recently 43, 45. Through extensive protein-DNA interaction studies, including gel shift competition assays, DNA footprint, binding site selection from random sequences, and chromatin immunoprecipitation, He et al. demonstrated unequivocally that BZR1 specifically binds to CGTG(T/C)G sequence, which was named BR response element (BRRE) 43. BRRE was found enriched in promoters of BR-repressed genes, including CPD, DWF4, and a large number of BR repressed genes identified in microarray studies. A BZR1-GAL4 fusion construct repressed the expression of a reporter gene containing GAL4 binding sites. Using chromatin immunoprecipitation, they showed that BZR1 binds to CPD and DWF4 promoters in vivo and the binding was increased by BR treatment. Furthermore, BRRE was shown to be essential for BR regulation of the CPD promoter, because mutation of the BRRE in the CPD-GUS reporter gene abolished its repression by BZR1. These results demonstrate that BZR1 binds to the BRRE in BR responsive promoters to regulate their expression, thereby inhibiting BR biosynthesis as well as promoting growth responses 43. It was proposed that BZR1 might indirectly activate BR-induced genes by repressing other transcription repressors 43, which is consistent with microarray data showing that most BR-induced genes respond slowly to BR treatment 56.
BES1 has also been shown to bind to DNA 45. In this case, BES1 was shown to interact with three homologous basic-helix-loop-helix (bHLH) transcription factors (BIM1-3) in yeast two-hybrid assays 45. Gel shift assays demonstrated that BES1 could bind to the E-box (CANNTG) elements in the promoter of SAUR-AC1, a gene showing early induction by both BR and auxin 56, 57. The BIM1 protein was also shown to bind the same E-box, and synergism between BES1 and BIM1 was observed for both DNA binding in vitro and activation of SAUR-AC1 promoter in transient assays. Overexpression of BIM1 partially suppressed a weak bril allele, and triple mutant of BIM1-3 showed reduced cell elongation, suggesting a role of the BIM1-3 proteins in mediating BR responses. It was proposed that BES1 and the BIM1-3 proteins synergistically bind to E-box elements to activate BR-induced gene expression 45 .
It is surprising that BZR1 and BES1 showed different DNA-binding specificities and opposite transcriptional activities, given their high sequence homology (88% overall amino acid sequence identity). Within the 83 amino acid region of BZR1 that is sufficient to bind DNA 43, only six amino acids differ between BZR1 and BES1, and only two of these six are in the N-terminal most conserved 69 amino acid region containing the predicted HLH domain 45. It is possible that BZR1 and BES1 have similar DNA binding specificities. Although different DNA motifs were shown to bind to BZR1 and BES1, the binding specificities may not be as distinct as it appears. The BRRE element (CGTGTG) differs from the E-box (CANNTG) by only one base pair. In fact one of the mutated BRREs used in competition gel shift assays (m8, CATGTG) matches perfectly the E-box sequence 43. However this mutated BRRE showed the weakest binding to BZR1 among all the DNAs with single nucleotide mutation of the core sequence, though it showed better binding than the one with all six base pairs mutated 43. Therefore BZR1 is likely to discriminate against E-box among sequences similar to BRRE. On the other hand, whether BES1 binds BRRE remains unknown. In fact, the optimal binding site sequence of BES1 has yet to be determined, for example, by binding site selection from random sequences. Without the optimal binding site sequence for BES1 or direct comparison of E-box with BRRE for their binding affinities to BES1, it is difficult to conclude whether BES1 and BZR1 indeed have different DNA-binding specificity.
Is BZR1 a transcription repressor while BES1 an activator, or can they both activate and repress different promoters? Although BZR1 has been shown to repress the CPD promoter and BES1 to activate the SAUR-Ac promoter, it is not clear if the different transcriptional activities are due to different promoters used in the studies or intrinsic differences between BZR1 and BES1 proteins. The reduced CPD and DWF4 expression in the bes1-D mutant 13, 47 suggests that BES1 might function as a transcription repressor on these promoters, possibly acting through BRRE. It is also possible that BZR1 might function as an activator for some BR induced genes, although BZR1 can repress transcription of BR down regulated genes and an artificial promoter 43. The opposite cell elongation phenotypes of light-grown bzr1-1D and bes1-D are consistent with opposite transcriptional activities of the BZR1 and BES1 proteins, but such difference in phenotype could also be caused by different tissue-specific or light-regulated expression of the BZR1 and BES1 genes. Indeed transgenic plants overexpressing the mutant bzr1-1D gene show a long hypocotyl phenotype similar to bes1-D (Y Sun and Wang, unpublished data). Further studies will clarify the similarities and differences between BZR1 and BES1 at the biochemical and physiological levels.
Conclusion and prospect
Our understanding of BR signal transduction has greatly improved in recent years. The demonstration of BR perception by BRI1 and BR-induced BRI1-BAK1dimerization, the identification of phosphorylation sites and an autoregulatory domain of BRI1, and the discovery of DNA binding activities of BZR1 and BES1 have established the BR pathway as one of the best-understood signal transduction pathways in plants. Yet there are still many questions to be answered. The major gap in our knowledge of the BR signaling cascade is between BRI1/BAK1 and BIN2. The mechanisms by which BR signaling regulates BIN2 kinase and the BSU1 phosphatase will be a focus of future studies. In addition, the components that regulate the degradation or localization of phosphorylated BZR1 and BES1 are yet to be identified. While genetic studies have been fruitful in identifying the major components of the pathway, biochemical approaches are likely to play an increasingly important role in revealing the details and filling the gaps of the BR signaling pathway in future studies.
References
Mitchell JW, Mandava NB, Worley JF, Plimmer JR, Smith MV . Brassins: A new family of plant hormones from rape pollen. Nature 1970; 225:1065–1066.
Grove MD, Spencer GF, Rohwedder WK, et al. Brassinolide, a plant growth-promoting steroid isolated from Brassica napus pollen. Nature 1979; 281:216–217.
Mandava NB . Plant growth-promoting brassinosteroids. Ann Rev Plant Physiol Plant Mol Bio 1988; 39:23–52.
Clouse SD, Sasse JM . BRASSINOSTEROIDS: essential regulators of plant growth and development. Annu Rev Plant Physiol Plant Mol Biol 1998; 49:427–451.
Ohashi-Ito K, Fukuda H . HD-zip III homeobox genes that include a novel member, ZeHB-13 (Zinnia)/ATHB-15 (Arabidopsis), are involved in procambium and xylem cell differentiation. Plant Cell Physiol 2003; 44:1350–1358.
Symons GM, Davies C, Shavrukov Y, et al. Grapes on steroids. Brassinosteroids are involved in grape berry ripening. Plant Physiol 2006; 140:150–158.
Li J, Nagpal P, Vitart V, McMorris TC, Chory J . A role for brassinosteroids in light-dependent development of Arabidopsis. Science 1996; 272:398–401.
Szekeres M, Nemeth K, Koncz-Kalman Z, et al. Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell 1996; 85:171–182.
Clouse SD . Molecular genetic studies confirm the role of brassinosteroids in plant growth and development. Plant J 1996; 10:1–8.
Clouse SD, Langford M, McMorris TC . A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol 1996; 111:671–678.
Li J, Chory J . A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 1997; 90:929–938.
Friedrichsen DM, Joazeiro CA, Li J, Hunter T, Chory J . Brassinosteroid-insensitive-1 is a ubiquitously expressed leucine-rich repeat receptor serine/threonine kinase. Plant Physiol 2000; 123:1247–1256.
Vert G, Nemhauser JL, Geldner N, Hong F, Chory J . Molecular mechanisms of steroid hormone signaling in plants. Annu Rev Cell Dev Biol 2005; 21:177–201.
He Z, Wang ZY, Li J, et al. Perception of brassinosteroids by the extracellular domain of the receptor kinase BRI1. Science 2000; 288:2360–2363.
Wang ZY, Seto H, Fujioka S, Yoshida S, Chory J . BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature 2001; 410:380–383.
Li J, Lease KA, Tax FE, Walker JC . BRS1, a serine carboxypeptidase, regulates BRI1 signaling in Arabidopsis thaliana. Proc Natl Acad Sci U S A 2001; 98:5916–5921.
Yang XH, Xu ZH, Xue HW . Arabidopsis membrane steroid binding protein 1 is involved in inhibition of cell elongation. Plant Cell 2005; 17:116–131.
Kinoshita T, Cano-Delgado A, Seto H, et al. Binding of brassinosteroids to the extracellular domain of plant receptor kinase BRI1. Nature 2005; 433:167–171.
Cano-Delgado A, Yin Y, Yu C, et al. BRL1 and BRL3 are novel brassinosteroid receptors that function in vascular differentiation in Arabidopsis. Development 2004; 131:5341–5351.
Zhou A, Wang H, Walker JC, Li J . BRL1, a leucine-rich repeat receptor-like protein kinase, is functionally redundant with BRI1 in regulating Arabidopsis brassinosteroid signaling. Plant J 2004; 40:399–409.
Wang X, Goshe MB, Soderblom EJ, et al. Identification and functional analysis of in vivo phosphorylation sites of the Arabidopsis BRASSINOSTEROID-INSENSITIVE1 receptor kinase. Plant Cell 2005; 17:1685–1703.
Oh MH, Ray WH, Huber SC, et al. Recombinant brassinosteroid insensitive 1 receptor-like kinase autophosphorylates on serine and threonine residues and phosphorylates a conserved peptide motif in vitro. Plant Physiol 2000; 124:751–766.
Nam KH, Li J . BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 2002; 110:203–212.
Li J, Wen J, Lease KA, et al. BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 2002; 110:213–222.
Russinova E, Borst JW, Kwaaitaal M, et al. Heterodimerization and endocytosis of Arabidopsis brassinosteroid receptors BRI1 and AtSERK3 (BAK1). Plant Cell 2004; 16:3216–3229.
Massague J . TGF-beta signal transduction. Annu Rev Biochem 1998; 67:753–791.
Huse M, Muir TW, Xu L, et al. The TGF beta receptor activation process: an inhibitor- to substrate-binding switch. Mol Cell 2001; 8:671–682.
Ehsan H, Ray WK, Phinney B, et al. Interaction of Arabidopsis BRASSINOSTEROID-INSENSITIVE 1 receptor kinase with a homolog of mammalian TGF-beta receptor interacting protein. Plant J 2005; 43:251–261.
Wang X, Li X, Meisenhelder J, et al. Autoregulation and homodimerization are involved in the activation of the plant steroid receptor BRI1. Dev Cell 2005; 8:855–865.
Nam KH, Li J . The Arabidopsis transthyretin-like protein is a potential substrate of BRASSINOSTEROID-INSENSITIVE 1. Plant Cell 2004; 16:2406–2417.
Shiu SH, Karlowski WM, Pan R, et al. Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell 2004; 16:1220–1234.
Clay NK, Nelson T . VH1, a provascular cell-specific receptor kinase that influences leaf cell patterns in Arabidopsis. Plant Cell 2002; 14:2707–2722.
Shah K, Gadella TW Jr, van Erp H, Hecht V, de Vries SC . Subcellular localization and oligomerization of the Arabidopsis thaliana somatic embryogenesis receptor kinase 1 protein. J Mol Biol 2001; 309:641–655.
Zhao DZ, Wang GF, Speal B, Ma H . The excess microsporocytes1 gene encodes a putative leucine-rich repeat receptor protein kinase that controls somatic and reproductive cell fates in the Arabidopsis anther. Genes Dev 2002; 16:2021–2031.
Albrecht C, Russinova E, Hecht V, Baaijens E, de Vries S . The Arabidopsis thaliana SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASES1 and 2 control male sporogenesis. Plant Cell 2005; 17:3337–3349.
Karlova R, Boeren S, Russinova E, et al. The Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE1 Protein Complex Includes BRASSINOSTEROID-INSENSITIVE1. Plant Cell 2006; 18:626–638.
Yamamuro C, Ihara Y, Wu X, et al. Loss of function of a rice brassinosteroid insensitive1 homolog prevents internode elongation and bending of the lamina joint. Plant Cell 2000; 12:1591–1606.
Nomura T, Bishop GJ, Kaneta T, et al. The LKA gene is a BRASSINOSTEROID INSENSITIVE 1 homolog of pea. Plant J 2003; 36:291–300.
Montoya T, Nomura T, Farrar K, et al. Cloning the tomato curl3 gene highlights the putative dual role of the leucine-rich repeat receptor kinase tBRI1/SR160 in plant steroid hormone and peptide hormone signaling. Plant Cell 2002; 14:3163–3176.
Wang ZY, He JX . Brassinosteroid signal transduction--choices of signals and receptors. Trends Plant Sci 2004; 9:91–96.
Nakamura A, Fujioka S, Sunohara H, et al. The role of OsBRI1 and its homologous genes, OsBRL1 and OsBRL3 in rice. Plant Physiol 2006; 140:580–590.
Wang ZY, Nakano T, Gendron J, et al. Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev Cell 2002; 2:505–513.
He JX, Gendron JM, Sun Y, et al. BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science 2005; 307:1634–1638.
Yin Y, Wang ZY, Mora-Garcia S, et al. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 2002; 109:181–191.
Yin Y, Vafeados D, Tao Y, et al. A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell 2005; 120:249–259.
He JX, Gendron JM, Yang Y, Li J, Wang ZY . The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis. Proc Natl Acad Sci U S A 2002; 99:10185–10190.
Mora-Garcia S, Vert G, Yin Y, et al. Nuclear protein phosphatases with Kelch-repeat domains modulate the response to brassinosteroids in Arabidopsis. Genes Dev 2004; 18:448–460.
Peng P, Li J . Brassinosteroid Signal Transduction: A Mix of Conservation and Novelty. J Plant Growth Regul 2003; 22:298–312.
Cohen P, Frame S . The renaissance of GSK3. Nat Rev Mol Cell Biol 2001; 2:769–776.
Jope RS, Johnson GV . The glamour and gloom of glycogen synthase kinase-3. Trends Biochem Sci 2004; 29:95–102.
Bijur GN, Jope RS . Glycogen synthase kinase-3 beta is highly activated in nuclei and mitochondria. Neuroreport 2003; 14:2415–2419.
Li J, Nam KH, Vafeados D, Chory J . BIN2, a new brassinosteroid-insensitive locus in Arabidopsis. Plant Physiol 2001; 127:14–22.
Li J, Nam KH . Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science 2002; 295:1299–1301.
Perez-Perez JM, Ponce MR, Micol JL . The UCU1 Arabidopsis gene encodes a SHAGGY/GSK3-like kinase required for cell expansion along the proximodistal axis. Dev Biol 2002; 242:161–173.
Li J . Brassinosteroid signaling: from receptor kinases to transcription factors. Curr Opin Plant Biol 2005; 8:526–531.
Goda H, Sawa S, Asami T, et al. Comprehensive comparison of auxin-regulated and brassinosteroid-regulated genes in Arabidopsis. Plant Physiol 2004; 134:1555–1573.
Nemhauser JL, Mockler TC, Chory J . Interdependency of brassinosteroid and auxin signaling in Arabidopsis. PLoS Biol 2004; 2:E258.
Acknowledgements
This work was supported in part by grants from National Natural Science Foundation of China (No. 30328004, No.30571269), National Institutes of Health (R01 GM66258-01).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wang, ZY., Wang, Q., Chong, K. et al. The brassinosteroid signal transduction pathway. Cell Res 16, 427–434 (2006). https://doi.org/10.1038/sj.cr.7310054
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.cr.7310054