Abstract
Assessment of the network of toxicity pathways by Omics technologies and bioinformatic data processing paves the road toward a new toxicology for the twenty-first century. Especially, the upstream network of responses, taking place in toxicant-treated cells before a point of no return is reached, is still little explored. We studied the effects of the model neurotoxicant 1-methyl-4-phenylpyridinium (MPP+) by a combined metabolomics (mass spectrometry) and transcriptomics (microarrays and deep sequencing) approach to provide unbiased data on earliest cellular adaptations to stress. Neural precursor cells (LUHMES) were differentiated to homogeneous cultures of fully postmitotic human dopaminergic neurons, and then exposed to the mitochondrial respiratory chain inhibitor MPP+ (5 μM). At 18–24 h after treatment, intracellular ATP and mitochondrial integrity were still close to control levels, but pronounced transcriptome and metabolome changes were seen. Data on altered glucose flux, depletion of phosphocreatine and oxidative stress (e.g., methionine sulfoxide formation) confirmed the validity of the approach. New findings were related to nuclear paraspeckle depletion, as well as an early activation of branches of the transsulfuration pathway to increase glutathione. Bioinformatic analysis of our data identified the transcription factor ATF-4 as an upstream regulator of early responses. Findings on this signaling pathway and on adaptive increases of glutathione production were confirmed biochemically. Metabolic and transcriptional profiling contributed complementary information on multiple primary and secondary changes that contribute to the cellular response to MPP+. Thus, combined ‘Omics’ analysis is a new unbiased approach to unravel earliest metabolic changes, whose balance decides on the final cell fate.
Similar content being viewed by others
Main
Omics technologies combined with systems biology approaches and quantitative assessment of the network of toxicity pathways are at the heart of worldwide efforts to develop a new toxicology for the twenty-first century.1, 2, 3 The success of these technologies has already been demonstrated in biomedicine by the discovery of new pathways and drug targets not evident from classical examinations.4, 5, 6 Such investigations have not yet been reported in neurotoxicology, but transcriptomics and metabolomics profiling are being increasingly used in related fields such as developmental toxicology.7, 8, 9, 10 Re-examination of established toxicants is essential to test the feasibility of new approaches and to gain knowledge about how and when they are best applied.11, 12
One of the best-characterized neurotoxicants is 1-methyl-4-phenylpyridinium (MPP+). This compound is the active metabolite of 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) that triggers specific dopaminergic degeneration and parkinsonism in mice and humans.13 MPP+ is accumulated in its target cells by the dopamine transporter. Once inside the cells, MPP+ inhibits complex I of the respiratory chain, and is assumed to cause cell death by energy failure.14, 15 However, the compound also triggered cell death in mouse mesencephalic neurons lacking a functional complex I.16 Moreover, survival of human dopaminergic cells after accumulation of MPP+ has been shown to be uncoupled from ATP depletion,17 and some neurons were even protected by MPP+ treatment from apoptosis triggered by other stimuli.18 An alternative primary mechanism contributing to the toxicity may be the generation of reactive oxygen species (ROS), possibly through altered electron flow toward oxygen at a subunit of complex I.19
Besides the primary upstream mechanisms, MPP+ toxicity has also been linked to a plethora of downstream steps, comprising protease activation, protein translocations and phosphorylation events.20, 21 In this light, it is astonishing that there is a dearth of studies examining which upstream adaptations precede the final decision on cell death. Such information would be required to quantitatively model and predict cytotoxicity.22, 23
Few Omics studies have addressed MPP+ toxicity. Transcriptome analysis of murine N2a cells revealed changes related to transamination, transporters and G-protein-coupled receptors.24 A proteomics study with the same cells indicated changes in transaminase and other mitochondrial proteins.25 Transcriptome mapping in mouse striatum suggested three waves of gene expression following MPTP treatment: early upregulation of oxidative stress genes (Gadd45, Ddit4), intermediate (24 h) regulation of proinflammatory genes and late responses (72 h) characterized by stress response pathways (Nrf-2, Atf6, Zic1).26 Proteomics and transcriptomics studies of mice treated with MPTP for 7 days (ongoing tissue degeneration) showed changes in over 500 gene products, many of them associated with dopamine signaling, mitochondrial dysfunction, protein degradation, calcium signaling, the oxidative stress response and apoptosis.27
Although transcriptomics data indicate potential changes in enzymatic pathways, only direct data on metabolite levels may substantiate these findings. Therefore, we performed here the first combined metabolomics and transcriptomics study in the field of neurotoxicology. Information on the metabolic changes and the resultant network of early adaptations triggered by MPP+ within dopaminergic neurons is hard to obtain by analysis of complex tissues consisting of lots of different neuronal and glial cell populations. To address this issue, and to provide direct information on human neurons, we made use of LUHMES cells. This human dopaminergic precursor cell line allows the generation of homogeneous, fully postmitotic dopaminergic neuron cultures.28, 29
Results
Metabolome changes in MPP+-exposed dopaminergic neurons
After 6 days of differentiation (d6), LUHMES cells are dopaminergic and postmitotic as characterized by high expression levels of Fox3 (NeuN), tyrosine hydroxylase (TH) and SLC6A3 (dopamine transporter (DAT)).28 The cells were exposed to a toxicologically relevant17, 30 concentration of MPP+ (5 μM; Figure 1a). Cell death was assessed by measurements of LDH release, by counting of viable cells (calcein positive) and by quantification of resazurin reduction. Significant cytotoxicity required at least 48 h of MPP+ exposure, and most cells were dead after 72 h (Figures 1b and c). Cellular ATP and glutathione (GSH) levels, frequently used indicators of MPP+ toxicity, were maintained for at least 24 h, and showed a significant decrease after 36 h of treatment (Figure 1c).
Time course of MPP+-induced cell death events and metabolome changes. (a) Experimental scheme for cell differentiation, MPP+ exposure and sampling. In all experiments of this study, an MPP+ concentration of 5 μM was used, and cells were analyzed on day 8 (d8) of differentiation (green arrow). Red arrows mark time points used for Omics analysis. Blue arrows mark time points that were analyzed in follow-up experiments. (b and c) Cell viability data: resazurin reduction and lactate dehydrogenase (LDH) release were measured and calcein-positive/-negative cells were counted. Changes of ATP and total cellular glutathione (GSH) were measured in parallel cultures and all data were normalized to untreated controls. (d) Samples obtained after 24 or 36 h of treatment with MPP+ or solvent control were analyzed by quadrupole time-of-flight liquid chromatography-mass spectroscopy (Q-TOF LC-MS). A principal component analysis (PCA) of all metabolite data (labeled by length of exposure) was performed and the first two dimensions are displayed. (e) Cells were stained with tetramethylrhodamine ethyl esther (TMRE, green) and calcein-AM (red) to identify energized mitochondria. Representative micrographs display cells treated with solvent (control) or MPP+ (24 and 48 h). (f) The number of TMRE-positive pixels in all neurites of the field was determined by an unbiased image processing algorithm. Data are means±S.D. from 3 independent experiments, and 30 fields per experiment (*P≤0.05)
These basic model parameters indicated that toxicant stress was compensated to a large extent for up to 24–36 h after initiation of MPP+ exposure. After that time, key functions could not be maintained. To broadly characterize the metabolic adaptations before cell death, we performed an untargeted metabolomics analysis: 190 metabolites were significantly altered and 59 were assigned to molecular structures (Supplementary Table S1). A principal component analysis (PCA) of the total quantified metabolite patterns indicated large and highly reproducible differences between control cells and 24 and 36 h samples (Figure 1d and Supplementary Figure S1). Some of the data corroborated known effects of MPP+ exposure. For instance, the altered energy metabolism was indicated by a strong decrease in intracellular glucose (and other sugars) accompanied by an increase of pyruvate and lactate. Consumption of phosphocreatine and a parallel accumulation of creatine suggested an exhaustion of the cellular energy buffer (Figure 2 and Supplementary Figure S2). A cellular struggle to maintain energy supplies was also indicated by a gradual increase of ADP, AMP and adenine, although the mitochondrial membrane potential (Figures 1e and f) and levels of ATP (Figures 1c and 2) were more or less maintained for at least 24 h. In addition, cell death-associated events, such as the release of cytochrome c into the cytosol or regulation of Bcl-2 family proteins, were only measurable at times later than 24 h (Supplementary Figures S3A and B). Increases in methionine sulfoxide (Figure 2 and Supplementary Figure S2) suggested an increased oxidative stress in the system,30 and increased ROS production, as well as several further markers of oxidative stress, were detected few hours after addition of MPP+ (Supplementary Figure S4).
Metabolic adaptations in MPP+-treated neurons. LUHMES cells were treated with 5 μM MPP+ for different times, and samples were taken at day 8. Metabolite concentrations were determined by Q-TOF LC-MS in four independent experiments. Data were normalized to untreated controls and are displayed as means±S.D. Metabolites that changed significantly (P≤0.05, FDR adjusted) are displayed. D-Gluc, D-glucose; UDP-gal, uridinediphosphate galactose; UDP-gluc, uridinediphosphate glucose; P-creatine, phosphocreatine; Met-SO, methionine sulfoxide; SAM, S-adenosylmethionine; SAH, S-adenosylhomocysteine
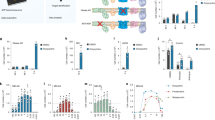
The broad metabolite profiling also allowed new insights. For instance, the strong increase of S-adenosyl-methionine (SAM, Supplementary Figure S2), S-adenosyl-homocysteine (SAH) and cystathionine pointed to alterations of the methionine/cysteine metabolism (Figure 2), possibly to replenish the redox buffer GSH. In a targeted analysis, we therefore specifically looked for cellular cysteine levels. After 18 h, the levels of this amino acid were significantly increased, whereas its oxidized form, cystine, decreased (Figure 3a). In addition, other profound changes in amino acid homeostasis became evident. The untargeted metabolomics analysis showed lower levels of alanine, glutamate and asparagine, and degradation of branched amino acids was indicated by the strong increase in 2-oxo-isovalerate. Cellular concentrations of the poorly gluconeogenic amino acids arginine, lysine, tryptophan and tyrosine increased upon MPP+ treatment, and the increase in glycine was matched by a similar decrease of sarcosine (Figure 2). More such shifts in metabolism were observed: for instance, the conversion of ornithine to putrescine decreased, and this was followed by a decrease of spermidine and spermine (Figure 3b). The major shifts were expected for central energy metabolism. However, alterations in the citric acid cycle (TCA cycle), an assumed primary mode of action of MPP+, could not be unambiguously deduced from our standard metabolomics data. Therefore, we analyzed changes in stable-isotopic enrichment patterns of TCA cycle intermediates to measure the effect of MPP+ on relative glucose oxidation in the TCA cycle. By using uniformly labeled [U-13C]glucose we found that MPP+ treatment did indeed nearly abolish the relative flux through the pyruvate dehydrogenase complex (Figure 3c).
Multiple secondary metabolic changes triggered early after exposure to MPP+. Cells were exposed to MPP+ (5 μM) for different times. (a and b) Using a targeted analysis approach, the absolute levels of (a) cysteine (2.98 pmol/106 control cells) and cystine (3.6 pmol/106 control cells) as well as of the (b) polyamines putrescine (1.06 nmol/106 control cells), spermidine (0.17 nmol/106control cells) and spermine (0.28 nmol/106 control cells) were measured in three independent experiments. Data are displayed after normalization to controls (*P≤0.05). For background information, a scheme of ornithine–polyamine metabolism is displayed (SAM, S-adenosylmethionine; Arg, arginine; arrow indicates direction of regulation by MPP+). (c) Relative glucose oxidation in the TCA cycle was determined by using uniformly labeled [U-13C6]glucose and by determination of mass isotopomer distribution of citrate. M2 mass isotopomers of citrate indicate relative carbon influx from glycolysis via the pyruvate dehydrogenase complex in the TCA cycle. The dopamine transport inhibitor GBR-12935 (1 μM) was used in some experiments as specificity control together with MPP+ to prevent all intracellular effects of MPP+ (*P≤0.05). (d) The absolute cellular concentrations of phosphatidylcholines, plasmalogens and lysophosphatidylcholines were determined in the same experiments as in (a and b). Data were normalized to those of control cells. Numbers below the bars indicate number of total acyl/carbon atoms and double bonds
In addition, many changes outside the core energy and amino acid metabolism were observed. Large numbers of phosphatidylcholines, plasmalogens and lysophosphatidylcholines were increased, but it is at present unclear how such possibly secondary changes relate to toxicity pathways or cellular stress adaptation (Figure 3d). To better understand the relevance of metabolite changes for conclusions on altered pathways, we complemented metabolomics analyses with an orthogonal approach, that is, transcriptome analysis.
Transcriptome changes preceding cell death in MPP+-exposed neurons
According to Affymetrix DNA microarray analysis (Affymetrix, Santa Clara, CA, USA), 411 probe sets (PSs) were changed by MPP+ treatment (Supplementary Table S2). Heatmap presentation, clustering analysis and PCA (Figure 4a and Supplementary Figure S5) suggested a high reproducibility of the response across different cell preparations. When the PSs were sorted with respect to the time course of gene regulation, four major clusters were apparent (Figure 4b). Clusters 1 and 2 contained the PSs monotonously down- or up-regulated over time. Cluster 3 contained PSs upregulated only after long exposure, and cluster 4 contained PSs first downregulated and then compensated back to base level at later time points. In a different grouping approach, we combined all PSs that were already significantly downregulated at 24 h to cluster 5 (n=64), or upregulated at 24 h to cluster 6 (n=116).
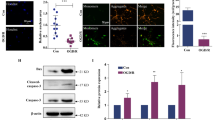
MPP+-induced transcriptome changes and their functional annotation. (a) Cells were treated with MPP+ (5 μM) for different times before samples were taken for DNA microarray-based transcriptome analysis. Probe sets significantly altered at least at one time point are displayed (FDR adjusted P-value of ≤0.05; fold change values ≥2). Colors represent Z-scores of the row-wise normalized expression values for each probe set. The Spearman’s correlations of the samples are indicated above the heatmap. Gene clusters (1–4) consist of probe sets with similar expression profiles. (b) Graphs display fold changes of the top 80 regulated genes for clusters 1 and 2 and of all genes of clusters 3 and 4. The black solid line represents the mean tendency of all genes of the cluster. (c) Overrepresented gene ontology (GO) terms are displayed as wordclouds for every cluster separately. For cluster 1 (downregulated) and cluster 2 (upregulated), only the top 30 GOs with a P-value of ≤0.001 are displayed (remaining GOs can be found in Supplementary Table S4). For cluster 5 (all genes downregulated significantly after the 24 h time point) and cluster 6 (upregulated at 24 h), all overrepresented GOs are displayed. (d) Representative images of cells with labeled paraspeckles component 1 protein (PSPC1, red) are displayed. Cells were treated with 5 μM MPP+ for 24 or 48 h and fixed for immunostaining. Compared with the nuclear counterstain (green), PSPC1 strongly decreased over time. (e) Cells were treated as in (a), and samples were analyzed by RNA sequencing (RNAseq). Overrepresented GOs were identified, and the ones that were not contained in the microarray data are displayed. A complete list is supplied in Supplementary Table S5. Calibration of wordcloud displays (indicated in dark blue): the height of the letters reflects the P-value of the GO
Significantly overrepresented gene ontology (GO) terms were identified for the transcript clusters to gain information on adaptive responses and stress pathways potentially triggered in the cells (Figure 4c and Supplementary Table S4). The GO of cluster 1 strongly pointed to changes of chromatin organization and related processes (mitosis, DNA conformation/packaging). Among the PSs of the related/overlapping cluster 5 (early downregulation), only one GO term, paraspeckles, was overrepresented. We verified this exemplary finding on the protein level by immunostaining and western blotting, and our data corroborated the downregulation of the paraspeckle-associated paraspeckles component 1 (PSPC1) protein (Figure 4d and Supplementary Figure S3C). Overrepresented GOs within upregulated genes (clusters 2 and 6) pointed to changes in metabolic processes related to amino acid and carboxylic acid turnover, but surprisingly not to, for example, glycolysis or the pentose phosphate cycle (Figure 4c and Supplementary Table S4).
As second approach to explore changes in gene expression, we used RNA deep sequencing (RNAseq) (Supplementary Table S3). At 24 h, 376 transcripts were altered (Figure 5a). The genes that had been identified by microarray analysis were confirmed by RNAseq, and the quantitative results were correlated to a high degree (Figure 5b). The additional transcripts identified by RNAseq, but not microarray, yielded information on the expression of genes encoded by mitochondrial DNA: MPP+ exposure significantly reduced the levels of transcripts of complex I and III subunits of the respiratory chain, whereas it did not have an effect on nuclear-encoded subunits of these complexes (Figure 5c). To capture all additional information contributed by RNAseq, we identified all overrepresented GOs among this data set, and compared them with those of the microarray analysis. The additional GOs among the sequencing data pointed to ‘alterations of ion transport’ (among upregulated genes) and to ‘disturbances in electron transport coupled to ATP synthesis’ and to ‘spindle/microtubule cytoskeleton organization’ (among downregulated genes) (Figure 4e and Supplementary Table S5).
Time course of transcriptome changes identified by RNA sequencing and RT-qPCR. (a) Cells were treated with MPP+ (5 μM) for different times before samples were taken for RNA sequencing (RNAseq) analysis. Differentially expressed transcripts were identified (FDR adjusted P-value of ≤0.05; fold change values ≥2). The numbers of upregulated genes are highlighted in red and the downregulated genes in blue. (b) Scatter plot of fold changes as determined by microarray or RNAseq. Each data point corresponds to one MPP+-regulated transcript. (c) Expression values for transcripts coded by mitochondrial genes were selected from RNAseq data set. Regulated complex I subunits are highlighted in orange. The scheme of complex I illustrates the location of these subunits (orange) in the protein complex. (d) Cells were treated as in (a) and mRNA was prepared after 2–48 h. The samples were analyzed by RT-qPCR for selected marker genes. Data are means±S.E.M. of three independent differentiations. Color coding indicates biological processes that the genes are involved with. ASNS, asparagine synthase; ASS1, argininosuccinate synthase 1; CBS, cystathionine-β-synthase; CCNB1, cyclin B1; CTH, cystathionase (cystathionine γ-lyase); DDIT3, DNA damage-inducible transcript 3 (CHOP, GADD153); DDIT4, DNA damage-inducible transcript 4; GADD34, growth arrest and DNA damage-inducible protein (PPP1R15A); HNRNPM, heterogeneous nuclear ribonucleoprotein M; MLF1IP, centromere protein U (MLF1 interacting protein); NOXA, phorbol-12-myristate-13-acetate-induced protein 1 (PMAIP1); NQO1, NAD(P)H dehydrogenase, quinone 1; PHGDH, phosphoglycerate dehydrogenase; PPA2, pyrophosphatase (inorganic) 2; PSAT1, phosphoserine aminotransferase 1; PSPC1, paraspeckles component 1; PSPH, phosphoserine phosphatase; SFPQ, Splicing factor proline/glutamine-rich; SHMT2, Serine hydroxymethyl-transferase; SLC3A2, solute carrier family 3 (amino acid transporter heavy chain), member 2; SLC7A11, solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11; TXNIP1, thioredoxin-interacting protein; TYMS, thymidylate synthetase
Confirmation of transcriptome data by detailed PCR analysis
To obtain more information on the time course of transcriptome changes, expression of several genes identified by the two Omics approaches was followed by reverse transcription quantitative PCR (RT-qPCR) analysis (Figure 5d). Many of the transcripts were changed as early as 2–12 h after treatment. Thioredoxin interacting protein 1 expression was reduced already after 2 h, thereby allowing improved antioxidant defenses. Genes playing roles in chromosomal stability, such as HNRNPM (heterogeneous nuclear ribonucleoprotein), were also downregulated after as little as 6 h. Genes involved in adaptive central carbon metabolism (argininosuccinate synthase (ASS1) and serine-hydroxy-methyl-transferase) or in cysteine synthesis via the transsulfuration pathway (cystathionase (CTH) and cystathionine-β-synthase) were up-regulated between 12 and 24 h (Figure 5d). The same applied to protein levels of CTH and ASS1 and the activity of the cystine transporter (Supplementary Figure S6). We also explored paraspeckle-related genes further, and several of these, such as PSPC1, SFPQ (splicing factor proline/glutamine-rich) and HNRNPM, were coordinately downregulated. The corresponding proteins all contribute to paraspeckle structures that are presumed to play a role in mRNA retention in the nucleus.31, 32 As these rapid and distinct regulations had not been expected by us, we used a different, but mechanistically related, damage model for confirmation: the cells were exposed to the complex I inhibitor rotenone (100 nM) and very similar transcriptional changes were observed (Supplementary Figure S7). Thus, the transcriptome response we observed for MPP+ may reflect a general response to mitochondrial inhibition in human neurons.
Identification of ATF-4 as superordinate regulator of transcriptome changes
Some superordinate biological regulators should be identifiable, if the transcriptome response reflects indeed a cellular adaptation program in the early phase of cell death. We calculated statistical overrepresentation of pathways among our gene expression data to identify putative upstream regulators. Transcriptome data were linked to known transcription factor–gene networks, and the transcription factor ATF4 (activating transcription factor 4) signaling network emerged as the only significant pathway from this analysis (Figure 6a). ATF4 was thus identified by this completely nonbiased bioinformatics approach as very likely (P<10−25) upstream regulator, even though the gene itself was not among the hits of the original Omics analyses. We used PCR as a more sensitive method (rigorous statistical criteria had been applied to filter hits of the Omics approaches) to examine the regulation of ATF4. A 2.5-fold increase by RT-qPCR was indeed detected (Figure 6b). As the activity of ATF4 is mainly regulated on the translational level, we also examined protein levels. A strong and early increase was identifiable by western blot, and high levels were maintained for at least 2 days (Figure 6c). To further verify the initiation of ATF-4 signaling, we examined some upstream and downstream components of this pathway. The eukaryotic initiation factor 2α (eIF2α) inhibits the translation of ATF4 mRNA, and this block is released by phosphorylation of eIF2α. We observed here such phosphorylation at 6–12 h after exposure to MPP+, consistent with the strong rise of ATF4 protein levels during this time (Figure 6c). As downstream target of ATF4, we examined GADD34, and this was continuously upregulated over time (Figure 6c). Thus, the biochemical observation of ATF-4 pathway activation on several levels corroborated the suggestion from our initial bioinformatic analysis. To gain further insight on metabolic pathways, transcriptomics data were also mapped in the next step together with metabolite data onto known human metabolism networks.
Bioinformatic identification of ATF4 as upstream regulator of transcriptional upregulation. (a) Bioinformatic analysis with IPA software identified ATF4 as regulator of genes that were upregulated (cluster 2). The genes in cluster 2 that are known to be ATF4 targets are indicated, together with their extent of regulation (relative fold change of 24 h versus 0 h) according to microarray analysis. Pathways, in which the ATF4 target genes are involved, are indicated in dark blue (pw, pathway; AA, amino acid). (b) Cells were treated with MPP+ (5 μM), and ATF4 mRNA levels were analyzed by RT-qPCR (relative to GAPDH expression) after different times. (c) Cell lysates were prepared after different times following MPP+ treatment. They were analyzed by western blot for key elements of the ATF4 pathway. Data are representative for 3–4 experiments. eIF2a[pS52], eukaryotic initiation factor 2 α-phosphorylated at serine 52
Identification of transsulfuration changes by combined Omics pathway analysis
Metabolomics and transcriptomics data were used on a multi-Omics platform for combined pathway enrichment analysis (Figure 7 and Supplementary Figure S8). Serine emerged as an important knot. Its absolute levels were only regulated to a minor degree, but its generation from glucose was increased. Its downstream metabolites glycine and cystathionine were upregulated, as were the transcripts for the corresponding enzymes (SHMT2 and CBS). Moreover, three sequential enzymes linking glycolytic intermediates and serine were also induced. This appears to be rather a coordinate response than a random finding, as all these enzymes are known downstream targets of ATF-4. Changes of this metabolic pathway were linked for example, to altered C1 metabolism, and by this way to DNA turnover. However, the most significant link was to the ‘transsulfuration pathway’ and some of its upstream elements. The upregulation of this metabolic route indicated that rate-limiting glutathione precursors (glycine and cysteine) were generated by MPP+-exposed cells at accelerated rates. Our data suggest that the increased levels of the methionine-derived thiols SAM, SAH and homocysteine, as well as the serine biosynthesis pathway acted as precursors for the transsulfuration process. The biological significance of this alteration is underlined by the additional finding that the transporter responsible for replenishing cellular cysteine from extracellular sources was also upregulated.
Combined metabolomics–transcriptomics identification of pathways affected by MPP+. Transcriptomics and metabolomics data of the 24 h treatment sample with 5 μM MPP+ were used for pathway mapping. Enzymes (corresponding mRNA) and metabolites that were upregulated are displayed in red. Blue font indicates decreased levels. ATF4 targets are encircled in orange (SLC7A11 has been identified by RNAseq only, the other target genes were identified on both transcriptomics platforms). Underlying biological processes affected by the indicated changes are displayed in green. CBS, cystathionine-β-synthase; CTH, cystathionase; DHFR, dihydrofolatereductase; dTMP, deoxythymidine monophosphate; dUMP, deoxyuridine monophosphate; GSH, glutathione; MTHFD2, methylenetetrahydrofolate dehydrogenase; PHGDH, phosphoglycerate dehydrogenase; PSAT1, phosphoserine aminotransferase 1; PSPH, phosphoserine phosphatase; ROS, reactive oxygen species; SHMT2, serine hydroxymethyl-transferase; TYMS, thymidylate synthetase. M2 citric acid and M2 fumaric acid are downregulated mass isotopomers representing the relative downregulation of glucose oxidation in the tricarboxylic acid (TCA) cycle
Finally, we addressed the question of higher GSH levels as result of the aforementioned regulations. GSH was measured 24 h after exposure to different MPP+ concentrations. Significant increases (by 10%) were observed at 1 μM, but not at 5 μM (Figure 8). These data showed the augmentation of GSH levels by metabolic stress, but the effect may have been masked by simultaneously increased demand. To allow measurements under conditions of less demand, we used a slightly modified model: day 5 (d5) neurons are much less sensitive to MPP+ toxicity, although they already express DAT and accumulate the toxicant30 (Figure 8). Under these conditions, 1 μM MPP+ raised cellular GSH levels by ∼50% at 24 h, and the effect started after few hours and lasted for 3 days. These metabolic regulations were accompanied by upregulation of ATF-4 and the transsulfuration enzymes CTH and CBS. Knockdown of ATF-4 reduced GSH levels in MPP+-challenged cells, and siRNA against CTH completely abolished the MPP+-driven GSH increase (Figure 8). In summary, these experiments fully corroborated the early upregulation of GSH supply by MPP+ treatment, and they provide evidence that the altered transsulfuration pathway, as identified here, indeed changes cellular properties.
Separation of MPP+ toxicity and GSH counterregulation in immature cells. Mature (d8) or immature cells (d5) were analyzed after treatment with MPP+ for the times indicated. (a) Intracellular GSH concentrations were determined for mature LUHMES cells (d8) treated with various concentrations (0.01, 0.5, 1, 5 and 25 μM) of MPP+ for 24 h (*P-value ≤0.05). (b) Intracellular ATP levels (as measure of overall viability) were determined in d5 LUHMES cells treated with 1, 5 or 25 μM MPP+ for the indicated times. No significant changes were observed. (c) LUHMES (d5) were treated as in (a), (MPP+ for 24 h) and intracellular GSH was determined (*P-value ≤0.05). (d) Intracellular GSH concentrations were determined for d5 cells treated for various durations with 1 μM MPP+ (*P-value ≤0.05). (e) Lysates of d5 cells were prepared after exposure for indicated time periods to 1 or 5 μM MPP+. Proteins were analyzed by western blot. ATF4 was visualized by immunoblotting and GAPDH was used as loading control. (f) The transsulfuration pathway enzyme cystathionase (CTH) and cystathionine-β-synthase (CBS) mRNA levels were evaluated by RT-qPCR in d5 cells treated with 1 μM (blue) and 5 μM (red) MPP+ for different times. (g) Intracellular GSH concentrations were determined for d5 cells treated with siRNA against ATF4 or CTH for 24 h (d2–d3) and subsequently with MPP+ (d3–d5). Scrambled siRNA (scr) was used as experimental negative control. Data are means±S.E.M. (n=4), and they are presented relative to untreated cells in each experiment. NS, nonsignificant difference between solvent and MPP+-treated cells in the presence of CTH siRNA. **P-value ≤0.01 (increase of GSH by MPP+ in scr-treated cells; reduction of GSH by specific siRNAs)
Discussion
A comprehensive description of cell death events requires knowledge of all processes taking place on the level of protein modifications as well as protein interactions. For modeling and quantitative predictions, all these changes, and their interdependence, will need quantification. In death receptor or cytokine-driven apoptosis, such studies are on the way,33, 34 whereas, for example, neurodegenerative events have only been described very roughly.35, 36, 37, 38 Models in this field require knowledge of changes of cellular metabolites and their fluxes through different pathways. Indeed, first quantitative models have been developed concerning metabolic stress and mitochondrial dysfunction, but their scope is extremely narrow,22, 39 and it is not clear which pathways need to be covered at all. For instance, there is still little information on the adaptive cellular changes taking place before the point of no return in MPP+-triggered neuronal death, although this toxicant has been the subject of thousands of papers. To open a door to capturing early pathways, we used combined analysis of global metabolite and transcriptome changes. The first pathway discovered by this new approach in neurotoxicology was ‘transsulfuration’, a cellular adaptation to provide additional GSH precursors. An unexpected prediction from this finding was that MPP+ exposure would lead to an initial increase of total GSH levels, instead of the depletion commonly observed in late phases of this model.30, 40, 41 This was indeed confirmed experimentally. Our finding now also provides a mechanistic basis for occasional reports in the literature that reported early increases of GSH upon neuronal stress.42, 43 The second major finding that emerged in an unbiased way from our study was identification of ATF4 as superordinate regulator of early transcriptome changes. We verified upregulation of the transcription factor and proofed the activation of the ATF4 pathway as early as 6 h after exposure to MPP+. Our two major findings were coherent, as ATF4 target genes are involved in providing better cellular access to the GSH precursor cysteine.
The central role of ATF4 and GSH to counteract increasing cell stress are displayed in Supplementary Figure S9 in the context of all our findings of early adaptations before cellular homeostasis breaks down. This opens a new perspective to the complex regulation of cell death. In the future, the interconnections between metabolic changes and the better known protein posttranslational changes need to be explored. ATF4 has in the past often been closely associated with ER stress signaling, and it is also a key regulator of altered amino acid metabolism under situations of metabolic demand.44 Elegant work has shown that it is activated by proteasome inhibitors or during starvation to provide cells with the cysteine essential for their survival.45 Oxidative death in ATF4−/− fibroblasts was rescued by treatments that increased cysteine levels and restored GSH synthesis.46 Furthermore, upregulation of ATF4 in hippocampal HT22 cells resulted in higher expression of the cystine transporter (SLC7A11), higher GSH levels and better resistance toward oxidative stress.47 Here, we observed strong upregulation of SLC7A11 and the transsulfuration pathway. The different time courses may explain, for example, an initial drop in cystine levels, and a return to baseline upon increased expression of the cystine–glutamate transporter. Functional importance of this carrier, that is, higher cystine-uptake rates, has already been demonstrated in LUHMES cells treated with the oxidative stress inducer methamphetamine48 and was confirmed here (Supplementary Figure S6).
The different roles of ATF4, cell death versus amino acid metabolism, may explain the discrepant findings in the literature. A role for ER stress and ATF4 in dopaminergic neuron death has been suggested 10 years ago,49 but could not be confirmed.50, 51 Later, the protein was rather associated with pharmacological protection,48 then again with enhanced damage,52 and most recently with neuroprotection.53 Such apparently contradictive findings are frequently encountered for signaling factors that affect several pathways. In the future, it will be more adequate to describe their role as nodes of a network, and based on the relative contributions of the responses they trigger. The research into the interaction of metabolite changes and signaling cascades necessary for this is likely to provide entirely new perspectives.49, 54 In a first modeling approach, the interaction of neurodegenerative protein aggregation and metabolism has been addressed. This research pointed to a switch of energy supply from glycolysis and alterations of serine–glycine turnover.36 Such adaptations are similar to those observed here experimentally.
The contribution of new cellular events, not recognized hitherto, needs to be better understood for predictive models. For instance, the change of paraspeckles by MPP+ was entirely unexpected. These structures may be related to RNA retention into the nucleus.55 In addition, the suggestion from RNAseq data of an altered microtubule cytoskeleton did not fit with canonical MPP+ signaling, but the data confirmed earlier findings of reduced mitochondrial mobility in LUHMES cells or PC12 cells upon MPP+ exposure.29, 56 Thus, our work does not only underline the importance of endogenous metabolism for a systems biology understanding of neurotoxicity, but also points to gaps in knowledge and available technologies. We show here the general potential of using metabolite data in combination with transcriptomics/proteomics data to comprehensively describe a systems response. The focus on early cellular adaptations, as opposed to the death execution, provides a basis for new neuroprotective strategies, based on strengthening endogenous defense strategies.
Materials and Methods
Dibutyryl-cAMP (cAMP), fibronectin, hoechst bisbenzimide H-33342, resazurin sodium salt, tetracycline, tetramethylrhodamine ethyl ester (TMRE) and MPP+ were from Sigma (Steinheim, Germany). Recombinant human FGF-2 and recombinant human GDNF were from R&D Systems (Minneapolis, MN, USA). Tween-20 and sodium dodecyl sulfate (SDS) were from Roth (Karlsruhe, Germany). All culture reagents were from Gibco (Carlsbad, CA, USA) unless otherwise specified.
Cell culture
Handling of LUHMES human neuronal precursor cells was performed as previously described in detail. Briefly, the conditionally immortalized cells (expressing a v-myc transgene under the control of a tet-off system28, 30, 48), maintained in proliferation medium, consisting of advanced DMEM/F12 containing 2 mM L-glutamine, 1 × N2 supplement (Invitrogen, Darmstadt, Germany) and 40 ng/ml FGF-2 in a 5% CO2/95% air atmosphere at 37 °C, were passaged every other day. For differentiation, 8 million cells were seeded in a Nunclon T175 tissue culture flask (Nunc A/S, Rosklide, Denmark) in proliferation medium. After 24 h, medium was changed to differentiation medium (DM II) consisting of advanced DMEM/F12 supplemented with 2 mM L-glutamine, 1 × N2, 2.25 μM tetracycline (to switch off the transgene), 1 mM dibutyryl 3′,5′-cyclic adenosine monophosphate (cAMP) and 2 ng/ml recombinant human glial cell-derived neurotrophic factor (GDNF). After 48 h, cells were trypsinized and seeded in a density of 1.84 × 105 cells/cm2 on dishes precoated with 50 μg/ml poly-L-ornithine (PLO) and 1 μg/ml fibronectin in advanced DMEM/F12 containing 2 mM L-glutamine, 1 × N2 and 2.25 μM tetracycline but without cAMP and GDNF (DM). On day 4 of differentiation, medium was exchanged with tetracycline-free DM.
Standard experimental setup
Cells were seeded at a density of 3.5 × 105 cells per well in 500 μl DM on PLO/fibronectin coated 24-well dishes. At day 6 of differentiation the time series of MPP+ exposure started in a reverse manner by adding 5 μM MPP+ to the media at different initiation times (e.g., treatment for 48 h started on day 6, treatment for 24 h started on day 7). Analyses were performed on day 8 of differentiation.
Cell viability measurement
Resazurin
Metabolic activity was detected by a resazurin assay. Briefly, resazurin solution was added to the cell culture medium to obtain a final concentration of 10 μg/ml. After incubation for 30 min at 37°C, the fluorescence signal was measured at an excitation wavelength of 530 nm using a 590 nm long-pass filter to record the emission. Fluorescence values were normalized by setting fluorescence values of untreated wells as 100% and the values from wells containing <5% calcein-positive cells as 0%.
LDH release
LDH activity was detected separately in the supernatant and cell homogenate. Cells were lysed in PBS/0.1% Triton X-100 for 2 h. Then, 20 μl of sample was added to 180 μl of reaction buffer containing NADH (100 μM) and sodium pyruvate (600 μM) in KPP buffer. Absorption at 340 nm was measured at 37°C in 1 min intervals over a period of 15 min. The slope of the absorption intensity was calculated. The ratio of LDHsupernatant/LDHtotal was calculated using the slopes of supernatant and homogenate. LDH release was expressed in percent. Control data were subtracted from LDH values. Basic release of untreated cells was ∼7% in 24 h.
Calcein-AM/TMRE staining
Calcein-AM staining, labeling live cells, and TMRE staining, labeling all intact mitochondria, were performed with 1 μM Calcein-AM/50 nM TMRE/1 μg/ml H-33342 for 30 min at 37°C. Images were collected in three different fluorescent channels using an automated microscope (Array-Scan VTI HCS Reader (Thermo Fisher, Pittsburgh, PA, USA). Using an imaging software (vHCS SCAN, Thermo Fisher), nuclei were identified in channel 1 (365±50/461±15 nm) as objects according to their size, area, shape and intensity. Calcein signal was detected in channel 2 (475±40/525±15 nm). An algorithm quantified all calcein-positive cells as viable and only H-33342-positive nuclei as ‘not viable’ cells.
For evaluating the mitochondrial mass, nuclei masks, determined in channel 1, were expanded and transferred to channel 3. All TMRE-positive pixels (575±25/640±35 nm) outside of these masks were counted as mitochondrial mass.
ATP determination
To determine intracellular ATP, cells grown in 24-well plates were scratched and sonicated in PBS buffer and boiled at 95°C for 10 min followed by centrifugation at 10 000 × g for 5 min for the removal of cell debris. For the detection of ATP levels, a commercially available ATP assay reaction mixture (Sigma) containing luciferin and luciferase was used. To a black 96-well plate, 50 μl of sample and 100 μl of assay mix were added. Standards were prepared by serial dilutions of ATP disodium salt hydrate (Sigma) to obtain final concentrations ranging from 1000 to 7.8 nM.
GSH determination
For GSH determination, cells were washed twice with PBS and lysed in 200 μl of 1% sulfosalicylic acid (w/v). The lysates were collected, sonicated 5 times and centrifuged at 12 000 × g for 5 min at 4 °C to remove cell debris. Total GSH content was determined by a DTNB (5,5′-dithiobis(2-nitrobenzoic acid)) reduction assay. Supernatants were diluted 1 : 10 in H2O, and 100 μl sample was mixed with 100 μl assay mixture containing 300 μM DTNB, 1 U/ml GSH reductase, 400 μM NADPH and 1 mM EDTA in 100 mM sodium phosphate buffer, pH 7.5 (all from Sigma). DTNB reduction was measured photometrically at 405 nm in 5 min intervals over 30 min. GSH standard curves were performed by serial dilutions ranging from 1000 to 7.8 nM, respectively.
Western blot analysis
Cells were lysed in RIPA buffer (50 mM Tris base, 150 mM NaCl, 1 mM EDTA, 0.25% sodium deoxycholate, 1% NP-40, 1 mM Na3VO4 and 50 mM NaF, pH 7.5) containing 1 × protease inhibitor (Roche, Basel, Switzerland) and 0.5% phosphatase inhibitor cocktail 2 (Sigma). Determination of protein concentration was performed by using a BCA protein assay kit (Pierce/Thermo Fisher Scientific, Rockford, IL, USA). Next, 35 μg of total protein was loaded onto 12% SDS gels. Proteins were transferred onto nitrocellulose membranes (Amersham, Buckinghamshire, UK). Loading and transfer were checked by brief Ponceau staining. Washed membranes were blocked with either 5% milk or 5% BSA, dependent on the primary antibody used, in TBS–Tween (0.1%) for 1 h. Primary antibodies were incubated at 4°C overnight. Following washing steps with TBS–Tween (0.1%), horseradish peroxidase-conjugated secondary antibodies were incubated for 1 h at RT. For visualization, ECL Western blotting substrate (Pierce/Thermo Fisher Scientific) was used. For the detailed list of antibodies used, see Supplementary Figure 10.
Immunocytochemistry
Cells were grown on 13 mm glass cover slips (Menzel, Braunschweig, Germany) in 24-well plastic cell culture plates (Nunclon) and fixed with 4% paraformaldehyde. After incubation with the primary antibody overnight and with the appropriate secondary antibody for 1 h, Hoechst-33342 (1 μg/ml Molecular Probes, Eugene, OR, USA) was added for 10 min before the final washing step. Cover slips were mounted on glass slides with Fluorsave reagent (Calbiochem, Merck, Darmstadt, Germany). For visualization, a confocal microscope (Zeis LSM510Meta) was used. For image processing, Photoshop (Adobe, San Jose, CA, USA) was used. The antibody against PSPC1 was from Sigma (rabbit, 1 : 200). As secondary antibody, anti-rabbit Alexa-488 (1 : 1000, Molecular Probes) was applied.
Quantitative PCR
For RT-qPCR analysis, RNA was extracted with the PureLink RNA mini Kit (Invitrogen) according to the manufacturer’s instructions. For transcript analyses of LUHMES, primers (Eurofins MWG Operon, Ebersberg, Germany) were designed using AiO (All in One) bioinformatics software57 and can be found in Supplementary Figure 11. All RT-qPCRs were based on the SsoFast EvaGreen detection system and were run in a CFX96 Cycler (Bio-Rad, München, Germany) and analyzed with Bio-Rad iCycler software. The threshold cycles (Ct) were determined for each gene and gene expression levels were calculated as relative expression compared with GAPDH (2−(ΔCt)) or as fold change relative to control (2−(ΔΔCt)). ΔCt and ΔΔCt were calculated using following formulas:


The siRNA-mediated knockdown of ATF4 and CTH
Knockdown experiments were performed essentially as described earlier58 in 24-well dishes using 15 pmol/well siRNA (siATF4: sc-35112 Santa Cruz Biotechnology Inc., Santa Cruz, CA, USA; siCTH: sc-78973 Santa Cruz Biotechnology Inc.; scrambled siRNA: 1027280 Qiagen, Hilden, Germany) and 1 μl lipofectamine (Lipofectamine 2000, Life Technologies, Carlsbad, CA, USA). The siRNA/lipofectamine mix was preincubated in 75 μl Opti MEM (Life Technologies) for 25 min. Then, 3.5 × 105 pre-differentiated LUHMES (d2) cells were added in 425 μl LUHMES differentiation medium. Medium was exchanged 24 h after seeding (d3), and cells were treated with 1 μM MPP+ or solvent for 48 h. GSH levels were determined at d5. Knockdown of the siRNAs used was confirmed by WB (at least 50% on protein level).
Affymetrix gene chip analysis
Analysis was performed as described earlier.7 Briefly, samples from ∼5 × 106 cells were collected using RNA protect reagent from Qiagen. The RNA was quantified using a NanoDrop N-1000 spectrophotometer (NanoDrop, Wilmington, DE, USA), and the integrity of RNA was confirmed with a standard sense automated gel electrophoresis system (Experion, Bio-Rad, Hercules, CA, USA). The samples were used for transcriptional profiling when the RNA quality indicator (RQI) number was >8. First-strand cDNA was synthesized from 100 ng total RNA using an oligo-dT primer with an attached T7 promoter sequence, followed by the complementary second strand. The double-stranded cDNA molecule was used for in vitro transcription (IVT, standard Affymetrix procedure) using Genechip 30 IVT Express Kit. During synthesis of the aRNA (amplified RNA, also commonly referred to as cRNA), a biotinylated nucleotide analog was incorporated that serves as a label for the message. After amplification, aRNA was purified with magnetic beads and 15 μg of aRNA was fragmented with fragmentation buffer as per the manufacturer’s instructions. Then, 12.5 μg fragmented aRNA was hybridized with Affymetrix Human Genome U133 plus 2.0 arrays as per the manufacturer’s instructions. The chips were placed in a GeneChip Hybridization Oven-645 for 16 h at 60 r.p.m. and 45°C. For staining and washing, Affymetrix HWS kits were used on a Genechip Fluidics Station-450. For scanning, the Affymetrix Gene-Chip Scanner-3000-7G was used, and the image and quality control assessments were performed with Affymetrix GCOS software. All reagents and instruments were acquired from Affymetrix. The generated CEL files were used for further statistical analysis. The authors declare that microarray data were produced according to MIAME guidelines and will be deposited in ArrayExpress upon acceptance of the manuscript.
Statistics and data mining
The microarray data analysis workflow was assembled using the Konstanz Information Miner open source software. The raw data were preprocessed using Robust Multiarray Analysis (RMA). Background correction, quantile normalization and summarization were applied to all expression data samples using the RMA function from the affy package of Bioconductor.59 Low-expression genes with a signal below an intensity of 64 in any one of the conditions were filtered out. The limma package (R; http://www.r-project.org/ and Bioconductor; http://www.bioconductor.org/) was used to identify differentially expressed genes, with untreated cells set as control group. The moderated t-statistics was applied and was used for assessing the raw significance of differentially expressed genes. Then, final P-values were derived by using the Benjamini–Hochberg method to control the false discovery rate (FDR) due to multiple hypothesis testing. Transcripts with FDR adjusted P-value of ≤0.05 and fold change values of ≥2 were considered significantly regulated. The hierarchical clustering analysis was performed as previously described.60 Average linkage was used as agglomeration rule for the clustering analysis. Distances based on the Pearson’s correlation coefficient was used to group together transcripts with similar expression patterns across samples (rows of the heatmap). Distances based on Spearman’s rank correlations of the gene expression values were used to measure the similarity between samples. Then, expression values within each row were normalized as Z-factors and color coded accordingly. The colors represent Z-scores of the row-wise normalized expression values for each gene, where the highest Z-score is in bright yellow and the lowest in dark red.
RNA sequencing
Illumina library preparation and sequencing
The sequencing library preparation has been done using 200 ng of total RNA input with the TrueSeq RNA Sample Prep Kit v2-Set B (RS-122–2002, Illumina Inc., San Diego, CA, USA) producing a 275-bp fragment including adapters in average size. In the final step before sequencing, eight individual libraries were normalized and pooled together using the adapter indices supplied by the manufacturer. Pooled libraries have then been clustered on the cBot Instrument form Illumina using the TruSeq SR Cluster Kit v3-cBot-HS (GD-401-3001, Illumina Inc.). Sequencing was then performed as 50 bp, single reads and 7 bases index read on an Illumina HiSeq2000 instrument using the TruSeq SBS Kit HS-v3 (50-cycle) (FC-401-3002, Illumina Inc.). Approximately 20–30 million reads per sample were sequenced.
RNAseq computational analysis
Illumina reads were converted to the industry standard FASTQ format and aligned to the Human GRCh37 Ensembl 70 reference genome using the STAR program on default settings (http://www.ncbi.nlm.nih.gov/pubmed/23104886). For increased alignment accuracy, the STAR genome index was generated to include splice junction annotations with the options ‘—sjdbGTFfile Homo_sapiens.GRCh37.70.primary_assembly.gtf —sjdbOverhang 49’. The SAM output from the STAR aligner was converted to BAM format using the Picard tools suite (http://picard.sourceforge.net). For gene expression estimation and differential expression analysis, the programs Cufflinks and Cuffdiff version 2.0.2 were used with the following options ‘-u —max-bundle-frags 1000000 —no-effective-length-correction —compatible-hits-norm’ (http://www.ncbi.nlm.nih.gov/pubmed/23222703). The quality of the RNAseq experiments was verified with RNASeQC version 1.17 (http://www.ncbi.nlm.nih.gov/pubmed/22539670). Pre-processed data from Cuffdiff 2.0.2 were further analyzed with CummeRbund 2.0.0 (http://compbio.mit.edu/cummeRbund/). A significance threshold of FDR (Benjamini–Hochberg multiple testing) corrected P-values was set at 0.05. The (base 2) log of the fold change y/x (FKPM) was used as measure for differential gene expression. For comparison with Microarray data, the ENSEMBL gene identifiers were converted to HGNC symbols. To test the correlation between the two platforms, the log2 fold changes of overlapping genes for the 48 h samples were plotted and Spearman’s correlation was calculated.
GO enrichment—wordclouds
To identify individual GOs for the altered genes of the transcriptomics studies, we entered the gene names into the query of gProfiler (http://biit.cs.ut.ee/gprofiler). Only GO terms consisting of <1000 genes were used to create GO wordclouds. If more than 30 GOs were identified, only the top 30 GOs with the lowest P-values were displayed. All identified GOs can be found in Supplementary Tables S4 and S5. The wordclouds were produced on http://www.wordle.net/advanced. Scaling of character size is linearly proportional to the negative log of the P-value of the respective GO category.
Metabolomics analysis—untargeted
After MPP+ treatment, cells were washed with ice cold PBS. Dry-ice cold 80 : 20 MeOH/water solution was added immediately to the wells. Cells were scraped and collected in a 1.5 ml Eppendorf tube (Eppendorf, Hamburg, Germany). Wells were washed again with MeOH/water and this solution was combined with the previous solution. Tubes were stored at −80°C for at least 2 h to precipitate the proteins. For metabolite extraction, tubes were placed on dry ice for 15 min and centrifuged at 14 000 × g for 5 min at 4°C. The supernatant was transferred to a new 1.5 ml tube and placed on dry ice. Then, 300 μl of 80 : 20 MeOH/water was added to the pellet and a second extraction was performed. The combined supernatants were evaporated to dryness at room temperature in a Speedvac concentrator (Savant, Thermo Fisher Scientific, Waltham, MA, USA). The dried samples were reconstituted with 60 μl of 60% MeOH with 0.1% FA and clarified by centrifugation at 14 000 × g for 5 min. The clarified samples were transferred to HPLC vials for LC-MS measurements.
Liquid chromatography
Chromatographic separations were performed using an Agilent 1260 high performance liquid chromatography system with a wellplate autosampler (Agilent, Santa Clara, CA, USA). For the reverse phase (RP) separation, a TARGA (Higgins, Mountain View, CA, USA) C18 column (50 × 2.1 mm i.d., 3 μm particle size, 120 Å pore size) was used on the LC system. The LC parameters for RPLC analysis were as follows: autosampler temperature, 4°C; injection volume, 5 μl; column temperature, 35°C; and flow rate, 0.3 ml/min. The solvents and optimized gradient conditions for LC were: Solvent A, water with 1 mM ammonium fluoride; Solvent B, 100% acetonitrile; elution gradient: 0 min 2% B; 20–25 min 98% B; post-run time for equilibration, 5 min in 2% B. For aqueous normal phase (ANP) separation, a Cogent Diamond Hydride (MicroSol, Eatontown, NJ, USA) column (150 × 2.1 mm i.d., 4 μm particle size, 100 Å pore size) was used for separation of metabolites. The LC parameters were as follows: autosampler temperature, 4°C; injection volume, 5 μl; column temperature, 35°C; and flow rate, 0.4 ml/min. The solvents and optimized gradient conditions for LC were: Solvent A, 50% methanol/50% water/0.05% formic acid; Solvent B, 90% acetonitrile with 5 mM ammonium acetate; elution gradient: 0 min 100% B; 20–25 min 40% B; post-run time for equilibration, 10 min in 100% B. The LC system was coupled directly to the Q-TOF mass spectrometer. A blank injection was run after every three samples and a QC sample was run after every five samples to identify the sample carryover and check for stability.
Mass spectrometry
A 6520 accurate-mass Q-TOF LC-MS system (Agilent) equipped with a dual electrospray (ESI) ion source was operated in negative-ion mode for metabolic profiling. The optimized ESI Q-TOF parameters for MS experiments were: ion polarity, negative; gas temperature, 325°C; drying gas, 10 l/min; nebulizer pressure, 45 psig; capillary voltage, 4000 V; fragmentor, 140 V; skimmer, 65 V; mass range, 70–1100 m/z; acquisition rate, 1.5 spectra/s; instrument state, extended dynamic range (1700 m/z, 2 GHz). MS/MS experiments were carried out to confirm the putative identification of metabolites based on mass accuracy. Nitrogen was employed as collision gas and collision energy was adjustable from 10 to 40 eV. Spectra were internally mass calibrated in real time by continuous infusion of a reference mass solution using an isocratic pump connected to a dual sprayer feeding into an electrospray ionization source. Data were acquired with MassHunter Acquisition software (Agilent Technologies, Santa Clara, CA, USA).
Statistics and data mining
For the data processing and chemometric analysis of the LC-MS data, the acquired raw data files (.d files) were processed with MassHunter Qualitative Analysis software (Agilent, version 5.0). Reproducibility of chromatograms was first inspected by overlaying the Total Ion Chromatograms (TICs) of all samples. Data files that showed extraneous peaks were excluded for further processing. Initially, putative metabolite identification was achieved by searching the accurate m/z values of the peaks against an in-house built database derived from HMDB, KEGG, METLIN and other public databases. At the same time, the Extracted Ion Chromatograms (EICs) for these matched putative metabolites were generated by performing ‘Find by Formula’ function integrated into the software. The abundance of the EICs was calculated by summing the intensities of all compound-related peaks (e.g., isotopic peaks, adduct peaks and so on). The preprocessed data files were exported as ‘cef’ formatted files that contain a table of mass and retention time pairs with associated intensities. These ‘cef’ files were imported into Mass Profiler Professional software (Agilent, version 12.1) for further data processing. For example, peak alignment, background noise subtraction and other data reduction processes could be done by using this software. The optimized parameters for these data processing steps were set as follows: minimum absolute abundance, 5000 counts; retention time range, 0–25 min; mass range, 70–1100 m/z; minimum of ions, 2; multiple charge state forbidden; retention time window, 1 min, mass window, 15 p.p.m.+2.0 mDa. To treat all extracted compounds equally regardless of their intensities, each entity was baselined to median of intensity of all samples. An ANOVA statistical test (P<0.05) followed by a Benjamini–Hochberg multiple test correction was performed with the normalized data for differential analysis. PCA was used for modeling the difference between the controls and MPP+-treated samples. A significant metabolite list was generated after ANOVA test and used for later pathway analysis.
MS/MS spectra acquired from reference metabolites were used for confirmation of the identification of statistically significant metabolites. More specifically, the exact m/z values and intensities of fragment ions from the acquired MS/MS spectra of putative metabolites must have a reasonable match with that of reference metabolites or the fragment ions from public databases (e.g., METLIN, MassBank), if available.
Metabolomics analysis—targeted
Sample preparation was made in the same manner as for the untargeted analysis with some minor modification, as there was no second extraction performed. Dried samples were stored at −80°C and send on dry ice to BIOCRATES Life Sciences AG (Innsbruck, Austria) to further process the samples. The frozen cell pellets were resuspended in 60 μl chilled phosphate buffer. Cell lysis was done by freezing the cell suspension in liquid nitrogen and thawing it in an ultrasonic bath (4°C). This freezing–thawing cycle was repeated 3 times. After this, the cell suspension was centrifuged at 2°C and the supernatant was directly use for analysis. The targeted metabolomics approach was based on measurements with the AbsoluteIDQTM p180 kit and the oxidative status assay (BIOCRATES Life Sciences AG). The p180 kit allows simultaneous quantification of 186 metabolites, consisting of amino acids, acylcarnitines, sphingomyelins, phosphatidylcholines, hexose (glucose) and biogenic amines. The fully automated assay was based on PITC (phenylisothiocyanate) derivatization in the presence of internal standards followed by FIA-MS/MS (acylcarnitines, lipids and hexose) and LC/MS (amino acids, biogenic amines) using an AB SCIEX 4000 QTrap mass spectrometer (AB Sciex, Darmstadt, Germany) with electrospray ionization. The experimental metabolomics measurement technique is described in detail by patent US 2007/0004044 (accessible online at http://www.freepatentsonline.com/y2007/0004044.html). For the oxidative status assay, three thiol amino acid redox couples (reduced and oxidized forms of homocysteine, cysteine and glutathione) were assayed by LC/MS/MS using a API 5500 mass spectrometer (AB Sciex). The analytes were separated on a porous graphitic carbon column (PGC) using gradient elution. The total run time of the analysis is 15 min and the injection volume was 20 μl. The analytes were quantified by positive ion tandem electrospray ionization mass spectrometry in multiple reaction monitoring mode using internal standard calibration.
Stable isotope-assisted metabolomics: GC/MS sample preparation and procedure
Cells were grown in six-well plates and incubated with 1 μM GBR-12935 and 12.5 mmol/l [U-13C6]glucose (total glucose concentration was 25 mmol/l (w/v)) on d6, and after 30 min, 5 μM MPP+ was added for 18 h. Cells were washed with 1 ml of 0.9% NaCl and quenched with 0.4 ml −20°C methanol. After adding an equal volume of 4°C cold water, cells were collected with a cell scraper and transferred in tubes containing 0.4 ml −20°C chloroform. The extracts were shaken at 1400 r.p.m. for 20 min at 4°C (Thermomixer Eppendorf) and centrifuged at 16 000 × g for 5 min at 4°C. Then, 0.3 ml of the upper aqueous phase was collected in GC/MS glass vials with microinserts and evaporated to dryness under vacuum at −4°C using a refrigerated CentriVap Concentrator (Labconco, Kansas City, MO, USA). Metabolite derivatization was performed using a Gerstel MPS. Dried polar metabolites were dissolved in 15 μl of 2% methoxyamine hydrochloride in pyridine at 40°C under shaking. After 30 min, an equal volume of 2,2,2-trifluoro-N-methyl-N-trimethylsilyl-acetamide was added and held for 30 min at 40°C under continuous shaking. Then, 1 μl sample was injected into an SSL injector at 270°C. GC/MS analysis was performed using an Agilent 7890A GC equipped with a 30 m DB-35MS+5m Duraguard capillary column (Agilent Technologies). Helium was used as carrier gas at a flow rate of 1 ml/min. The GC oven temperature was held at 80°C for 6 min and increased to 300°C at 6 °C/min. After 10 min, the temperature was increased to 325°C at 10°C/min for 4 min. The GC was connected to an Agilent 5975C inert XL MSD, operating under electron ionization at 70 eV. The MS source was held at 230°C and the quadrupole at 150°C. The detector was operated in scan mode with mass range m/z 70–800. The total run time of one sample was 59.167 min. All GC/MS chromatograms were processed by using MetaboliteDetector.61 To determine relative glucose oxidation in the TCA cycle, we calculated the mass isotopomer distribution (MID) for fragment 465 of citric acid, fragment 337 of malic acid and fragment 245 of fumaric acid.
Statistics and data mining
Cytotoxicity data (ATP, GSH, LDH, resazurin) and qPCR are presented as means of independent experiments, and statistical differences were tested by ANOVA with post hoc tests as appropriate using GraphPad Prism 5.0 (Graphpad Software, La Jolla, CA, USA).
Abbreviations
- GO:
-
gene ontology
- GSH:
-
glutathione
- MPP+:
-
1-methyl-4-phenylpyridinium
- MPTP:
-
methyl-phenyl-tetrahydropyridine
- PSPC1:
-
paraspeckles component 1
- ROS:
-
reactive oxygen species
- SAM:
-
S-adenosyl-methionine
- Omics:
-
generic term used for, for example, transcriptomics, metabolomics and so on
References
Tice RR, Austin CP, Kavlock RJ, Bucher JR . Improving the human hazard characterization of chemicals: a Tox21 update. Environ Health Perspect 2013; 121: 756–765.
Collins FS, Gray GM, Bucher JR . Toxicology. Transforming environmental health protection. Science 2008; 319: 906–907.
Krewski D, Acosta D, Andersen M, Anderson H, Bailar JC, Boekelheide K et al. Toxicity testing in the 21st century: a vision and a strategy. J Toxicol Environ Health B Crit Rev 2010; 13: 51–138.
Kwong LN, Costello JC, Liu H, Jiang S, Helms TL, Langsdorf AE et al. Oncogenic NRAS signaling differentially regulates survival and proliferation in melanoma. Nat Med 2012; 18: 1503–1510.
Carreras Puigvert J, von Stechow L, Siddappa R, Pines A, Bahjat M, Haazen LC et al. Systems biology approach identifies the kinase Csnk1a1 as a regulator of the DNA damage response in embryonic stem cells. Sci Signal 2013; 6: ra5.
Lee MJ, Ye AS, Gardino AK, Heijink AM, Sorger PK, MacBeath G et al. Sequential application of anticancer drugs enhances cell death by rewiring apoptotic signaling networks. Cell 2012; 149: 780–794.
Krug AK, Kolde R, Gaspar JA, Rempel E, Balmer NV, Meganathan K et al. Human embryonic stem cell-derived test systems for developmental neurotoxicity: a transcriptomics approach. Arch Toxicol 2013; 87: 123–143.
Balmer NV, Weng MK, Zimmer B, Ivanova VN, Chambers SM, Nikolaeva E et al. Epigenetic changes and disturbed neural development in a human embryonic stem cell-based model relating to the fetal valproate syndrome. Hum Mol Genet 2012; 21: 4104–4114.
Meganathan K, Jagtap S, Wagh V, Winkler J, Gaspar JA, Hildebrand D et al. Identification of thalidomide-specific transcriptomics and proteomics signatures during differentiation of human embryonic stem cells. PLoS One 2012; 7: e44228.
Theunissen PT, Robinson JF, Pennings JL, de Jong E, Claessen SM, Kleinjans JC et al. Transcriptomic concentration-response evaluation of valproic acid, cyproconazole, and hexaconazole in the neural embryonic stem cell test (ESTn). Toxicol Sci 2012; 125: 430–438.
Thomas RS, Wesselkamper SC, Wang NC, Zhao QJ, Petersen DD, Lambert JC et al. Temporal concordance between apical and transcriptional points of departure for chemical risk assessment. Toxicol Sci 2013; 131: 180–194.
Andersen ME, Clewell HJ, Carmichael PL, Boekelheide K . Can case study approaches speed implementation of the NRC report: ‘toxicity testing in the 21st century: a vision and a strategy?’. ALTEX 2011; 28: 175–182.
Langston JW, Langston EB, Irwin I . MPTP-induced parkinsonism in human and non-human primates—clinical and experimental aspects. Acta Neurol Scand Suppl 1984; 100: 49–54.
Bezard E, Przedborski S . A tale on animal models of Parkinson’s disease. Mov Disord 2011; 26: 993–1002.
Nicklas WJ, Vyas I, Heikkila RE . Inhibition of NADH-linked oxidation in brain mitochondria by 1-methyl-4-phenyl-pyridine, a metabolite of the neurotoxin, 1-methyl-4-phenyl-1,2,5,6-tetrahydropyridine. Life Sci 1985; 36: 2503–2508.
Choi WS, Kruse SE, Palmiter RD, Xia Z . Mitochondrial complex I inhibition is not required for dopaminergic neuron death induced by rotenone, MPP+, or paraquat. Proc Natl Acad Sci USA 2008; 105: 15136–15141.
Poltl D, Schildknecht S, Karreman C, Leist M . Uncoupling of ATP-depletion and cell death in human dopaminergic neurons. Neurotoxicology 2012; 33: 769–779.
Volbracht C, Leist M, Nicotera P . ATP controls neuronal apoptosis triggered by microtubule breakdown or potassium deprivation. Mol Med 1999; 5: 477–489.
Freeman BA, Crapo JD . Free-radicals and tissue-injury. Lab Invest 1982; 47: 412–426.
Saporito MS, Thomas BA, Scott RW . MPTP activates c-Jun NH(2)-terminal kinase (JNK) and its upstream regulatory kinase MKK4 in nigrostriatal neurons in vivo. J Neurochem 2000; 75: 1200–1208.
Schulz JB . Anti-apoptotic gene therapy in Parkinson's disease. J Neural Transm Suppl 2006; 70: 467–476.
Kolodkin A, Simeonidis E, Balling R, Westerhoff HV . Understanding complexity in neurodegenerative diseases: in silico reconstruction of emergence. Front Physiol 2012; 3: 291.
Geenen S, Taylor PN, Snoep JL, Wilson ID, Kenna JG, Westerhoff HV . Systems biology tools for toxicology. Arch Toxicol 2012; 86: 1251–1271.
Mazzio E, Soliman KF . Whole genome expression profile in neuroblastoma cells exposed to 1-methyl-4-phenylpyridine. Neurotoxicology 2012; 33: 1156–1169.
Burte F, De Girolamo LA, Hargreaves AJ, Billett EE . Alterations in the mitochondrial proteome of neuroblastoma cells in response to complex 1 inhibition. J Proteome Res 2011; 10: 1974–1986.
Pattarini R, Rong Y, Qu C, Morgan JI . Distinct mechanisms of 1-methyl-4-phenyl-1,2,3,6-tetrahydropyrimidine resistance revealed by transcriptome mapping in mouse striatum. Neuroscience 2008; 155: 1174–1194.
Zhang X, Zhou JY, Chin MH, Schepmoes AA, Petyuk VA, Weitz KK et al. Region-specific protein abundance changes in the brain of MPTP-induced Parkinson's disease mouse model. J Proteome Res 2010; 9: 1496–1509.
Scholz D, Poltl D, Genewsky A, Weng M, Waldmann T, Schildknecht S et al. Rapid, complete and large-scale generation of post-mitotic neurons from the human LUHMES cell line. J Neurochem 2011; 119: 957–971.
Schildknecht S, Karreman C, Poltl D, Efremova L, Kullmann C, Gutbier S et al. Generation of genetically-modified human differentiated cells for toxicological tests and the study of neurodegenerative diseases. ALTEX 2013; 30: 427–444.
Schildknecht S, Poltl D, Nagel DM, Matt F, Scholz D, Lotharius J et al. Requirement of a dopaminergic neuronal phenotype for toxicity of low concentrations of 1-methyl-4-phenylpyridinium to human cells. Toxicol Appl Pharmacol 2009; 241: 23–35.
Passon DM, Lee M, Rackham O, Stanley WA, Sadowska A, Filipovska A et al. Structure of the heterodimer of human NONO and paraspeckle protein component 1 and analysis of its role in subnuclear body formation. Proc Natl Acad Sci USA 2012; 109: 4846–4850.
Marko M, Leichter M, Patrinou-Georgoula M, Guialis A . hnRNP M interacts with PSF and p54(nrb) and co-localizes within defined nuclear structures. Exp Cell Res 2010; 316: 390–400.
Neumann L, Pforr C, Beaudouin J, Pappa A, Fricker N, Krammer PH et al. Dynamics within the CD95 death-inducing signaling complex decide life and death of cells. Mol Syst Biol 2010; 6: 352.
Janes KA, Albeck JG, Gaudet S, Sorger PK, Lauffenburger DA, Yaffe MB . A systems model of signaling identifies a molecular basis set for cytokine-induced apoptosis. Science 2005; 310: 1646–1653.
Fujita KA, Ostaszewski M, Matsuoka Y, Ghosh S, Glaab E, Trefois C et al. Integrating pathways of Parkinson’s disease in a molecular interaction map. Mol Neurobiol 2013; 49: 88–102.
Vazquez A . Metabolic states following accumulation of intracellular aggregates: implications for neurodegenerative diseases. PLoS One 2013; 8: e63822.
Miller JA, Horvath S, Geschwind DH . Divergence of human and mouse brain transcriptome highlights Alzheimer disease pathways. Proc Natl Acad Sci USA 2010; 107: 12698–12703.
Zhang B, Gaiteri C, Bodea LG, Wang Z, McElwee J, Podtelezhnikov AA et al. Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer’s disease. Cell 2013; 153: 707–720.
Zhou L, Aon MA, Almas T, Cortassa S, Winslow RL, O'Rourke B . A reaction-diffusion model of ROS-induced ROS release in a mitochondrial network. PLoS Comput Biol 2010; 6: e1000657.
Desole MS, Esposito G, Fresu L, Migheli R, Enrico P, Miele M et al. Correlation between 1-methyl-4-phenylpyridinium ion (MPP+) levels, ascorbic acid oxidation and glutathione levels in the striatal synaptosomes of the 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP)-treated rat. Neurosci Lett 1993; 161: 121–123.
Youdim MB, Drigues N, Mandel S . Oxidative stress indices in Parkinson’s disease: biochemical determination. Methods Mol Med 2001; 62: 137–153.
Drechsel DA, Liang LP, Patel M . 1-methyl-4-phenylpyridinium-induced alterations of glutathione status in immortalized rat dopaminergic neurons. Toxicol Appl Pharmacol 2007; 220: 341–348.
Seyfried J, Soldner F, Kunz WS, Schulz JB, Klockgether T, Kovar KA et al. Effect of 1-methyl-4-phenylpyridinium on glutathione in rat pheochromocytoma PC 12 cells. Neurochem Int 2000; 36: 489–497.
Han J, Backa SH, Hur J, Lin YH, Gildersleeve R, Shan JX et al. ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nature Cell Biology 2013; 15: 481.
Suraweera A, Munch C, Hanssum A, Bertolotti A . Failure of amino acid homeostasis causes cell death following proteasome inhibition. Mol Cell 2012; 48: 242–253.
Dickhout JG, Carlisle RE, Jerome DE, Mohammed-Ali Z, Jiang H, Yang G et al. Integrated stress response modulates cellular redox state via induction of cystathionine gamma-lyase: cross-talk between integrated stress response and thiol metabolism. J Biol Chem 2012; 287: 7603–7614.
Lewerenz J, Sato H, Albrecht P, Henke N, Noack R, Methner A et al. Mutation of ATF4 mediates resistance of neuronal cell lines against oxidative stress by inducing xCT expression. Cell Death Differ 2012; 19: 847–858.
Lotharius J, Falsig J, van Beek J, Payne S, Dringen R, Brundin P et al. Progressive degeneration of human mesencephalic neuron-derived cells triggered by dopamine-dependent oxidative stress is dependent on the mixed-lineage kinase pathway. J Neurosci 2005; 25: 6329–6342.
Vander Heiden MG, Cantley LC, Thompson CB . Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 2009; 324: 1029–1033.
Bernstein AI, O'Malley KL . MPP+-induces PUMA- and p53-dependent, but ATF3-independent cell death. Toxicol Lett 2013; 219: 93–98.
Holtz WA, O'Malley KL . Parkinsonian mimetics induce aspects of unfolded protein response in death of dopaminergic neurons. J Biol Chem 2003; 278: 19367–19377.
Lange PS, Chavez JC, Pinto JT, Coppola G, Sun CW, Townes TM et al. ATF4 is an oxidative stress-inducible, prodeath transcription factor in neurons in vitro and in vivo. J Exp Med 2008; 205: 1227–1242.
Sun X, Liu J, Crary JF, Malagelada C, Sulzer D, Greene LA et al. ATF4 protects against neuronal death in cellular Parkinson’s disease models by maintaining levels of parkin. J Neurosci 2013; 33: 2398–2407.
Kaminski MM, Sauer SW, Kaminski M, Opp S, Ruppert T, Grigaravicius P et al. T cell activation is driven by an ADP-dependent glucokinase linking enhanced glycolysis with mitochondrial reactive oxygen species generation. Cell Rep 2012; 2: 1300–1315.
Nakagawa S, Hirose T . Paraspeckle nuclear bodies—useful uselessness? Cell Mol Life Sci 2012; 69: 3027–3036.
Cartelli D, Ronchi C, Maggioni MG, Rodighiero S, Giavini E, Cappelletti G . Microtubule dysfunction precedes transport impairment and mitochondria damage in MPP+ -induced neurodegeneration. J Neurochem 2010; 115: 247–258.
Karreman C . AiO, combining DNA/protein programs and oligo-management. Bioinformatics 2002; 18: 884–885.
Scholz D, Chernyshova Y, Leist M . Control of Abeta release from human neurons by differentiation status and RET signaling. Neurobiol Aging 2013; 34: 184–199.
Gautier L, Cope L, Bolstad BM, Irizarry RA . affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 2004; 20: 307–315.
Berry MP, Graham CM, McNab FW, Xu Z, Bloch SA, Oni T et al. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature 2010; 466: 973–977.
Hiller K, Hangebrauk J, Jäger C, Spura J, Schreiber K, Schomburg D . MetaboliteDetector: comprehensive analysis tool for targeted and nontargeted GC/MS based metabolome analysis. Anal Chem 2009; 81: 3429–3439.
Acknowledgements
We are indebted to many colleagues for valuable contributions and insightful discussions during the course of this work. In particular, we thank M Berthold for help with bioinformatics analysis and colleagues from Boehringer-Ingelheim for help with NGS sequencing. The metabolomics work for pathway of toxicity mapping by LZ and TH was supported by NIH transformative research grant ‘Mapping the Human Toxome by Systems Toxicology’ (RO1ES020750) and FDA grant ‘DNTox-21c Identification of pathways of developmental neurotoxicity for high throughput testing by metabolomics’ (U01FD004230). This work was supported by grants and support from the Doerenkamp-Zbinden foundation, the BMBF and the German Research Foundation (DFG).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by G Melino
Supplementary Information accompanies this paper on Cell Death and Disease website
Supplementary information
Rights and permissions
Cell Death and Disease is an open-access journal published by Nature Publishing Group. This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Krug, A., Gutbier, S., Zhao, L. et al. Transcriptional and metabolic adaptation of human neurons to the mitochondrial toxicant MPP+. Cell Death Dis 5, e1222 (2014). https://doi.org/10.1038/cddis.2014.166
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cddis.2014.166
Keywords
This article is cited by
-
Mitochondrial complex I inhibition triggers NAD+-independent glucose oxidation via successive NADPH formation, “futile” fatty acid cycling, and FADH2 oxidation
GeroScience (2024)
-
Transcriptomic-based evaluation of trichloroethylene glutathione and cysteine conjugates demonstrate phenotype-dependent stress responses in a panel of human in vitro models
Archives of Toxicology (2023)
-
Transcriptional landscape of mitochondrial electron transport chain inhibition in renal cells
Cell Biology and Toxicology (2023)
-
Mapping the cellular response to electron transport chain inhibitors reveals selective signaling networks triggered by mitochondrial perturbation
Archives of Toxicology (2022)
-
Functional alterations by a subgroup of neonicotinoid pesticides in human dopaminergic neurons
Archives of Toxicology (2021)