Abstract
Circular RNAs (circRNAs) have been identified from various tissues and species, but their regulatory functions during developmental processes are not well understood. We examined circRNA expression profiles of two developmental stages of bovine skeletal muscle (embryonic and adult musculus longissimus) to provide first insights into their potential involvement in bovine myogenesis. We identified 12 981 circRNAs and annotated them to the Bos taurus reference genome, including 530 circular intronic RNAs (ciRNAs). One parental gene could generate multiple circRNA isoforms, with only one or two isoforms being expressed at higher expression levels. Also, several host genes produced different isoforms when comparing development stages. Most circRNA candidates contained two to seven exons, and genomic distances to back-splicing sites were usually less than 50 kb. The length of upstream or downstream flanking introns was usually less than 105 nt (mean≈11 000 nt). Several circRNAs differed in abundance between developmental stages, and real-time quantitative PCR (qPCR) analysis largely confirmed differential expression of the 17 circRNAs included in this analysis. The second part of our study characterized the role of circLMO7—one of the most down-regulated circRNAs when comparing adult to embryonic muscle tissue—in bovine muscle development. Overexpression of circLMO7 inhibited the differentiation of primary bovine myoblasts, and it appears to function as a competing endogenous RNA for miR-378a-3p, whose involvement in bovine muscle development has been characterized beforehand. Congruent with our interpretation, circLMO7 increased the number of myoblasts in the S-phase of the cell cycle and decreased the proportion of cells in the G0/G1 phase. Moreover, it promoted the proliferation of myoblasts and protected them from apoptosis. Our study provides novel insights into the regulatory mechanisms underlying skeletal muscle development and identifies a number of circRNAs whose regulatory potential will need to be explored in the future.
Similar content being viewed by others
Main
Circular RNAs (circRNAs) were first described in 1991 for rodent and human tumor cells,1 and only few additional circRNAs were uncovered during the following 20 years.2, 3, 4 The covalently closed loop structure of circRNAs—which neither show 5′-to-3′-polarity nor a polyadenylated tail—prevents the application of several analytical methods that are widely used in RNA biology. Non-linear reads were regularly interpreted as aberrant by-products resulting from spliceosome-mediated splicing errors and were not considered in large-scale RNA-sequencing studies.5 It appears as if circRNAs are generated from back-spliced exons, represent intron-derived RNAs. With the advent of RNA deep sequencing technologies and bioinformatic tools enabling the analysis of extensive data-sets, large numbers of circRNAs could be identified in the transcriptomes of a variety of eukaryotic organisms.6, 7, 8, 9, 10 Ribosomal RNA-depleted total RNA libraries (rRNA− libraries) and libraries that were additionally treated with RNase R (rRNA−+RNase R+ libraries) are most frequently used to identify circRNAs.11
Only recently have studies begun to consider the potential function(s) of circRNAs. While our current knowledge is limited, the regulatory functions of some circRNAs have been well characterized. For example, ciRS-7 and Sry inhibit miRNAs in murine tissue by sponging miR-7 and miR-138, suggesting that circRNAs may indeed play important roles in post-transcriptional gene regulation.7, 12 Moreover, circRNAs may sponge RNA-binding proteins. As another example, circMbl is derived from the muscleblind locus (MBL/MBNL1)—a splicing factor in Drosophila melanogaster—and contains MBL binding sites. Interestingly, overexpression of MBL induces circMbl production, and circMbl, in turn, reduces the production of MBL1 messenger RNA (mRNA).13 More recently, a novel subclass of circRNAs, named exon-intron circRNAs (EIciRNAs), has been shown to act as transcriptional regulators in human cells. EIciRNAs are predominantly localized in the nucleus, interact with RNA Polymerase II and U1 small nuclear ribonucleoproteins, and function as cis-inducers of host-gene transcription.14
Our present study was intended to serve as a starting point for future studies examining the role played by circRNAs in bovine muscle development. To this end, we characterized circRNA expression profiles in different developmental stages of bovine muscle tissue. In light of the current problems faced by the Chinese beef cattle industry in terms of a shortage of high-quality beef, we decided to use Chinese Qinchuan cattle for our study, as the results of our study might be implemented in future breeding schemes, in which breeding is informed by molecular information of potential breeding stock. Qinchuan cattle rank among the top five Chinese yellow beef cattle breeds and has an excellent meat quality, good tolerance to roughage feeding and a remarkable resistance to stress.15, 16 The aim of our study was to elucidate the potential role of circRNAs during the process of muscle growth and development in Qinchuan cattle to better understand the molecular mechanisms underlying myogenesis. We generated rRNA− libraries of embryonic and adult muscle samples using the TopHap-Fusion method. In order to reduce false positives arising from the RNA library treatment and the applied method to detect circRNAs, the RNAs of several samples were pooled and afterwards prepared as rRNA−+ RNase R+ libraries to investigate circRNA contents.
Results
Expression profiles of circRNAs in embryonic and adult bovine muscle tissue
We identified a considerable number of RNAs in embryonic and adult bovine muscle tissue when considering total RNA libraries (Table 1). In the libraries that were depleted of ribosomal RNA (rRNA−), we identified 12 981 circRNAs candidates, 1287 of which contained at least one unique back-spliced read (Supplementary Table S2), including 530 circular intronic RNAs (ciRNAs). Only 589 of those candidate circRNAs were detected in the libraries we prepared following the rRNA−+RNase R+ method (Supplementary Table S3). We found 3308 and 7273 circRNAs to be specific to the embryo and adult libraries, respectively (Figure 1b). Our results confirm that RNase R treatment depleted the samples of linear RNAs, thereby increasing the accuracy of circRNA identification. The percentage of mapped sequence reads that could be aligned to exonic regions were markedly lower in embryonic (42%) than adult samples (83%; Figure 1c).
Identification of circular RNAs in bovine skeletal muscle tissue. (a) Workflow for the preparation and analysis of circRNA libraries, (b) Venn diagram depicting different circRNAs uncovered at two developmental stages (embryonic and adult tissues), and (c) origin of circRNAs described in this study in the bovine genome
Standard metrics to characterize the length of circRNAs detected in this study (minimum, maximum, mean and median length, N50-value, and total length) are provided in Table 2. As illustrated in Figure 2a, the length of most circRNAs (n=12,127) was <2000 nucleotides (nt), and the mean length was 822 nt, which was shorter than the mean transcript length of protein-coding genes (2373 nt, Figure 2b).
Profiling of circular RNAs in bovine skeletal muscle. (a) Size distribution of circRNAs and (b) of the respective host genes, (c) Circos plot showing the distribution of circRNAs in different chromosomes, (d) numbers of back-spliced reads in circRNAs, (e–g) lengths of flanking introns, and (h) circRNAs that contained varying numbers of exons. (i) Genomic distances of back-splicing sites of most circRNAs ranged within 50 kb, with only few circRNAs spanning 100–300 kb
We found the genomic loci from which circRNAs are derived to be widely distributed across chromosomes except the Y chromosome (Figure 2c, Supplementary Figure S3). However, the distribution of circRNAs detected in this study was not uniform among different chromosomes, even though there was a general trend that numbers of circRNAs with back-spliced reads per chromosome increased with absolute chromosome length (Figure 2c, Supplementary Figure S3). Notably, the expression levels of most circRNAs (n=12 512) were not higher than 50 back-spliced reads (FPKM, Figure 2d). According to the histogram depicting flanking intron lengths, the length of flanking intron regions of most circRNAs was no longer than 105 nt, and the mean length of upstream or downstream flanking intron regions was about 11,000 nt (Figures 2e–g).
There was no noticeable difference in exon numbers of highly expressed circRNAs (n=9659) between the libraries from embryonic and adult muscle tissues, and most circRNAs contained two to seven exons, while only about 7% of circRNAs contained one exon (Figure 2h). This observation suggests that the formation of circRNAs is regulated by different way. Genomic distances of back-splicing sites were <50 kb in most circRNAs (n=12 116), and only a few back-splicing sites spanned 100–300 kb, suggesting that circRNAa are likely generated within the same gene region, and may arise from RNA splicing throughout development and growth of skeletal muscle (Figure 2i). The latter findings suggest that the formation of circRNAs is regulated through (a) specific pathway(s) and does not merely represent transcriptomic noise, that is, random byproducts of canonical splicing.
Identification of differentially expressed circRNA
On the basis of expression of circRNA analysis, we found 828 circRNAs to be significantly (P<0.05) differently expressed when comparing the libraries derived from embryonic and adult tissues (Supplementary Table S4). The ten most up-regulated and down-regulated circRNAs in adult muscle tissue compared to the embryonic stage are presented in Tables 3 and 4, respectively. Among differentially expressed circRNAs, circRNA2388 had the highest overall expression level (FPKM=1869.8) of all up-regulated circRNA, and circRNA941 had the highest expression level (FPKM=704.6) of all down-regulated circRNAs (Table 5).
To further explore the potential functions of circRNA, we prepared a clustered heatmap (Figure 3a). While several circRNAs showed similar expression patterns, it became evident that a considerable number of circRNAs was differentially expressed not only in the comparison of embryonic and adult muscle tissues, but also among different samples from the same developmental stage (Figure 3b). According to this analysis, at least 624 circRNAs were up-regulated in adult samples compared to embryonic samples, while at least 204 circRNAs were down-regulated (Supplementary Table S4). Generally, embryonic tissues displayed a clear propensity for high circRNA expression (Figure 3b).
Differentially expressed circular RNAs in bovine skeletal muscle. (a) Clustered heat map of the top 100 most differentially expressed circRNAs when comparing embryonic and adult muscle tissues, and (b) scatter plot showing the correlation between abundances of individual circRNAs at the embryonic and adult stage
Correlation between circRNAs and their parental linear transcripts
In order to shed light on the relationship between circRNA biogenesis and the expression of linear transcripts from the respective genes, we calculated Pearson correlation coefficients (rP) between expression levels of circRNAs and their host mRNAs, assuming that a strict linear relationship between both would indicate that circRNAs are generated as (stochastic) by-products of linear mRNA (Figure 4a). We found a low rP of 0.34 between circRNAs and their parental mRNAs in embryonic stage to adult stage. Moreover, we discovered that one parental gene could generate multiple (on average 2.92) circRNA isoforms; specifically, the 12 981 circRNAs detected in our study arose from only 4446 host genes (Figure 4b). A striking example was the ATRX gene, from which six different circRNA isoforms were detected (>1 back-spliced read; Figure 4c). However, only one or two circRNA isoforms were usually expressed at higher levels, while the majority of isoforms showed low expression (Figures 4d and e).
Characteristics of circular RNA in bovine skeletal muscle. (a) Clustered heat map showing abundances of the corresponding linear host transcripts of the top 100 most differentially expressed circRNAs. (b) Numbers of circRNAs produced by the same gene. (c) Exemple of circATRX, which showed six alternative circRNA isoforms. (d, e) Box plots showing abundances of differentially expressed circRNA isoforms. The first four circRNAs are presented
To further examine the potential functions of circRNAs in bovine muscle development, we compared embryonic and adult libraries and classified differentially expressed circRNAs (≥2 back-spliced reads) as up- or down-regulated, and we asked if the corresponding (linear) mRNAs show similar patterns of up- and down-regulation (or, alternatively, similar expression patterns) when comparing both developmental stages (Table 6). We considered circRNAs with markedly different patterns of altered expression levels compared with their paternal mRNAs potential candidates in regulating muscle development.
We found 32 circRNAs that were down-regulated when comparing embryonic and adult samples to be accompanied by similar changes in the expression of the corresponding parental mRNA (for example, ECH1, EXOSC9, COQ3, COQ3, EPRS, FAM173B). When considering up-regulated circRNAs, two corresponding paternal mRNAs were similarly up-regulated (IGDCC4 and TULP4). These circRNAs may be byproducts arising from canonical splicing variants, or they function as miRNA sponges to release and promote target gene expression during muscle development.
Approximately 67 circRNAs had opposing patterns of differential mRNA expression between developmental stages, or mRNA expression patterns did not differ (for example, SVIL, ANGEL2, MYL1, MYOM1, ALKBH8, CAAP1, CSNK1G3 and MIER1). Those circRNAs, or the corresponding mRNAs, could function as promoters or suppressors during bovine muscle development and represent candidates for future studies exploring the regulatory functions of circRNAs and their host genes.
Delineation of gene ontology and KEGG pathway analysis
As a first step to investigate whether circRNAs might regulate the transcription of their host genes, we performed GO-enrichment analysis of those genes that produced differently expressed circRNAs. The 20 most significant functional annotations are shown in Figure 5a. We found significant enrichment of 107 functional groups (P<0.05, Supplementary Table S5).
We then employed KEGG pathway enrichment analysis to further understand the biological functions and molecular interactions of genes hosting significantly differentially expressed circRNAs, assuming that the identified pathways may be involved in the development and growth of bovine skeletal muscle. The top 20 most significant KEGG pathways are shown in Figure 5b. We found 152 pathways to be significantly enriched, and the highest level of significance was found for ‘calcium signaling pathway’ (ko04020) with 41 annotated genes (Supplementary Table S8).
Co-expression of circRNAs/mRNAs
We used the annotations of the functions of mRNAs to predict the potential functions of co-expressed circRNAs (Supplementary Figure S1). The co-expression network revealed that one circRNA or mRNA correlates with one to dozens of differentially expressed circRNAs and suggests that the regulatory relationships between circRNAs and mRNAs may play important roles in muscle development. We found that up-regulation of circRNA1039 was negatively correlated with expression levels of AARSD1, while down-regulation of circRNA1711 was positively correlated with expression levels of AARSD1, which is involved in ‘nucleotide binding’.
Competing endogenous RNA network
To construct a competing endogenous RNA network, we selected mRNAs that are related to muscle development and predicted their binding miRNAs from our high-throughput sequencing data. Based on shared binding sites in mRNAs and corresponding circRNAs, we constructed a mRNA-miRNA-circRNA network with a total of seven circRNAs, 200 mRNAs, and ten miRNAs (Supplementary Figure S2).
Validation of differentially expressed circRNAs by qPCR
We confirmed stage-specific differences in the abundance of certain circRNAs when comparing embryonic and adult tissue samples using qPCR. We randomly selected 17 differentially expressed circRNAs and amplified their junction regions using specific qPCR primers (Figure 6a). Normalized read counts of those circRNAs from our circRNA-Seq analysis are shown in Figure 6b. We verified the amplified PCR products with specific circRNA junctions by Sanger sequencing (Figure 6c). Our qPCR analysis found the presumed patterns of up- and down-regulation of the 17 circRNAs to be remarably similar in both types of analysis (Figure 6d), suggesting that circRNA-Seq data provided reliable information about the relative abundance of circRNAs.
Validation of putative circular RNA by quantitative real-time PCR (qPCR). (a) Schematic view illustrating the design of primers for circRNAs used in qPCR. (b) 17 circRNAs that were selected as they exhibited significantly different expression patterns (assessed from our RNA-sequencing approach) when comparing two development stages. (c) Representative examples of PCR products purified and sequenced to confirm circRNA junction sequences. (d) Validation of differential expression of circRNAs using qPCR, whereby 17 circRNAs confirmed the predicted pattern, while two circRNAs Data are presented as means±S.E.M. for three individuals. *P<0.05
circLMO7 acts as a competing endogenous RNA for miR-378a-3p
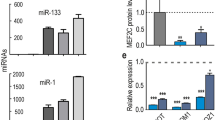
In the following, we focused on the most down-regulated circRNA (circRNA42), which we renamed circLMO7 based on its host gene LMO7, which is located on chromosome 12. Comparing different embryonic tissues, we found circLMO7 to be predominantly expressed in muscle tissue (Figure 7a), suggesting a potential role in muscle development. We transfected bovine primary myoblasts with pcDNA-circLMO7 vectors to explore the putative role of circLMO7 in muscle development. As predicted, overexpression of circLMO7 led to a more than 30-fold increase in the abundance of circLMO7 RNA (Figure 7b). Our analysis of potential miRNA recognition sequences revealed the presence of a putative binding site of miR-378a-3p in circLMO7. In support of this finding, overexpression of circLMO7 significantly decreased the abundance of miR-378a-3p (P<0.05; Figures 7c and d). Moreover, our luciferase-based assay found miR-378a-3p to inhibit Rluc expression from psiCHECK2-circLMO7W vectors (Figure 7e).
Expression analysis of circLMO7 using qPCR. (a) Expression levels of circLMO7 in different tissues of embryonic cattle. (b) Visualization of the efficiency of circLMO7 overexpression vector pcDNA-circLMO7. (c) Effect of circLMO7 on the abundance of miR-378a-3p. (d) RNAhybrid predicted a miR-378a-3p binding site and provided ΔG values for its interaction with circLMO7. (e) Our miR-378a-3p overexpression vector was co-transfected with psiCHECK2-circLMO7W or psiCHECK2-circLMO7M into bovine primary myocytes. Renilla luciferase activity was normalized to Firefly luciferase activity. (f) mRNA expression of HDAC4 in primary bovine myocytes transfected with circLMO7 and/or miR-378a-3p mimic for 24 h was detected using qPCR. Data are shown as means±S.E.M. for three individuals. *P<0.05
Our previous research found miR-378a-3p to target the 3′UTR of the HDAC4 gene to inhibit its expression.17 To confirm the involvement of circLMO7 in the miR-378a-3p-mediated regulation of HDAC4 expression, we transfected bovine primary myoblasts with pcDNA-circLMO7 and/or miR-378a-3p mimic. As expected, circLMO7 overexpression increased the abundance of HDAC4 mRNA, but this effect was inhibited by miR-378a-3p overexpression (Figure 7f). Altogether, these findings suggest that circLMO7, by binding miR-378a-3p, acts as a decoy to mitigate the inhibiting effect of miR-378a-3p during the expression of HDAC4.
Effects of circLMO7 on myoblast differentiation, proliferation, and apoptosis
We tested for an involvement of circLMO7 in myoblast differentiation. Our immunofluorescence assay suggests that miR-378a-3p overexpression promoted the expression of the myoblast determination factor MyoD and induced myotube formation (Figure 8a), while this effect was not observed after circLMO7 overexpression. Likewise, circLMO7 overexpression significantly decreased the expression of well-established markers of bovine myogenesis, namely MyoD and myogenin (MyoG), which was detectible at both mRNA and protein levels (Figures 8b and c and Supplementary Fig. S7). More marker gene detection results showed in Supplementary Fig. S4. These results suggest that circLMO7 inhibits myoblast differentiation by regulating concentrations of miR-378a-3p.
circLMO7 suppresses the differentiation of bovine primary myocytes. (a) Bovine primary myocytes were transfected with pcDNA-circLMO7 and/or miR-378a-3p mimic, and cell differentiation was detected by immunofluorescence. (b) mRNA of marker genes for myocyte differentiation were detected by qPCR. (c) The presence of proteins translated from marker genes of myocyte differentiation were analyzed using western blots
Next, we assessed the potential effect of circLMO7 on cell proliferation. Cell cycle analysis revealed that circLMO7 overexpression increased the number of myoblasts in the S- and G2-phases (2.01 and 2.55%), and decreased the proportion of cells in the G0/G1-phase (4.6%). Conversely, miR-378a-3p overexpression increased the number of myoblasts in the G0/G1-phase (Figures 9a and b). We found circLMO7 overexpression to promote cell proliferation according to the CCK-8 and EdU incorporation assays (Figures 9c,Supplementary Figure S5). It also increased the expression of our marker for cell proliferation (CyclinD1), which was detectible at the mRNA level (Figure 9d).We asked whether circLMO7 plays a role as a regulator of myoblast apoptosis. The Annexin V-FITC/PI staining assay showed that miR-378a-3p overexpression induced apoptosis in primary bovine myoblasts, while circLMO7 overexpression protected myoblasts from apoptosis induced by miR-378a-3p (Figures 10a and b). In support of these findings, circLMO7 overexpression significantly increased expression levels of three well-established markers of cell survival, BCL-2, BAX, and Caspase9, which was detectible at both the mRNA and protein levels (Figures 10c and d,Supplementary Fig. S6). In summary, our results suggest that circLMO7 inhibits bovine myoblast differentiation, promotes cell proliferation, and protects myoblasts from apoptosis by binding miR-378a-3p.
circLMO7 promotes the proliferation of bovine primary myocytes. (a, b) Bovine primary myocytes were transfected with pcDNA-circLMO7 and/or miR-378a-3p mimic, and cell phases were analyzed by flow cytometry. (c) Cell proliferation was assessed using the cell counting kit-8 (CCK-8) assay. (d) mRNA of the proliferation marker CyclinD1 was quantified using qPCR. (e) Cell proliferation indices were assessed after treatment with 5-Ethynyl-2′-deoxyuridine (EdU). The scale bar represents 200 μm. Data are presented as means±S.E.M. for three individuals. *P<0.05
circLMO7 inhibits apoptosis in bovine primary myocytes. (a, b) We transfected bovine primary myocytes with pcDNA-circLMO7 and/or miR-378a-3p mimic and determined apoptosis by Annexin V-FITC/PI binding followed by flow cytometry. (c) mRNA of apoptosis marker genes (BCL-2, BAX, caspase9) was detected using qPCR. (d) Proteins of apoptosis marker genes were detected using western blots. Data are presented as means±S.E.M. for three individuals. *P<0.05
Discussion
Most studies examining the molecular mechanisms underlying muscle development and growth in cattle investigated the roles played by protein-coding genes, and accordingly, studies using RNA-sequencing methods usually focused on mRNAs. The potential functions of circRNAs in bovine muscle development, however, remained elusive. Our present study for the first times provides an overview of the types and relative abundances of circRNAs that can be found in two developmental stages of bovine skeletal muscle tissue (that is, embryonic and adult musculus longissimus samples). Using high-throughput RNA-seq analysis, we identified and annotated a considerable number of circRNAs.
CircRNAs do not possess a poly(A) tail and are RNase R-resistant, which offers the opportunity to specifically enrich circRNAs from total RNA. In total, we identified 1287 circRNAs candidates that contained at least one unique back-spliced read in our rRNA-depleted RNA libraries, but only 589 circRNAs candidates were detected after additional RNase R treatment. This suggests that RNase R treatment effectively depleted the libraries of linear RNAs, which increases the accuracy of circRNA identification. However, it can also affect the enrichment of certain circRNAs: a previous study found that some circular-junction candidates—such as CDR1as/ciRS-7—were not enriched in RNase R-treated RNA libraries.6 Moreover, some circRNAs with long exons may be less likely to be identified after RNase R treatment.5, 6, 18
Our high-throughput RNA-seq data enabled us to detect specific changes in the relative abundances of different circRNAs when comparing circRNAs retrieved from embryonic and adult muscle tissue, and qPCR largely confirmed differential expression patterns. These findings suggest that the mechanisms regulating circRNA levels in muscle tissue are species- and developmental stage-specific and that circRNAs are probably not just by-products of regular transcription (that is, formation of linear mRNA). Interestingly, the expression patterns of several circRNAs were uncoupled in several cases when comparing circRNA abundances and those of linear transcripts of their host genes (resulting in low Pearson correlation coefficients), which supports the idea that circRNA biogenesis is a tightly regulated process just like canonical splicing. Likewise, when comparing circRNA abundances and host mRNA abundances between different stages of bovine muscle development, there were several cases in which massive changes in mRNA expression levels were accompanied by only moderate changes in circRNA expression.19 In support of our findings, a recent study found expression levels of circRNAs in porcine embryonic brain tissue to be moderately correlated with the abundance of linear transcripts of their host genes,9 which was also true in comparisons of multiple cell lines.8, 20 Thus, seemingly up-regulated circRNA abundances compared to their linear host mRNA levels may partly reflect the higher degradation rate of linear transcripts. We found the mean length of upstream or downstream flanking introns of circRNAs to be about 11 000 nt. One explanation for this general pattern is that longer introns may provide enough time for circRNA formation through back-splicing events by slowing down the alternative (that is, conventional) splicing process. Furthermore, longer introns are more likely to lead to complementary base pairing close to the borders of circularized exons, for example, when complementary transposable elements, such as SINEs9 or ALU repeats are present.6, 21, 22 Our results suggest that one parental gene can generate multiple circRNA isoforms, but only one or two circRNA isoforms were usually expressed at higher levels, while the majority of isoforms exhibited at exceedingly low expression levels. We speculate that the formation of alternative circRNA variants is under tight (temporal) control, and different variants could be involved in the regulation of muscle development. Future studies will need to elaborate on the question of what induces the differential expression of circRNA isoforms from one parental gene and whether different circRNA isoforms have distinct functions.
On the basis of our observation of several circRNAs being differentially expressed across developmental stages, we propose that studying circRNAs may lead to the discovery of a large number of regulatory circRNAs involved in bovine muscle development and growth. For example, circRNAs have been suggested to function as miRNA sponges, but the regulatory effect of such circRNAs has not yet been investigated in detail in previous studies. In mammalian cells, circRNA from the CDR1 gene acts as a sponge for miR-7, and a circRNA from the testes-specific Sry gene acts as a sponge for miR-138.7, 12 In our present study, we exemplify the involvement of one circRNA in regulating myoblast proliferation, differentiation, and apoptosis. Specifically, we demonstrate (using pcDNA-circLMO7 and miR-378a-3p overexpression vectors) that circLMO7 could serve as a modulator of cell differentiation and cell survival by sponging miR-378a-3p in bovine myoblasts. It appears that circLMO7—a differentially expressed circRNA when comparing embryonic and adult muscle tissues—acts as a decoy for miR-378a-3p, thereby mitigating the effects of the latter during muscle development. Our results call for additional investigation into the roles played by different miRNAs and circRNAs during bovine muscle development.
Conclusion
Our study is the first to provide an overview of circRNA expression in embryonic and adult bovine muscle tissues. Thousands of circRNAs were identified, several of which showed very different abundances when comparing both developmental stages. Our study may serve as a starting point for in-depth investigations into the roles played by several of those circRNAs during bovine myogenesis. We characterized and functionally evaluated the role played by one of the differentially expressed circRNA, circLMO7. The results of several independent experimental approaches suggest that circLMO7 inhibits cell differentiation of myoblasts and promotes cell survival by sponging miR-378a-3p. Our study adds to our knowledge on the genetic mechanisms underlying skeletal muscle formation in Qinchuan cattle and other cattle breeds (that is, economically important traits), which can be implemented in future breeding programs that are informed by molecular information of breeding stock.
Materials and methods
Identification of circRNAs in bovine muscle tissue through RNA-sequencing
Sample preparation
Qinchuan cattle from which samples were taken received humane care, as described in Proclamation No. 5 of the Ministry of Agriculture, China. All experimental procedures were approved by the Animal Care and Use Committee of the Northwest A&F University. We collected six musculus longissimus (LM) samples from two developmental states (embryonic, 90 days old; and adult, 24 months old) from Yiming abattoir, a local slaughterhouse in Xi'an. In addition, we collected subcutaneous adipose, heart, liver, spleen, lung, kidney, and small intestine tissues from embryos. All tissue samples were snap-frozen in liquid nitrogen and stored at –80 °C freezer until use.
Library preparation and Illumina sequencing
We subjected 2 μl of the RNA extract to electrophoresis on a denaturing agarose gel to assess total RNA yield and then quantified RNA concentrations using a NanoDrop spectrophotometer (NanoDrop, Wlinington, USA) and Agilent 2100 Bioanalyzer (Agilent, Santa Clara, CA, USA). We treated total RNA samples (3 μg) with the epicenter Ribo-ZeroTM Kit (Illumina, San Diego, CA, USA) to remove rRNA before constructing RNA-seq libraries (to obtain sequence information of linear transcripts). For RNase R treatment, ribosome-depleted RNA was incubated for 15 min at 37 °C with 5 units RNase R per μg RNA (Epicentre Technologies, Madison, WI, USA). We then fragmented the rRNA−+RNase R+ samples (which we used to obtain sequence information of circRNAs) and used the thusly treated samples to synthesize first- and second-strand complementary DNA (cDNA) with random hexamer primers, dNTPs and DNA Polymerase I using the PrimeScript RT regent Kit (TaKaRa, Japan). The cDNA fragments were cleaned and concentrated using AMPure XP beads, after which ends were repaired and modified with T4 DNA polymerase Klenow DNA polymerase to add -A and adapters at the 3′ end of the DNA fragments. We purified the ligated cDNA products with AMPure XP beads and treated them with uracil DNA glycosylase to remove second-strand cDNA. We subjected purified first-strand cDNA to PCR amplification. We checked the quality of the libraries with an Agilent 2100 Bioanalyzer and subjected them to sequencing using a HiSeq 2500 (Illumina, San Diego, CA, USA) on a 150 bp paired-end run.
Treatment of raw sequencing data
We removed adapters from the raw FASTQ files using Trim Galore. We aligned the filtered sequence data to the Bos taurus reference genome (bosTau7) from UCSC (http://genome.ucsc.edu/) with TopHat2 (version 2.0.14).20 We first assembled the linear transcripts and estimated their abundance using CuffLinks, after which we identified the reads of fusion transcripts (obtained from TopHat2) that did not align to the linear RNA sequences. Afterwards we ran the program TopHat-Fusion while incluyding both fusion transcripts and linear transcripts to identify back-spliced junctions reads. Only sequences of circRNAs with <100 kb length, with at least two supporting reads, and with no more than two mismatches were retained for further analysis. We normalized circRNA contents as the number of uniquely mapped fragments per kilobase of exon per million fragments mapped (FPKM).
Gene ontology and pathway analysis
We used Gene Ontology (GO) analysis (http://www.geneontology.org) to characterize circRNA-hosting genes. GO-terms provide information about the biological processes in which genes are involved, either a cellular component or metabolic pathway, and highlight the molecular function(s) while reducing complexity. We also performed Kyoto Encyclopedia of Genes and Genomes (KEGG; http://www.kegg.jp) pathway analysis to provide insights into the molecular interaction and reaction networks of circRNAs that were differentially expressed using Visualization and Integrated Discovery (DAVID, version 6.7; http://david.ncifcrf.gov). In both cases, the -log10 P-value denotes significant enrichment of a given GO term or pathway among up- and down-regulated entities.
Correlation analysis of putative co-expression
We investigated the relationship between expression levels of linear (mRNAs) and circular transcripts (circRNAs) based on normalized signal intensities of our circRNA and linear RNA profiling data using Pearson correlations. If the expression levels of two circRNAs exceeded a preselected threshold and were similar in the Pearson analysis, they would be considered to have co-expression relationship and connected by a string indicating a tight correlation. CircRNAs or genes importance in the network depends on the degree of correlation. The absolute value of Pearson correlation coefficient (rP)≥0.90, FDR<0.01 and P<0.01 were reserved for further analysis. We selected 20 significantly differentially expressed circRNAs and 184 differentially expressed mRNAs (that is, protein-coding genes) to build a coding-noncoding co-expression network according to the degree of correlation. The involved genes were involved in a number of biological processes, such as ‘skeletal muscle fiber development’, ‘negative regulation of smooth muscle cell proliferation’, and ‘calcium-mediated signaling’.
Network analysis of competing endogenous RNA
We asked whether there is a correlation between miRNA, mRNA and circRNA? To shed light on this question, we focused on those mRNAs and circRNAs whose expression levels exhibited a tight correlation in our previous analysis. We constructed an mRNA-miRNA-circRNA interaction network based on miRNA seed sequence binding sites that we detected in the mRNA and circRNA sequences. The interactions of miRNA–mRNA and miRNA–circRNA were predicted by TargetScan (http://www.targetscan.org/), miRcode (http://www.mircode.org/), and miRanda (http://www.microrna.org/microrna/home.do). We predicted miRNA binding sites using miRcode and miRanda, while the interactions of miRNA–mRNA and miRNA–circRNA were predicted by TargetScan.
RNA preparation and quantitative real-time PCR
We conducted quantitative real-time PCR (qPCR) to validate the results from our RNA-sequencing approach, focusing on these circRNAs which are significant difference in the expression levels. Specifically, we asked if different expression levels as inferred from our RNA-sequencing also become apparent when applying qPCR. We used the same cDNA libraries of circRNAs stemming from three embryonic and three adult muscle samples in our qPCR analysis. In addition, we prepared circRNA cDNA libraries from embryonic subcutaneous adipose, heart, liver, spleen, lung, kidney, and small intestine tissues. We used the SYBR Green PCR Master Mix (Takara, Japan), and qPCR cycling conditions followed previously described protocols.23 We amplified 17 circRNAs using the primers listed in Supplementary Table S1, while the β-actin gene was used as internal control (that is, as a housekeeping gene). Primers were designed usingsoftware Primer 5, and all primers were spanning the distal ends of circRNAs. We ascertained sequence specificity through BLAST searches. The 2−ΔΔCt method was used to analyze the relative expression levels of different circRNAs.
Role of circLMO7 in cell cycle, differentiation, and apoptosis
Vector construction
Afterwards, we concentrated on the role of CircLMO7 in bovine myogenesis. We first constructed vectors to experimentally alter the expression of circLMO7 in transfected bovine myoblasts. To this end, we amplified the genomic region hosting circLMO7 along with its flanking (intron) regions of the LMO7 gene (Supplementary Table S1) using the PrimerSTAR Max DNA Polymerase Mix (Takara, Dalian, China). We generated different vector types as follows: (1) full-length circLMO7 was amplified and cloned in the pcDNA3.1+ vector (Invitrogen, Carlsbad, CA, USA) to obtain their overexpression plasmid (pcDNA-circLMO7). (2) We generated another vector to detected the regulation of circLMO7 and miR-378a-3p. A psiCHECK2-circLMO7-W vector (pCH-circLMO7-W) was constructed by inserting full length circLMO7 fragments (containing the miR-378a-3p binding sequence) into the psiCHECK-2 vector (Promega, Maddison, WI, USA) at the 3′end of the Renilla gene using the restriction enzymes Xho I and Not I (TaKaRa, Dalian, China). (3) Finally, we constructed a mutant psiCHECK2-circLMO7-Mut vector (pCH-circLMO7-Mut) by mutating complementary to the seed region of miR-378a-3p using overlapping extension PCR. All constructs were verified by sequencing.
Cell culture and treatment
Primary bovine myoblasts were isolated and cultured from bovine longissimus muscle as described in previous studies.23, 24 We plated myoblasts at the stage of 80% confluence at a density of 5 × 105 cells per well in six-well plates in 2 ml culture medium per well or 1 × 104 cells per well in 96-well plates (NEST, Wuxi, China) in 100 μl culture medium per well and incubated them, as described previously.23 After growth to ~80% confluence, the cells were treated with: (1) miR-378a-3p mimic (2 μg/ml; Genepharma, Shanghai, China), (2) pcDNA-circLMO7 (2 μg/ml), or (3) miR-378a-3p mimic+pcDNA-circLMO7. After incubation, the myoblasts were used for the different assays outlined below. To induce differentiation of myoblasts, the culture medium was changed to high-glucose Dulbecco’s modified Eagle’s medium (DMEM) with 2% horse serum.23
Cell cycle assay
To gain insights into the effects of circLMO7 in different periods of cell cycle, we analyzed the cell cycle of different treatment groups ((1) miR-378a-3p mimic (2 μg/ml; Genepharma, Shanghai, China), (2) pcDNA-circLMO7 (2 μg/ml), or (3) miR-378a-3p mimic+pcDNA-circLMO7) using the Cell Cycle Testing Kit (Multisciences, Hangzhou, China). We collected cells that had been cultivated in six-well plates and centrifuged them at 800 g/min for 5 min. The supernatant was discarded, and the cells were washed once with cold phosphate buffered saline (PBS, pH=7.4). We resuspended the cells in 1 ml of kit reagent A and 10 μl of reagent B, followed by vortexing for 10 s and incubation for 30 min at room temperature, after which the cell suspension was used for flow cytometry (FACS CantoTM II, BD BioSciences, USA).
Cell proliferation assay
We examined cell proliferation using the Cell Counting Kit-8 (CCK-8) assay (Multisciences, Hangzhou, China). For the CCK-8 assay, we used cells plated in 96-well culture plates. We ran six independent replicates per treatment group. After 24 h of incubation at 37 ºC, 10 μl of CCK-8 reagent was added to each well, and cells were incubated at 37 °C for 2 h. The absorbance of each sample at 450 nm wavelength was detected using a microplate reader (Molecular Devices, Sunnyvale, USA). In addition, we assessed cell proliferation using the Cell-Light EdU DNA cell proliferation kit (Ribobio, Guangzhou, China) according to the manufacturer’s instructions. We ran three independent replicates for each treatment group.
Assessment of apoptosis
We assessed incidences of cell apoptosis using an Annexin V-FITC/PI staining assay. After incubation, cells from the different treatment groups (three independent replicates per treatment) were washed three times with PBS buffer (pH=7.4), collected by trypsinization, washed again with PBS and then resuspended in 500 μl 1 × binding buffer (Multisciences, Hangzhou, China). Afterwards, we incubated the cells for 10 min in the dark at room temperature in the presence of Annexin V-FITC (5 μl) and propidium iodide (PI) (10 μl,Multisciences, Hangzhou, China). Afterwards, cells were analyzed using flow cytometry (FACS CantoTM II, BD BioSciences, USA).
Western blot analysis
We asked if circLMO7 affect the translation of genes which were regulated with cell proliferation and differentiation. We collected cells from the different treatment groups, pelleted them by centrifugation (12 000 × g, 5 min) and lysed them in RIPA buffer (Solarbio, Beijing, China). We prepared total protein and determined protein concentrations using the Bradford method. Proteins were then separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and subsequently transferred to nitrocellulose membranes and blocked with 5% skim milk powder solution for 1.5 h at room temperature. We incubated membranes overnight with the primary antibodies. We purchased anti-MyoD, anti-MyoG, anti-BCL-2, anti-BAX, anti-Caspase9, and anti-β-actin from Abcam (Cambridge, MA, USA). Afterwards, we washed the membranes with PBS-tween and incubated them for 1.5 h with horseradish peroxidase-conjugated secondary antibodies (Abcam, Cambridge, MA, USA). Protein bands were detected after treatment with SuperSignal West Femto agent of Thermo (Thermo Scientific, Karlsruhe, Germany).
Assessment of differentiation using immunofluorescence
To provide information about the effects of circLMO7 in myoblasts differentiation, we used immunofluorescence assay. Primary bovine myoblasts at the stage of 95% confluence were washed three times with PBS buffer (pH 7.4), and permeabilized for 15 min in PBS containing 0.5% Triton X-100 before fixation in PBS containing 4% paraformaldehyde for 30 minutes. Immunostaining was carried out as follows: cells were incubated overnight at 4 °C with the primary MyoD-antibody (1:500; Abcam, Cambridge, MA, USA), diluted in 1% bovine serum albumin. Afterwards, we washed cells with PBS and incubated them at room temperature for 2 h with the corresponding secondary antibody goat anti-rabbit IgG H&L (1:1000; Abcam, Cambridge, MA, USA) diluted in 1% bovine serum albumin in PBS. We visualized DNA using 5 mg/ml DAPI. Finally, we washed the thusly prepared cells three times with PBS and observed them under a fluorescence microscope (DM5000B, Leica, Germany).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Nigro JM, Cho KR, Fearon ER, Kern SE, Ruppert JM, Oliner JD et al. Scrambled exons. Cell 1991; 64: 607–613.
Capel B, Swain A, Nicolis S, Hacker A, Walter M, Koopman P et al. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell 1993; 73: 1019–1030.
Zaphiropoulos PG . Circular RNAs from transcripts of the rat cytochrome P450 2C24 gene: correlation with exon skipping. Proc Natl Acad Sci USA 1996; 93: 6536–6541.
Pasman Z, Been M, Garcia-Blanco M . Exon circularization in mammalian nuclear extracts. RNA 1996; 2: 603.
Jeck WR, Sharpless NE . Detecting and characterizing circular RNAs. Nat Biotechnol 2014; 32: 453.
Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013; 19: 141–157.
Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013; 495: 333–338.
Rybak-Wolf A, Stottmeister C, Glažar P, Jens M, Pino N, Giusti S et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell 2015; 58: 870–885.
Venø MT, Hansen TB, Venø ST, Clausen BH, Grebing M, Finsen B et al. Spatio-temporal regulation of circular RNA expression during porcine embryonic brain development. Genome Biol 2015; 16: 1.
Abdelmohsen K, Panda AC, De S, Grammatikakis I, Kim J, Ding J et al. Circular RNAs in monkey muscle: age-dependent changes. Aging 2015; 7: 903–910.
Chen I, Chen CY, Chuang TJ . Biogenesis, identification, and function of exonic circular RNAs. Wiley Interdiscip Rev: RNA 2015; 6: 563–579.
Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK et al. Natural RNA circles function as efficient microRNA sponges. Nature 2013; 495: 384–388.
Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell 2014; 56: 55–66.
Li Z, Huang C, Bao C, Chen L, Lin M, Wang X et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol 2015; 22: 256–264.
He H, Liu X . Characterization of transcriptional complexity during longissimus muscle development in bovines using high-throughput sequencing. PLoS ONE 2013; 8: e64356.
Sun J, Li M, Li Z, Xue J, Lan X, Zhang C et al. Identification and profiling of conserved and novel microRNAs from Chinese Qinchuan bovine longissimus thoracis. BMC Genomics 2013; 14: 1.
Wei X, Li H, Zhang B, Li C, Dong D, Lan X et al. miR-378a-3p promotes differentiation and inhibits proliferation of myoblasts by targeting HDAC4 in skeletal muscle development. RNA Biol 2016; 13: 1300–1309.
Yang L, Duff MO, Graveley BR, Carmichael GG, Chen L-L . Genomewide characterization of non-polyadenylated RNAs. Genome Biol 2011; 12: 1.
Sun X, Li M, Sun Y, Cai H, Li R, Wei X et al. The developmental transcriptome landscape of bovine skeletal muscle defined by Ribo-Zero ribonucleic acid sequencing. J Anim Sci 2015; 93: 5648–5658.
Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO . Cell-type specific features of circular RNA expression. PLoS Genet 2013; 9: e1003777.
Liang D, Wilusz JE . Short intronic repeat sequences facilitate circular RNA production. Genes Dev 2014; 28: 2233–2247.
Zhang X-O, Wang H-B, Zhang Y, Lu X, Chen L-L, Yang L . Complementary sequence-mediated exon circularization. Cell 2014; 159: 134–147.
Sun X, Li M, Sun Y, Cai H, Lan X, Huang Y et al. The developmental transcriptome sequencing of bovine skeletal muscle reveals a long noncoding RNA, lncMD, promotes muscle differentiation by sponging miR-125b. BBA-Mol Cell Res 2016 2016; 1863: 2835–2845.
Li H, Wei X, Yang J, Dong D, Huang Y, Lan X et al. Developmental transcriptome profiling of bovine muscle tissue reveals an abundant GosB that regulates myoblast proliferation and apoptosis. Oncotarget 2017; 8: 32083.
Acknowledgements
This work was supported by the ‘National natural science foundation of China’ [grant no. 31772574], the ‘National 863 program’ of China [grant no. 2013AA102505], the ‘Program of the national industrial technology system for beef cattle and yak’ (CARS-37), ‘Bio-breeding capacity-building and industry-specific projects’ by the National Development and Reform Commission (2014–2573), ‘Specific projects in science and technology of Henan Province’ (141100110200), ‘Innovative engineering projects for science and technology coordination of Shaanxi Province’ (No. 2014KTZB02-02-02-02, No. 2015KTCL02-08), and the ‘Project for breeding and commercial use of Pinan Cattle.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited G Calin
Supplementary Information accompanies this paper on Cell Death and Disease website
Supplementary information
Rights and permissions
Cell Death and Disease is an open-access journal published by Nature Publishing Group. This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Wei, X., Li, H., Yang, J. et al. Circular RNA profiling reveals an abundant circLMO7 that regulates myoblasts differentiation and survival by sponging miR-378a-3p. Cell Death Dis 8, e3153 (2017). https://doi.org/10.1038/cddis.2017.541
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cddis.2017.541
This article is cited by
-
Identification of circRNA-associated ceRNA networks in the longissimus dorsi of yak under different feeding systems
BMC Veterinary Research (2024)
-
Genome-wide analysis of circular RNA-mediated ceRNA regulation in porcine skeletal muscle development
BMC Genomics (2023)
-
Melatonin promotes the development of the secondary hair follicles by regulating circMPP5
Journal of Animal Science and Biotechnology (2023)
-
Biological functions of circRNAs and their advance on skeletal muscle development in bovine
3 Biotech (2023)
-
Circ2388 regulates myogenesis and muscle regeneration
Cell and Tissue Research (2023)