ABSTRACT
A developmentally retarded mutant (drm1) was identified from ethyl methanesulfonate (EMS)-mutagenized M2 seeds in Columbia (Col-0) genetic background. The drm1 flowers 109 d after sowing, with a whole life cycle of about 160 d. It also shows a pleiotropic phenotype, e.g., slow germination and lower germination rate, lower growth rate, curling leaves and abnormal floral organs. The drm1 mutation was a single recessive nuclear mutation, which was mapped to the bottom of chromosome 5 and located within a region of 20-30 kb around MXK3.1. There have been no mutants with similar phenotypes reported in the literature, suggesting that DRM1 is a novel flowering promoting locus. The findings that the drm1 flowered lately under all photoperiod conditions and its late flowering phenotype was significantly restored by vernalization treatment suggest that the drm1 is a typical late flowering mutant and most likely associated with the autonomous flowering pathway. The conclusion was further confirmed by the revelation that the transcript level of FLC was constantly upregulated in the drm1 at all the developmental phases examined, except for a very early stage. Moreover, the transcript levels of two other important repressors, EMF and TFL1, were also upregulated in the drm1, implying that the two repressors, along with FLC, seems to act in parallel pathways in the drm1 to regulate flowering as well as other aspects of floral development in a negatively additive way. This helps to explain why the drm1 exhibits a much more severe late-flowering phenotype than most late-flowering mutants reported. It also implies that the DRM1 might act upstream of these repressors.
Similar content being viewed by others
INTRODUCTION
The transition from vegetative growth to reproduction is one of the most important developmental events in flowering plants since it is related to the competence and survivability of a particular species living in a particular environment. The flowering time, as the phenotypic indicator of this transition, is either induced by environmental factors or regulated by endogenous signals. Four major pathways controlling flowering time have been defined in Arabidopsis 1, 2, 3, 4.
The photoperiod flowering pathway regulates flowering time by responding to long-day conditions. Late-flowering mutants in this pathway flower late under long-day conditions but similarly or even identically to the wild type under short days. These mutants are weakly, or not at all, sensitive to vernalization. CO, CRY2, FHA, GI, FT and FWA are some components characteristic in this pathway 5. Regulation of flowering time in response to seasonal day length fluctuations is mediated by the interactions between light signals and intrinsic time-keeping mechanisms that are associated with the circadian clock 6, 7. CRY2 and PHYA, candidates for photoreceptors that perceive the photoperiod under long days, entrain the circadian clock to oscillate within a period of 24 h 8, 9. The components of the oscillator include TOC1, CCA1 and LHY genes, which are also implicated in the photoperiodic induction of flowering 10, 11.
The Gibberellin (GA) promotes flowering and is absolutely required for flowering in non-inductive short days. Mutations in genes involved in GA biosynthesis and signaling result in delayed flowering. A growing number of genes associated with this pathway have been identified, including SPY, PHOR1, RGLs, RGA, GAI, GA4, GA5 and FPF1 2, 12.
The vernalization responsive pathway regulates flowering in response to extended exposure to cold temperature (vernalization). Many naturally occurring mutants flower very late but flower early if exposed to low temperatures for 4 to 8 w. The vernalization response is mediated by dominant alleles of two genes, FRI and FLC, through reducing the expression of FLC 13, 14. Other genes involved in vernalization response include VRN1 and VRN2 15, 16, HOS1 17 and VIP1-7 18.
The autonomous pathway is defined by one group of late flowering mutants, such as fca, fpa, fve, fld, ld, and fy, which flower late under both long-day and short-day conditions. However, the late flowering phenotypes of these mutants can be overcome by vernalization or exposure to far red-enriched light 5. The characteristic feature of these mutants is that they all contain much higher levels of FLC transcript than the wild-type plant or late-flowering mutants associated with the photoperiod flowering pathway or the GA pathway 13, 19, 20.
Different flowering time pathways are known to interconnect and converge on the activation of the same flowering-time genes, which are termed as the flowering-time pathway integrators. Thus far, three genes have been identified: FT, SOC1 (AGL20) and LFY 21, 22, 23. These floral integrators then activate the expression of the downstream floral organ identity genes, AP3, PI and AG, leading to the development of floral organs 24, 25, 26, 27.
A large number of genes, functioning as floral repressors, have also been identified from early flowering mutants, including EMF1 and 2, TFL1 and 2, CLF, EBS1, EFS, ELF3, ELF4 ELF5, ESD4, FIE, SYD,SVP and TOE 28, 29, 30, 31. These floral repressor genes interconnect with the network of flowering pathways in one way or another to regulate flowering negatively. For example, ESD4 is involved in the autonomous floral promotion pathway 29, 32 and SVP interacts with the photoperiod pathway 33. Moreover, some of the repressor genes, such as EMF and TFL, also affect the development of inflorescences and floral organs in Arabidopsis 34, 35, 36.
In this study, a developmentally retarded mutant (drm1) was isolated in Arabidopsis, which flowers extremely late and shows a pleiotropic phenotype. DRM1 was located within a region of 20-30 kb around MXK3.1 on the bottom of chromosome 5 and appears to be a novel flowering promoting locus. It was further defined as a novel component of the autonomous flowering pathway. Repressors FLC, EMF and TFL1 were also found to be involved in the regulation of the severe phenotype of late flowering as well as floral abnormality in the drm1.
MATERIALS AND METHODS
Plant materials and growth conditions
All the wild-type and mutant Arabidopsis lines used in this study are in a Columbia (Col-0) background unless indicated otherwise. When grown in soil pots, seeds were sown in square pots (10 cm in length) with soil [v (peat soil): v (vermiculite): v (pearlite) = 3:9:0.5, Shanghai Institute of Landscape Science] presoaked with PNS medium. Plants were grown in a controlled room with 22 ± 2°C temperature and ∼100 μmol m-2 s-1 light intensity under standard long-day conditions (16 h light/8 h dark).
For photoperiod treatment, plants on soil pots were grown under various photoperiod conditions, including 24 h continuous light, 16 h light/ 8 h dark, 8 h light/ 16 h dark and 4 h light/20 h dark.
For vernalization treatment, imbibed seeds were placed at 4°C in the dark for 30 d before they were transferred to long-day conditions, as described above, whereas untreated seeds were kept at room temperature for 28 d and then sowed on pots and placed at 4°C in the dark for2 d before being transferred to long-day conditions.
Isolation and characterization of the drm1 mutant
Ethyl methanesulfonate (EMS) mutagenized M2 seeds in Col-0 genetic background were purchased from Lehle Seeds (Round Rock, TX, USA). For mutant screening, M2 seeds were sown on water-soaked soil, and treated at 4°C for 2 d before being transferred to the growth room. The drm1 mutant was isolated from M2 population for its abnormal phenotype. The phenotype of the drm1 was stable and reproducible in M3 and onward generations under the growth conditions described above.
The drm1 plants were backcrossed with the wild type for 3 times, and the resulting homozygous drm1 plants were used for all the analysis. The phenotype of the drm1 mutant was characterized under long-day photoperiod. Bolting time was measured as days from seed sowing to the first flower bud emerging while flowering time was recorded to the first flower opening. Rosette leaves were counted when a visible inflorescence of ∼3 cm was apparent.
Genetic analysis and mapping of drm1 locus
For genetic analysis, drm1 plants were crossed with wild-type plants reciprocally, and the resulting F1 seedlings were allowed to self-pollinate to produce F2 populations. The F1 and F2 seedlings were scored for either mutant or wild phenotype.
For mutation locus mapping, homozygous drm1 plants were crossed to wild-type plants in Landsberg background. From the segregating F2 population, 2300 homozygous drm1 plants were selected to make a mapping population, and DNA was extracted from each of these plants. The linkage between the mutation locus and molecular markers was determined by using simple sequence length polymorphism (SSLP) markers 37. For fine mapping, we designed a set of novel SSLP markers by using the Cereon arabidopsis polymorphism collection (Tab. 1). Mapping procedure was performed as described in Lukowitz and Jander 38, 39. All the primers were synthesized by TaKaRa Biotechnology Co.
Total RNA extraction and semi-quantitative RT-PCR
Total RNA was extracted from mixed rosette leaves of wild-type as well as drm1 plants using the TRI reagent (Invitrogen). First-stand cDNA synthesis was performed with 3 μg total RNA using a SuperScript kit (Gibco BRL), and the products were standardized for semi-quantitative RT-PCR using β-actin11 as a control.
The semi-quantitative RT-PCR was adopted to monitor the change in transcript levels of flowering time genes. Gene-specific primers and the amplification cycles used were as for each gene are as supplied in Supplemental Tab. 1. PCR conditions were as follows: 5 min at 94°C, then 20-40 cycles of 94°C for 45 sec, 55°C for 90 sec, 72°C for 45 sec, and then 72°C for 5 min. The amplified fragments were separated on a 1.2% agarose gel. Reproducible expression patterns were obtained for each of the genes with RNA samples extracted from different batches of seedlings.
RESULTS
Isolation and phenotypic characterization of the drm1 mutant
In a screening for stay-green mutants using a dark stressed approach, a number of developmentally retarded and / or late-flowering mutants were obtained from ethyl methane sulphonate (EMS) mutagenized M2 seeds of Columbia (Col) ecotype of Arabidopsis thaliana. Of them, an extremely slow-developing mutant, designated drm1 (developmentally retarded mutant) was selected for further analysis.
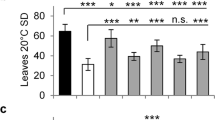
Under long-day conditions, all the developmental phases of the drm1 mutant are severely retarded, and a period of 109 d is needed from sowing to flowering compared to 42 d for the wild type. Consequently, the plant size of the mutant is much smaller at the early stage of development, but becomes comparable to that of the wild type towards flowering time. In fact it can produce as many as 58 rosette leaves compared to 24 of the wild-type at the time of bolting due to the extended period of vegetative growth. Besides, the process of senescence at plant level is also significantly delayed, and therefore an extremely long lifespan was recorded (Tab. 2 and Fig. 1).
Phenotype of the drm1. (A) A plantlet of the drm1 and (B) a plant of the wild-type grown under long-day conditions in soil supplemented with PNS medium 40 d after sowing. (C) A plant of the drm1 (left) and a plant of the wild-type (right) 48 d after sowing. (D) A plant of the drm1 with a visible flower bud and increased number of rosette leaves 90 d after sowing. (E) A plant of the drm1 (124 d-old) showing more multiple inflorescences compared to those of the (F) wild-type (60 d-old) at the same developmental phase.
The mutation displays a pleiotropic phenotype. Its germination rate is significantly lower and its germination speed is 3-4 d slower compared to the wild-type (Tab. 2). Its rosette leaves, thick and pale green at the seedling stage (Fig. 1A), curl up after fully-grown (Fig. 1D). Furthermore, the drm1 produces significantly more inflorescences, particularly more secondary, tertiary and quaternary ones (Fig. 1E, F; Fig. 2E). Although the inflorescences of the drm1 bear more flowers, the fertilities of many flowers are severely reduced due to unusual shorter stamens, which result in a number of infertile siliques and / or seeds (Fig. 2A, B). Abnormal numbers of petals (5 or 6) and stamens (5 or 3) were also observed (Fig. 2 C, D).
Floral and inflorescence characteristics of the drm1. (B) A primary inflorescence of the drm1 showing aberrantly developed siliques in comparison with that of the (A) wild-type. (D) A flower of the drm1 showing altered number of petals and abnormally shorter stamens compared to those of the (C) wild-type. (E) A graph displaying increased numbers of inflorescences of the drm1 at different hierarchical levels.
To analyze the inheritance of the drm1 mutation, crosses between drm1 plants and wild-type plants were made reciprocally. No mutant phenotype was observed in the resulting F1 plants and a 1 (drm1) to 3 (WT) segregation ratio was revealed in all F2 populations. Considering the differential germination rates between the drm1 and the wild type, χ2 test was conducted again based on the converted data and no substantial inconsistency was found between the two calculations (Tab. 3). These results strongly suggest that the mutant phenotype was caused by a single, recessive nuclear mutation.
Mapping of the drm1 locus
The DRM1 locus was mapped to the bottom of chromosome 5, flanked by the MBK5 (121.65cM) and MQN 23.1 (127.31cM), using SSLP markers 37. New SSLP markers were designed within this region for fine mapping using a mapping population of 2300 plants. The locus was further located within an interval of 80 kb between MXK3.2 and F1505.1 markers. As no recombinants were detected by using MXK3.1 marker, we speculated that the chromosomal location of the DRM1 should be within a region of 20-30 kb around MXK3.1. Sequencing the genomic DNA within this region is under way (Fig. 3).
Physiological and molecular characterization of the late flowering mutant phenotype
As the late flowering is the most obvious characteristics of the drm1, experiments were designed to characterize the late flowering mutant phenotype physiologically and molecularly.
Vernalization treatments were carried out as described in Materials and methods. A 30-d vernalization treatment significantly reduced the time needed for flowering in the drm1 mutant. Treated drm1 plants flowered on 75.7 d averagely whereas untreated flowered on 110.5 d after being transferred to normal long-day conditions from 4°C. Although not fully restored, the late flowering phenotype of the drm1 was significantly overcome by the vernalization treatment (Fig. 4).
Effect of vernalization treatment on the flowering time of the drm1. (A) The late flowering phenotype of the drm1 was significantly restored (left) by a 30 d vernalization treatment compared to that of untreated (right), both of which were grown under long-day conditions for 100 d. (B) A graph showing the effect of vernalization treatment on the flowering time of both the drm1 and the wild-type (WT).
Responses of the drm1 to different photoperiods were also determined in order to further define its late flowering phenotype. Although the wild-type always flowered earlier than the drm1 under all the photoperiods examined, both the drm1 and the wild-type exhibited photoperiod responses and delayed flowering as the period of illumination was shortened. The drm1 flowered 98.9 and 295.6 d after sowing under the photoperiods of 24 h continuous illumination and 4 h illumination/20 h dark, respectively (Fig. 5).
To understand the molecular mechanism underlying the late flowering phenotype of the drm1, transcript levels of some important genes known to play important roles in flowering pathways were examined using the semi-quantitative RT-PCR. It was found that the transcript levels of all the examined integrator genes, SOC1 (AGL20), FT and LFY, were significantly down-regulated in the drm1 compared with those in the wild-type at the similar phase of vegetative growth, whereas those of the flowering repressor genes, FLC, EMF1, EMF2 and TFL1, were significantly up-regulated (Fig. 6). The transcript levels of the above repressor genes were all found to be markedly up-regulated at all examined phases (vegetative, transitional and flowering) in the drm1 compared with those in the wild-type, except that that of the FLC was unexpectedly detected to be down-regulated in the 20 d-old drm1 (Fig. 7). These results strongly suggest that the DRM1 mutation do affect the expression of flowering genes and the drm1 can therefore be considered as a late flowering mutant. Considering the finding that a significant up-regulation of FLC expression was detected in most developmental phases, it was postulated that the mutation was most likely associated with the autonomous flowering pathway. This postulation is in accordance with the finding that no significant differences were detected in the transcript levels of other examined flowering genes, such as GI, CO (components of photoperiod flowering pathway), SPY (a component of the GA flowering pathway), FCA and LD (components of the autonomous flowering pathway upstream of FLC), which are all characteristic components of other flowering pathways (Fig. 6).
DISCUSSION
The phenotype of the drm1 is comparable to those of extended lifespan mutants in other model organisms
The flowering time is an important characteristic of a species in plants, and flowering pathways guaranteeing flowering at the right time under particular conditions have been identified both intensively and extensively in Arabidopsis thalinia. Mutations in genes involved in these flowering pathways result in either early flowering or late flowering 1. The flowering time in plants is a trait considered to be equivalent to the lifespan in animals, and a list of lifespan extending or shortening mutants resulted from the mutations of a conserved pathway have also been observed in other model organisms 40. Interestingly, the drm1, along with a large number of other late flowering mutants, shares a similar pleiotropic phenotype with most lifespan extending animal mutants, such as reduced fertilities, small sizes and lower growth rates, indicating that these crucial life processes are evolutionarily linked through certain signal pathways 40.
The DRM1 is likely a novel flowering promoting locus involved in the autonomous flowering pathway
To our best knowledge, no mutants with a similar phenotype to that of the drm1 have been reported in the literature. According to the rates of recombinants, the DRM1 mutation is located to an interval of 79.9 kb between MXK3.2 and F1505.1 markers on the bottom of chromosome 5. So far, no known genes in flowering pathways have been reported to lie in this region. These results indicate that the DRM1 is very likely a novel flowering promoting locus. However, a final conclusion cannot be made until the detection of the candidate gene and the completion of transgenic complementation.
Diverse flowering pathways have been elucidated through characterizing groups of distinctive mutants and cloning the related genes. The autonomous flowering pathway is defined by those mutants that flower late under both long-day and short-day conditions and by their responsiveness to vernalization 5. Molecularly, they all contain much higher levels of FLC transcript than the wild-type plant or late-flowering mutants associated with the photoperiod flowering pathway or the GA pathway 13, 19, 20. Our results of vernalization and photoperiod treatments, as well as part result of the RT-PCR, strongly suggest that the drm1 is a typical late flowering mutant and most likely associated with the autonomous pathway. The conclusion is further confirmed by the finding that the transcript level of FLC gene is constantly up-regulated while those of SOC1 (AGL20), FT and LEY genes are down-regulated in the drm1. In addition, no detection of significant differences in the transcript levels of GI, CO, SPY, FCA and LD helps to exclude other possible pathways associated. Elucidating the mechanism of how the DRM1 works together with other known components in the autonomous flowering pathway is under way.
The phenotypic severity of late flowering as well as vegetative growth and floral development in the drm1 is likely associated with the coordinate action of three repressors, FLC, EMF and TFL1
As indicated previously, FLC is a key repressor component involved in both the autonomous pathway and the vernerization pathway. EMF down regulates most of the flower organ genes and thus probably acts via global repression of the flower program rather than on a single flowering or flower organ identity gene 29. TFL1 functions to suppress flower formation at the apex and to delay the transition from vegetative to reproductive development 1, 36, 41. It has been proposed that TFL1 acts by influencing a central mechanism controlling the identity of shoot apical meristem and consequently by preventing the expression of floral meristem identity genes, such as AP1, LFY and CAL at all stages of development throughout the life cycle 35, 36. The over-expression of TFL1 greatly extends the vegetative and inflorescence growth phases, resulting in more highly branched plants which form flowers much late than the wild type 36, 41.
It has been demonstrated that FLC, EMF and TFL1 function in parallel pathways to regulate floral development and subsequent flowering process 29, 36. In our study, it was showed that the transcript levels of all the three important repressors were constantly up-regulated in the drm1 at all developmental phases, except for that of FLC in 20 d-old seedlings. It is probably due to some kind of inactivation of the FLC gene in such an early stage seedlings harvested for RNA extraction. These findings suggest that the DRM1 is likely a flowering activator through suppressing the repressors to control flowering time and to regulate the development of architectures of inflorescence and flower. The coordinate action of the three repressors in a negatively additive way helps to explain the much more severe phenotype of the drm1 than those of other related late flowering mutants. However, more genetic evidence is needed to establish the exact relationship of the DRM1 to these repressors as well as to other components of the autonomous flowering pathway.
References
Levy YY, Dean C . Control of flowering time. Curr Opin Plant Biol 1998; 1:49–54.
Mouradov A, Cremer F, Coupland G . Control of flowering time: interacting pathways as a basis for diversity. Plant Cell 2002; 14 Suppl: S111–30.
Simpson GG, Dean C . Arabidopsis, the Rosetta stone of flowering time? Science 2002; 296:285–9.
Boss PK, Bastow RM, Mylne JS, Dean C . Multiple Pathways in the Decision to Flower: Enabling, Promoting, and Resetting. Plant Cell 2004; 16:S18–31.
Koornneef M, Hanhart CJ, van der Veen JH . A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol Gen Genet 1991; 229:57–66.
Doyle MR, Davis SJ, Bastow RM, et al. The ELF4 gene controls circadian rhythms and flowering time in Arabidopsis thaliana. Nature 2002; 419:74–7.
Hayama R, Coupland G . Shedding light on the circadian clock and the photoperiodic control of flowering. Curr Opin Plant Biol 2003; 6:13–9.
Lin C . Plant blue-light receptors. Trends Plant Sci 2000; 5:337–42.
Mockler T, Yang H, Yu X, et al. Regulation of photoperiodic flowering by Arabidopsis photoreceptors. Proc Natl Acad Sci U S A 2003; 100:2140–5.
Schaffer R, Ramsay N, Samach A, et al. The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell 1998; 93:1219–29.
Wang ZY, Tobin EM . Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression.Cell 1998; 93:1207–17.
Olszewski N, Sun TP, Gubler F . Gibberellin signaling: biosynthesis, catabolism, and response pathways. Plant Cell 2002; 14:Suppl: S61–80.
Michaels SD, Amasino RM . FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 1999; 11:949–56.
Michaels SD, Amasino RM . Loss of FLOWERING LOCUS C activity eliminates the late-flowering phenotype of FRIGIDA and autonomous pathway mutations but not responsiveness to vernalization. Plant Cell 2001; 13:935–41.
Gendall AR, Levy YY, Wilson A, Dean C . The VERNALIZATION 2 gene mediates the epigenetic regulation of vernalization in Arabidopsis. Cell 2001; 107:525–35.
Chandler J, Wilson A, Dean C . Arabidopsis mutants showing an altered response to vernalization. Plant J 1996; 10:637–44.
Lee H, Xiong L, Gong Z, et al. The Arabidopsis HOS1 gene negatively regulates cold signal transduction and encodes a RING finger protein that displays cold-regulated nucleo—cytoplasmic partitioning. Genes Dev 2001; 15:912–24.
Zhang H, van Nocker S . The VERNALIZATION INDEPENDENCE 4 gene encodes a novel regulator of FLOWERING LOCUS C. Plant J 2002; 31:663–73.
Sheldon CC, Burn JE, Perez PP, et al. The FLF MADS box gene: a repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell 1999; 11:445–58.
Sheldon CC, Finnegan EJ, Rouse DT, et al.The control of flowering by vernalization. Curr Opin Plant Biol 2000; 3:418–22.
Kobayashi Y, Kaya H, Goto K, Iwabuchi M, Araki T . A pair of related genes with antagonistic roles in mediating flowering signals. Science 1999; 286:1960–2.
Blazquez MA, Weigel D . Independent regulation of flowering by phytochrome B and gibberellins in Arabidopsis. Plant Physiol 1999; 120:1025–32.
Lee H, Suh SS, Park E, et al. The AGAMOUS-LIKE 20 MADS domain protein integrates floral inductive pathways in Arabidopsis. Genes Dev 2000; 14:2366–76.
Blazquez MA, Weigel D . Integration of floral inductive signals in Arabidopsis. Nature 2000; 404:889–92.
Busch MA, Bomblies K, Weigel D . Activation of a floral homeotic gene in Arabidopsis. Science 1999; 285:585–7.
Krizek BA, Meyerowitz EM . The Arabidopsis homeotic genes APETALA3 and PISTILLATA are sufficient to provide the B class organ identity function. Development 1996; 122:11–22.
Wagner D, Sablowski RW, Meyerowitz EM . Transcriptional activation of APETALA1 by LEAFY. Science 1999; 285:582–4.
Noh YS, Bizzell CM, Noh B, Schomburg FM, Amasino RM . EARLY FLOWERING 5 acts as a floral repressor in Arabidopsis. Plant J 2004; 38:664–72.
Sung ZR, Chen L, Moon YH, Lertpiriyapong K . Mechanisms of floral repression in Arabidopsis. Curr Opin Plant Biol 2003; 6:29–35.
Hartmann U, Hohmann S, Nettesheim K, et al. Molecular cloning of SVP: a negative regulator of the floral transition in Arabidopsis. Plant J 2000; 21:351–60.
Aukerman MJ, Sakai H . Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes. Plant Cell 2003; 15:2730–41.
Reeves PH, Murtas G, Dash S, Coupland G . early in short days 4, a mutation in Arabidopsis that causes early flowering and reduces the mRNA abundance of the floral repressor FLC. Development 2002; 129:5349–61.
Scortecci K, Michaels SD, Amasino RM . Genetic interactions between FLM and other flowering-time genes in Arabidopsis thaliana. Plant Mol Biol 2003; 52:915–22.
Chen L, Cheng JC, Castle L, Sung ZR . EMF genes regulate Arabidopsis inflorescence development. Plant Cell 1997; 9:2011–24.
Bradley D, Ratcliffe O, Vincent C, Carpenter R, Coen E . Inflorescence commitment and architecture in Arabidopsis. Science 1997; 275:80–3.
Ratcliffe OJ, Amaya I, Vincent CA, et al. A common mechanism controls the life cycle and architecture of plants. Development 1998; 125:1609–15.
Bell CJ, Ecker JR . Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 1994; 19:137–44.
Lukowitz W, Gillmor CS, Scheible WR . Positional cloning in Arabidopsis. Why it feels good to have a genome initiative working for you. Plant Physiol 2000; 123:795–805.
Jander G, Norris SR, Rounsley SD, et al. Arabidopsis map-based cloning in the post-genome era. Plant Physiol 2002; 129:440–50.
Kenyon C . A conserved regulatory system for aging. Cell 2001; 105:165–8.
Ratcliffe OJ, Bradley DJ, Coen ES . Separation of shoot and floral identity in Arabidopsis. Development 1999; 126:1109–20.
Acknowledgements
The work was supported by a National Science Foundation of China grant (39870452) to Ben Ke Kuai. Mutant screening was conducted at Dong Lab, DCMB group, Department of Biology, Duke University. We are grateful to Profs. Dong X and Huang H for their valuable advices and help. We would also like to thank Zhang Q, Zhang W and Sung WJ for their technical assistances.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Rights and permissions
About this article
Cite this article
ZHU, Y., ZHAO, H., REN, G. et al. Characterization of a novel developmentally retarded mutant (drm1) associated with the autonomous flowering pathway in Arabidopsis. Cell Res 15, 133–140 (2005). https://doi.org/10.1038/sj.cr.7290278
Received:
Revised:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/sj.cr.7290278
Keywords
This article is cited by
-
Overexpression of the CmJAZ1-like gene delays flowering in Chrysanthemum morifolium
Horticulture Research (2021)
-
Research progress on the autonomous flowering time pathway in Arabidopsis
Physiology and Molecular Biology of Plants (2017)
-
Cloning and characterization of a Doritaenopsis hybrid PRP39 gene involved in flowering time
Plant Cell, Tissue and Organ Culture (PCTOC) (2012)
-
Isolation and characterization of the FVE gene of a Doritaenopsis hybrid involved in the regulation of flowering
Plant Growth Regulation (2012)
-
A Pleiotropic Phenotype is Associated with Altered Endogenous Hormone Balance in the Developmentally Stunted Mutant (dsm1)
Journal of Plant Biology (2010)