Abstract
Hematopoiesis and commitment to a restricted lineage are guided by a timely expressed set of cytokine receptors and their downstream transcription factors. A member of the mRNA export complex, THOC5 (suppressors of the transcriptional defects of hpr1 delta by overexpression complex 5) is a substrate for several tyrosine kinases such as macrophage colony-stimulating factor (M-CSF) receptor and various leukemogenic tyrosine kinases, such as Bcr-Abl, or NPM-ALK. THOC5 tyrosine phosphorylation is elevated in stem cells from patients with chronic myeloid leukemia, suggesting that THOC5 may be involved in leukemia development. THOC5 is also an essential element in the maintenance of hematopoiesis in adult mice. In this report, we show that THOC5 is located in the nuclear speckles, and that it is translocated from the nucleus to cytoplasm during M-CSF-induced bone marrow-derived macrophage differentiation. Furthermore, we have identified THOC5 target genes by trancriptome analysis, using tamoxifen-inducible THOC5 knockout macrophages. Although only 99 genes were downregulated in THOC5-depleted macrophages, half of the genes are involved in differentiation and/or migration. These include well-known regulators of myeloid differentiation inhibitor of DNA binding (Id)1, Id3, Smad family member 6 (Smad6) and Homeobox (Hox)A1. In addition, a subset of M-CSF-inducible genes, such as Ets family mRNAs are THOC5 target mRNAs. Upon depletion of THOC5, unspliced v-ets erythroblastosis virus E26 oncogene homolog (Ets1) mRNA was accumulated in the nucleus. Furthermore, THOC5 was recruited to chromatin where Ets1 was transcribed and bound to unspliced and spliced Ets1 transcripts, indicating that THOC5 has a role in processing/export of M-CSF-inducible genes. In conclusion, regulation of immediate-early gene response by THOC5, a member of mRNA export complex contributes to the M-CSF-induced macrophage differentiation.
Similar content being viewed by others
Main
As most mature blood cells are short-lived, new blood cells continually arise from hematopoietic stem cells and become committed to the several hematopoietic lineages, such as the myeloid lineage. These differentiation processes are tightly regulated by timely expressed cytokines/growth factors and their receptors.1 Macrophage colony-stimulating factor (M-CSF, or CSF-1) and its receptor c-Fms tyrosine kinase have a key role in monocyte/macrophage differentiation.2, 3, 4 M-CSF induces the expression of different sets of immediate-early response genes, such as v-ets erythroblastosis virus E26 oncogene homolog (Ets) family genes, Myc, Fos, early growth response 1 (Egr1) or JunB and delayed-early response genes, such as D-type G1 cyclin, that ensure entry of macrophages into S phase.5 Transcriptional control mechanisms of the expression of these genes during differentiation were mainly studied by focusing on the cis- and trans-elements in promoters.
The THO complex, which is a sub-member of TREX (transcription/export), was originally identified in Saccharomyces cerevisiae as a five-protein complex (Tho2p, Hpr1p, Mft1p, Thp2p and Tex1)6, 7, 8, 9, 10, 11, 12 that has a role in transcriptional elongation, nuclear RNA export and genome stability. In higher eukaryotes such as Drosophila melanogaster13 or humans,14 three proteins (THOC1/hHpr1/p84, THOC2/hRlr1 and THOC3) and three additional unique proteins were identified, namely, suppressors of the transcriptional defects of hpr1 delta by overexpression complex 5 (THOC5)/Fms-interacting protein,15 THOC6 and THOC7, as members of the THO complex.
It has been shown that the depletion of THOC1 in cancer cells, such as human cervix carcinoma, HeLa or HEK293 cells, causes apoptosis; however, normal fibroblasts grow in the absence of THOC1,16 suggesting that the role of the THO complex differs in different cell types. The mammalian THO complex has been studied, however, mainly in HeLa cells.14, 17, 18 Consequently, the role of the THO complex during the normal cell differentiation process remains uncertain.
We have previously shown that THOC5 is a nuclear/cytoplasm shuttling protein and substrate for several tyrosine kinases such as M-CSF receptor, Fms,15 and various leukemogenic tyrosine kinases, such as Bcr-Abl or NPM-ALK.19 THOC5 phosphorylation is elevated in stem cells from patients with chronic myeloid leukemia (CML), suggesting that THOC5 may be involved in leukemia development.20 Furthermore, the ectopic expression of THOC5 in mouse myeloid progenitor cell line, FDC-P1Mac11 cells, alters M-CSF-induced macrophage differentiation. Instead, cells differentiate into granulocytes, suggesting that the level of THOC5 expression could form a threshold that determines differentiation either into macrophages or into granulocytes.15 Using interferon-inducible THOC5 knockout mice, we have previously shown that the depletion of the THOC5 gene causes rapid apoptosis of hematopoietic cells but not of any other organs. After bone marrow transplantation, mice survived for >3 months without any symptoms, suggesting that THOC5 has a key role in maintaining hematopoietic cells but not in the maintenance of terminally differentiated organs, such as liver.21 In this paper, we report on a study of the role of THOC5 during monocyte/macrophage differentiation induced by stimulation with M-CSF using primary bone marrow cells derived from tamoxifen-inducible THOC5 knockout mice. We show that although only 99 genes in bone marrow-derived macrophages were downregulated by the depletion of THOC5, depletion suppressed M-CSF-mediated macrophage differentiation and proliferation. In the absence of THOC5, mRNA export of M-CSF-inducible genes, such as Ets family transcription factor genes, and regulators of myeloid differentiation, such as inhibitor of DNA binding (Id)1, Id3, Smad family member 6 (Smad6) and Homeobox (Hox)A1, were impaired. These data imply that THOC5 has a role in myeloid differentiation.
Results
THOC5 is an essential element in the maintenance of hematopoiesis in adult mice
Using interferon-inducible THOC5 knockout mice, we have previously shown that the depletion of the THOC5 gene causes rapid apoptosis of hematopoietic cells but not of any other organs.21 However, as interferon is one of the important cytokines for hematopoiesis,22 the observed phenotype may be due to the synergistic effects of interferon and knockdown of THOC5. Furthermore, we are not able to knockdown THOC5 in a primary cell culture obtained from interferon-inducible THOC5 knockout mice.21 To study the role of THOC5 during hematopoietic differentiation at the molecular level, we generated tamoxifen-inducible THOC5 knockout (CreERT2 THOC5 (flox/flox)) mice. In these mice, the loxP sites are located before Exon 4 and after Exon 5 of THOC5. The deletion mutation of THOC5 was induced by 1 mg/20 g body weight of tamoxifen i.p. injection twice at 3-day intervals in 6-week-old CreERT2 THOC5 (flox/flox) and control ROSA26-CreERT2 mice. THOC5 exon 4/5 were deleted from bone marrow of all ERT2-Cre THOC5 (flox/flox), but not control mice, within 2 days after tamoxifen treatment (Supplementary Figures S1a and b). In agreement with previous data,21 upon treatment with tamoxifen in all CreERT2 THOC5 (flox/flox) mice, bone marrow cells that contain nuclei began to decrease in number 2 days after tamoxifen treatment, and on the sixth day only few cells containing nuclei were detected (Supplementary Figure S1c), indicating that THOC5 per se is an essential element in the maintenance of hematopoiesis.
THOC5 is required for M-CSF-induced growth of macrophages in vitro
To gain further insight into the molecular action of THOC5, we studied the role of THOC5 during M-CSF-induced macrophage differentiation in vitro using CreERT2 THOC5 (flox/flox) system. Bone marrow cells were isolated from CreERT2 THOC5 (flox/flox) and ROSA26-CreERT2 (Control) mice. Cells were incubated in the presence of L929-conditioned medium for 3 days in process shown to promote the formation of non-activated macrophages in vitro and were then either treated with tamoxifen (10 μM) or were left untreated in the presence of M-CSF. When tamoxifen was added, the amount of THOC5 protein in CreERT2 THOC5 (flox/flox) macrophages was reduced to 25% within 3 days and further decreased to 5% within 7 days after tamoxifen treatment (Figure 1a). In control macrophages, the amount of THOC5 product was not altered (Figure 1a). To examine the role of THOC5 in M-CSF-induced monocyte/macrophage growth, the numbers of adherent cells were counted in the presence or absence of tamoxifen. In the absence of tamoxifen, the cell numbers from both control and CreERT2 THOC5 (flox/flox) mice were increased approximately twofold within 3 days. The growth of cells from ROSA26-CreERT2 control mice was slightly reduced by treatment with tamoxifen (Figure 1b), indicating that tamoxifen treatment suppressed cell growth and/or adherence potency, which may be the result of oxidative stress induced by tamoxifen. The cells from CreERT2 THOC5 (flox/flox) mice did not grow after treatment with tamoxifen (Figure 1b); however, caspase 3 was not activated in THOC5-depleted cells (Figure 1a). These data suggest that THOC5 is required for M-CSF-induced growth of macrophages in vitro, or the decreased number is due to detachment as a consequence of an altered surface receptor attachment upon depletion of THOC5. To examine the effect of THOC5 depletion on macrophage development,23 the expression of markers of macrophage status were pursued. One such marker of development is the Mannose Receptor (CD206) that has been shown to associate with alternate pathway activation. Both ROSA26-CreERT2 control mice and CreERT2 THOC5 (flox/flox) developed increased expression of this marker (Figure 1c). However, loss of THOC5 did induce some cells to apparently decrease expression of this protein. Thus THOC5 certainly does not promote expression of a marker associated with macrophage activation. Interestingly, CD45 (a marker for hematopoietic cells), CD163 (hemoglobin scavenger receptor expressed by activated macrophages) and CD11c (also associated with activation) were lost too by induced depletion of THOC5 expression (Figure 1c). We conclude that the phenotype of the macrophages is markedly altered by THOC5 loss, but this does not align with altered activation status (Figure 1c).
THOC5 is required for growth of bone marrow-derived macrophages in vitro. (a) Bone marrow cells were isolated from CreERT2 THOC5 (flox/flox) and ROSA26-CreERT2 (control) mice. Cells were incubated for an additional 3 days in L929 cell-conditioned medium. Cells were then treated with (+) or without (−) tamoxifen (10 μM) (tamoxifen treatment). Total cell lysates from a sister culture of bone marrow-adherent cells on day 3, 5 and 7 (arrows) after tamoxifen treatment were analyzed by THOC5, GAPDH, cleaved (Cl.) caspase 3 and caspase 3-specific immunoblot. As control for Cl. caspase 3 blot, macrophages were treated with etoposide for 8 h. (b) Cell numbers of sister macrophage cultures isolated from CreERT2 THOC5 (flox/flox) and ROSA26-CreERT2 (CreERT2 control) mice were counted on days 3 and 5 (arrows) after tamoxifen treatment (+) in the presence of M-CSF. As control, cells were incubated for same period without tamoxifen treatment (−). Average values from three independent experiments±S.D. are shown. Probability (P)-value: Student t-test. (c) Bone marrow cells from CreERT2 and CreERT2 THOC5 mice treated with tamoxifen (3 days) were stained with CD206 Alexa Fluor (AF) 488, CD45 Brilliant Violet (BV) 421, CD11c AF 647 and CD163 PE. To control for autoflourescence, cells were analyzed with no antibody labelling (unstained). Cell surface fluorescence intensity was assessed using flow cytometry
Subcellular localization of THOC5 is altered during macrophage differentiation
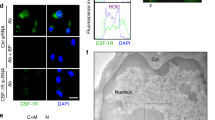
We next examined the subcellular distribution of THOC5 in macrophages at different differentiation stages. THOC5 was mainly detected in nuclear speckle-like domains at an early stage of macrophage culture (4 days after isolation),24 while at a later stage (8–10 days after isolation), THOC5 was detected diffusely in the nucleus and in the cytoplasm (Figure 2). As protein kinase C phosphorylation of THOC5 blocks the nuclear localization of THOC5,25 it could be that THOC5 is not imported sufficiently to the nucleus at a late stage by such a modification. These data suggest that THOC5 may have a role in mRNA export only at early stages of monocyte/macrophage development. Another member of TREX, Aly which has a wide range of roles in mRNA export,17 was detected predominantly in nuclear speckle-like domains.24 The nuclear location of Aly did not change in any of the differentiation periods (Figure 2). To examine which genes were THOC5 dependent at early macrophage stages, we next analyzed gene expression pattern in the presence and absence of THOC5 by transcriptome analysis.
Subcellular localization of THOC5 changes during macrophage differentiation. Bone marrow-adherent cells were prepared as described in Figure 1a, and further incubated in the presence of M-CSF. Four, 8, and 10 days after isolation, macrophages were fixed and stained with THOC5- or Aly-specific antibodies and FITC-conjugated secondary antibody, followed by DAPI (4,6-diamidino 2-phenylindole) staining
Only 99 transcripts were downregulated by the depletion of THOC5 in bone marrow macrophages
We analyzed the expression profile of cytoplasmic RNA in bone marrow macrophages derived from CreERT2 THOC5 (flox/flox) and ROSA26-CreERT2 control mice with or without tamoxifen in the presence of M-CSF for 0 and 3 days. We performed two independent experiments for expression profiling. In order to identify THOC5-dependent transcripts, whole data were subjected to a multistep filtering procedure as described in Supplementary Information (Materials and Methods). Using this filter, we have identified 65 transcripts that were upregulated and 100 transcripts (including THOC5 itself) that were downregulated by the depletion of THOC5 (Figure 3a). Of the 99 genes, 17 genes are transcriptional regulators (Figure 3b). A macrophage differentiation maker, ankyrin repeat and SOCS box-containing 2 (Asb2) that is known to be upregulated during monocyte/macrophage differentiation,26 was clearly upregulated in the presence of THOC5; however, Asb2 was only modestly upregulated in the absence of THOC5 (Figure 3b), suggesting that THOC5 influenced M-CSF-induced macrophage differentiation. Notably, Etv5, HoxA1, Id1 and Id3 were upregulated in the second experiment, not in the first experiment (in the absence of tamoxifen), but in both cases depletion of THOC5 caused downregulation of these transcription factors. As these genes are regulators of myeloid differentiation,27, 28, 29 macrophages which were examined in the first and second experiment may be at a slightly different stage of myelopoiesis. Interestingly, no single transcriptional regulator was found in the group of THOC5-dependent upregulated transcripts (Figure 3a).
Identification of THOC5-dependent genes in bone marrow-derived macrophages by transcriptome analysis. Bone marrow macrophages-derived from ROSA26-CreERT2 control and CreERT2 THOC5 (flox/flox) mice were treated with or without tamoxifen (Tam) in the presence of M-CSF for 3 days (3d samples). (a) Ratios from normalized fluorescence intensities were calculated from 3d samples /0d samples, and were finally converted into fold change values. Positive fold change values (color-coded in red) indicate an upregulation, whereas negative values (green color) indicate a downregulation during the 3 days of M-CSF-induced differentiation in two independent experiments (first experiment columns 1–4; second experiment columns 5–8). Directly adjacent lanes (columns 1+2, 3+4, 5+6 and 7+8) represent a comparison of non-treated, with tamoxifen-treated 3d samples. All samples were co-hybridized against the same common reference sample (0 day reference sample from the same mouse). In order to depict differences in mRNA expression between control and CreERT2 THOC5 (flox/flox) mice, ratios of ratios were calculated (as indicated in the header), converted into fold change values and depicted in a blue–grey color code. Blue color indicates an elevated relative expression with tamoxifen treatment, whereas grey indicates a decrease in expression with tamoxifen (first experiment, columns 9–10; second experiment columns 11–12). Finally, columns 13 and 14 represent the specificity of the THOC5 knockdown effects without tamoxifen side effects in experiments 1 and 2, respectively. (b) Transcriptional regulator genes that were downregulated by THOC5 depletion in macrophages. These transcripts were depicted along with color-coded fold-change values, accession numbers, Agilent probe IDs and official gene symbols. Classification was based on information from IPA
Half of the downregulated transcripts resulting from THOC5 depletion in macrophages are involved in differentiation and/or migration
To discover a biological significance of downregulated genes obtained from 3-day tamoxifen treatment, we uploaded the list of these genes to the Ingenuity Pathway Analysis (IPA) application for biological function and pathway analysis. Out of a total of 99 genes downregulated by depletion of THOC5, 68 genes were mapped to the IPA knowledge database for function/pathway analysis. Two top functional categories are ‘cellular development’ and ‘cellular movement’ (33 and 27 out of the 68 downregulated genes, respectively) (Table 1). More than 75% of the ‘cellular development’ genes were known to be involved in ‘differentiation’ (P=1.77E-04) and 12 out of the 25 genes belong to the category of transcriptional regulator genes (Table 1, bold). Moreover, >88% of ‘cellular movement’ genes (P=6.81E-05) are involved in ‘migration’, suggesting that THOC5 depletion also impairs macrophage migration.
Etv5, HoxA1, Id1, Id3, Smad6 and Ascl2 mRNAs are THOC5 target mRNAs
To confirm data obtained from transcriptome analysis, we examined the nuclear export of the downregulated mRNAs upon THOC5 depletion. We have chosen six ‘transcription regulator genes’, namely, Etv5, Id1, Id3, HoxA1, Smad family member 6 (Smad6) and achaete–scute complex homolog 2 (ASCL2) (Table 1, category: Differentiation), which are known to be induced upon M-CSF stimulation or to be involved in myeloid differentiation.5, 28, 29, 30, 31 We isolated nuclear and cytoplasmic RNA from THOC5-depleted bone marrow-derived macrophages, and actin mRNA was used as an internal control for equal amounts of cDNA used for each sample (Figures 4a and b, actin). As control for fractionation, aliquots of protein extracts from each sample were supplied for Histone H3 (nuclear fraction) and GAPDH (glyceraldehyde-3 phosphate dehydrogenase; cytoplasmic fraction) specific immunoblot (Blot). In this experiment, only spliced forms of mRNA can be detected, as we have chosen the primer pair for RT-PCR that is located at two different exons (Supplementary Table S1). For semi-quantification by RT-PCR, we first performed RT-PCR for different number of amplification cycles for each gene (Actin and Etv5 in Supplementary Figure S2). HoxA1, Id1, Id3, Smad6 and Ascl2 mRNAs were exported in the presence of THOC5; however, depletion of THOC5 drastically reduced the export (Figures 4a and b). Notably, the export of HoxA9, which is a functionally and structurally similar molecule to HoxA1, was not altered by the depletion of THOC5 (Figure 4a). Furthermore, one of Ets family genes, Etv5 mRNAs were not exported in the absence of THOC5 (Figures 4a and b), suggesting that immediate-early gene transcripts, such as Ets family mRNAs induced by M-CSF stimulation, may be THOC5 direct targets. We did not detect, however, any additional immediate-early genes. It may be due to that our microarray data were obtained from macrophages that had grown in the presence of M-CSF for the whole incubation period. We next, therefore, examined the kinetics of M-CSF-inducible immediate-early gene mRNAs, Ets1, Ets2 and Etv5. After serum starvation for 16 in the absence of L292-conditioned medium, macrophages were stimulated with human M-CSF (100 ng/ml) for 1–14 h. As expected, three known immediate-early genes, Ets1 and Ets2 were activated within 2 h and Etv5 was activated only 4 h after M-CSF stimulation (Figure 4c). Ets1 and Ets2, but not Etv5, were downregulated 8 h after M-CSF treatment (Figure 4c). Id1 and Id3 genes were not upregulated but slightly downregulated after M-CSF stimulation (Figure 4c). We next examined whether M-CSF-inducible immediate-early gene mRNAs are THOC5 target mRNAs. As we were not able to induce genes by M-CSF stimulation in THOC5 knockout macrophages after 16 h serum starvation, we utilized a more robust cell line.
Nuclear export of THOC5 target genes in bone marrow macrophages. (a) Bone marrow macrophages were isolated from CreERT2 THOC5 (flox/flox) or ROSA26-CreERT2 (control) mice as described (Figure 1a). Adherent cells were incubated with (+) and without (−) tamoxifen (10 μM) for 3 days, and nuclear (Nuc) and cytoplasmic (Cyt) RNAs were then isolated and applied for Etv5-, HoxA1-, HoxA9-, Id1-, Id3-, Smad6-, Ascl2-, THOC5- or actin-specific reverse transcriptase (RT)-PCR using primers as described in Supplementary Table S1 (RT-PCR, spliced). cDNA from all samples were standardized by adjusting equal levels of actin mRNA in both fractions (Supplementary Figure S2). Protein extracts were supplied for GAPDH- and Histone H3-specific immunoblot (Blot). We performed three independent experiments, and we show one example of representative data. (b) Signal intensity from Etv5-, HoxA1-, Id1- and actin-specific RT-PCR products was quantified using TINA 2.0 software. The percentage of signal intensity from the nuclear (Nuc) or the cytoplasmic (Cyt) fraction of total intensity (Nuc+Cyt) is shown. Average values±S.D. from three independent experiments are shown. (c) Control macrophages (5 days after isolation) were incubated in the absence of serum and M-CSF for 16 h and then stimulated with human M-CSF (100 ng/ml) for 1, 2, 4, 8 and 14 h. RNAs were isolated and supplied for Ets1-, Ets2-, Etv5-, Id1-, Id3- and GAPDH-specific RT-PCR. (a and c): the number of amplification cycles is shown in ‘cycles (n)’
THOC5 is required for Ets1 but not Egr1 mRNA processing upon stimulation with M-CSF
To study the role of THOC5 in immediate-early gene response, we generated a THOC5 (flox/flox) mouse embryonic fibroblast (MEF)32 cell line that expressed human M-CSF receptor cDNA (M-CSFR THOC5 (flox/flox) MEF; Figure 5a). We first examined whether the ectopically expressed human M-CSF receptor in MEF cells is activated by stimulation with its ligand. Upon stimulation with human M-CSF, two major downstream signal molecules, Erk1/2 and Akt were phosphorylated within 30 min (Figure 5a), and the immediate-early gene response in M-CSFR THOC5 (flox/flox) MEF cells were comparative in the macrophage system (Figure 5b), indicating that the ectopic expressed human M-CSF receptor in MEF cells is functional. To examine regulation of immediate-early genes by THOC5, we infected M-CSFR THOC5 (flox/flox) MEF with adenovirus carrying GFP and Cre-recombinase genes (Ad-GFP-Cre). THOC5 is downregulated by >95% within 4 days at the protein level (Figure 5c). As control, we infected the same cells with adenovirus carrying only the GFP gene (Ad-GFP). The level of THOC5 did not change after control virus infection. Virus-infected cells were incubated without serum for 24 h and were then stimulated with M-CSF for 1 and 2 h, and total RNAs were isolated and subjected to Ets1-, Ets2-, Egr1-, GAPDH- and THOC5-specific RT-PCR. In agreement with protein data, upon infection with Ad-GFP-Cre, exon 4/5 of THOC5 was deleted in all samples (Figure 5c, delta E4/5). Depletion of THOC5 inhibited M-CSF-induced upregulation of Ets1 and Ets2, but not Egr1 (Figures 5c and d, Ade-GFP-Cre), indicating that THOC5 has a key role in the processing of Ets1 and Ets2 but not Egr1 mRNA. We then examined the nuclear export of THOC5 target mRNAs. RNAs were isolated from the nuclear and the cytoplasmic fractions of 5000 M-CSFR THOC5 (flox/flox) MEF cells infected with Ade-GFP or Ade-GFP-Cre. RNA samples were then subjected to RT-PCR. For GAPDH and Egr1, the mRNAs were normally exported upon depletion of THOC5 (Figure 5e), while less spliced Ets1 transcripts were detected in both cytoplasmic and nuclear fractions; however, the unspliced Ets1 transcripts were accumulated within 20 min in the nucleus (Figure 5f). These data imply that THOC5 has a role in Ets1 mRNA processing after M-CSF stimulation.
THOC5 is required for the processing of Ets1. (a) Human c-Fms cDNA was expressed in MEF (M-CSFR THOC5 (flox/flox) MEF) cells. After serum starvation for 24 h, cells were stimulated with human M-CSF (100 ng/ml) for 0.5, 2 and 4 h and cell extracts were then subjected to Fms-, phosphoErk (p-Erk1/2)-, Erk-, phosphoAkt (p-Akt)-, Akt-, and GAPDH-specific immunoblot. (b) M-CSFR (flox/flox) MEF cells were treated as described in (a) and were stimulated with M-CSF for 1, 2 and 4 h. Total RNAs were isolated and subjected to Ets1-, Ets2-, Etv5-, Egr1-, Id1-, Id3-, HoxA1- and GAPDH-specific reverse transcriptase (RT)-PCR. (c) Sister cultures of M-CSFR THOC5 (flox/flox) MEF were infected with Ad-GFP or Ad-GFP-Cre virus for 3 days. After serum starvation for 24 h, cells were stimulated with M-CSF for 1 and 2 h. Protein extract from cells before stimulation (0 h) were supplied for THOC5- and GAPDH-specific immunoblot (Blot). Total RNAs were isolated from each preparation and supplied for Ets1-, Ets2-, Egr1-, GAPDH- and THOC5-specific RT-PCR (RT-PCR). (d) Aliquots of cDNA samples from (c) were applied for quantitative RT-PCR analysis of Ets1 mRNA (TaqMan RT-PCR). Relative expression levels compared with GAPDH were normalized. Average values from four independent PCR reactions+S.D. are shown. (e) Cells were prepared as described in (c), but RNA and protein were isolated from the nuclear and the cytoplasmic fractions obtained from 5000 cells and then supplied for Ets1-, Ets2-, Egr1-, and GAPDH-specific RT-PCR. Protein extracts were supplied for GAPDH- and Histone H3-specific immunoblot. (f) RNA was isolated from the nuclear fraction as described in (a) and subjected to Ets1- and GAPDH-specific RT-PCR using an exon–intron primer pair as indicated. RNA samples were reverse transcribed using oligo dT or random hexamer (random) primer as indicated (Supplementary Table S1). As negative controls, RNA samples were supplied for PCR without reverse transcription (−). We performed independent experiments, and we show one example of representative data (a–f)
THOC5 was recruited to the last exon of Ets1 gene but not to the promoter region
It has been shown that under heat-shock conditions the THO complex is recruited to the chromatin where genes are activated.12, 33 We next examine whether THOC5 is recruited to the Ets1 gene. THOC5 was recruited to chromatin where Ets1 was transcribed (Figure 6a Ets1, Exon 8) but not to Egr1 gene. THOC5 was not recruited, however, to the Ets1 promoter region (Figure 6a, Ets1, promoter). Both genes were precipitated using RNA polymerase II CTD Ser 5P antibody, while the control antibody did not precipitate any of these (Figure 6a).
THOC5 moves to the chromatin where the Ets1 gene was active and binds its target mRNAs. (a) pCTAP (TAP) and pCTAP carrying THOC5 cDNA (TAP-THOC5) were transfected into M-CSFR-THOC5 (flox/flox) MEF cells and were incubated for 24 h in the presence of 10% FCS. After serum starvation for 24 h, cells were stimulated with (+) or without (−) M-CSF for 2 h. After cross-linking by adding formaldehyde, protein and DNA were extracted and the chromatin was sheared by sonication. Cell extracts (Input 1/20) and binding fractions with Streptavidin Sepharose or immunoprecipitates using pS5 CTD (pS5 Ab) or control IgG were analyzed by Ets1 (promoter region and exon 8), and Egr1 (promoter region and exon 2) specific PCR (Supplementary Table S1;ChIP). The promoter region of each gene was described by Zhao et al.43 Numbers represent nucleotide numbers from the initiation site for each gene.44 (b) pCTAP and pCTAP carrying THOC5 cDNA (TAP-THOC5) were transfected into M-CSFR THOC5 (flox/flox) MEF cells and were then incubated further for 24 h in the presence of 10% FCS. After serum starvation for 24 h, cells were stimulated with (+) or without (−) M-CSF for 4 h. Cell extracts (Input) and binding fractions with Streptavidin Sepharose (Bound) were analyzed by THOC5-specific immunoblot (Blot). The same samples were analyzed by Ets1-, Ets2-, Etv5-, Egr1- and GusB-specific reverse transcriptase (RT)-PCR (RT-PCR). endo-THOC5, endogenous THOC5. (c) The same samples without M-CSF stimulation were analyzed by Id1-, Id3-, HoxA1- and HoxA9-specific RT-PCR (35 cycles) (RT-PCR). We performed three independent experiments and we show one example of representative data (a–c)
Both spliced and unspliced transcripts of members of Ets family bind THOC5
We next examined whether immediate-early gene mRNAs bind THOC5 by isolating the THOC5–mRNA complex. TAP-THOC5 was expressed in M-CSFR THOC5 (flox/flox) MEF cells and the THOC5 containing mRNP was purified with Streptavidin beads from cell extracts (Figure 6b). Two bands of 89 and 79 kDa proteins were detected from TAP-THOC5 transfectants by THOC5-specific immunoblot, whereby the upper band represents exogenous TAP-THOC5 and the lower band represents endogenous THOC5 (Figure 6b, Input). After purification with Streptavidin beads (Bound), the RNA was then isolated from the THOC5–mRNA complex. As shown in Figure 6b, unspliced and spliced Ets1, Ets2 and Etv5 but not Egr1 or GusB mRNAs were bound to THOC5 4 h after stimulation with M-CSF. These data suggest that THOC5 has a role in not only 3′processing,34 but may also contribute in splicing of its target mRNAs. We further examined the binding of HoxA1, Id1 and Id3 mRNAs to THOC5. Indeed, THOC5 binds to HoxA1, Id1 and Id3 mRNAs (Figure 6c). In agreement with our data obtained from the transcriptome analysis (GEO series accession number GSE41170) and the export assay (Figure 4a), HoxA9 did not bind to THOC5 (Figure 6c).
Discussion
It has been shown that only a subset of genes (approximately 100–200 genes) were downregulated upon depletion of THOC5 in MEF,32 Hela34 cells or macrophages (this report); however, we have previously shown that conventional THOC5 knockout mice are embryonic lethal, and upon depletion of THOC5 adult mice died from anemia and leukocytopenia.21 These facts raise the question of why downregulation of such a few genes causes such a strong phenotype. The aim of this study was to identify the molecular mechanism of THOC5-dependent cell proliferation/differentiation. In this study, we applied the bone marrow macrophage system. Using conditional THOC5 knockout cells, we show here that THOC5 has a role in the processing of known regulators of myeloid differentiation, Id1, Id3 and HoxA1 and a subset of M-CSF-inducible immediate-early gene mRNAs, which in turn have a key role in the decision of cells to enter the S phase. Notably, some population of immediate-early genes such as Egr1 is not dependent upon THOC5. In addition, we show that upon stimulation with M-CSF, THOC5 is recruited to the target gene and then binds to the target mRNAs (Figure 7). Furthermore, in mature macrophages, most THOC5 are translocated into the cytoplasm, suggesting the THOC5 has a lesser role in mRNA processing at this stage (Figure 7).
Scheme of summary. Upon stimulation with M-CSF, numbers of immediate-early response genes (IEG) are induced. We propose that co-transcriptionally spliced or intron-less immediate-early genes, such as Jun, Fos, or Egr1 are further processed without THOC5, while THOC5 is required for the processing of a subset of immediate-early genes, whereby THOC5 is recruited to chromatin where genes were activated, and participate in further processing of these mRNAs. THOC5-dependent genes have a key role in maintenance of the differentiation/proliferation at early stage of differentiation process. When macrophages are terminally differentiated, THOC5 is located in the cytoplasm and does not have a role in mRNA processing
Although it has been clearly demonstrated that THOC5 is highly phosphorylated in CD34-positive CML cells and that THOC5 may be involved in leukemic cell behavior,20 the biological consequence of tyrosine phosphorylation of THOC5 during macrophage differentiation is still unclear. The replacement of tyrosine phosphorylation site 225 by phenylalanine abolished the binding of THOC5 with target mRNAs;20 however, although we detected tyrosine phosphorylation of THOC5 in several leukemic cells,19, 20 we failed to detect tyrosine phosphorylation of THOC5 in the macrophage system. This may be due to the very transient tyrosine phosphorylation in normal cells.15 Notably, the N-terminal half of THOC5 exhibits potency of the complex formation with its target mRNAs.35 Interestingly, this region contains not only tyrosine phosphorylation site Y225,19 but also contains a typical conjugation site for the small ubiquitin-like modifier (SUMO), K223, or putative WW domain binding site, suggesting that post-translational modification of THOC5 by extracellular stimuli by cytokines/growth factors or heat shock may influence RNA-binding potency and/or selectivity of THOC5.
We have identified novel direct target mRNAs of THOC5, namely, Ets1, Ets2, Etv5, HoxA1, Smad6, Ascl2, Id1 and Id3. To study target selectivity of THOC5, we recently performed transcriptome analysis of immediate-early genes in the presence and absence of THOC5. Our analysis revealed that 50% of all serum-inducible genes were still upregulated in the absence of THOC5. Interestingly, all rapid-inducible genes, such as Fos or Jun, which are identified in macrophages stimulated by lipopolysaccharides,36 are exported in THOC5-depleted cells (Tran and Tamura, unpublished data). Moreover, unspliced THOC5-dependent immediate-early genes were accumulated in the nucleus. Similarly, lipopolysaccharide induced full-length nascent RNAs that contain introns accumulated on chromatin before release of functional mRNA for export.36 These data imply that THOC5 may have a role in the release of these immediate-early gene transcripts from chromatin. Recently, Katahira et al.34 proposed that THOC5 controls alternative polyadenylation site choice by recruiting CFIm68 on target genes; however, the known THOC5 target mRNA, Hsp7013, 17, 32, 37 and some of our newly identified THOC5 target immediate-early genes do not contain this element. Moreover, 70–75% of human mRNAs contains an alternative polyadenylation site,38 suggesting that THOC5 dependency may also associate with different RNA element(s).
IPA analysis of our transcriptome data reveals that two top functional categories are ‘cellular development’ and ‘cellular movement’. In agreement with these data, it has been shown that CXCL12 induced phosphorylation of THOC5 in Ba/F cells, and phosphorylated THOC5 inhibits cell migration,20 suggesting that THOC5 may also be involved in macrophage movement.
In conclusion, regulation of immediate-early gene response by THOC5, a member of mRNA export complex, contributes to M-CSF-induced macrophage differentiation.
Materials and Methods
Generation and genotyping of THOC5 (flox/flox), ROSA26-CreERT2 and CreERT2 THOC5 (flox/flox) mice
Generation of THOC5 (flox/flox) mice was previously described.21 To generate tamoxifen-inducible THOC5 knockout mice, THOC5 (flox/flox) and Cre-‘deleter’ Rosa 26 mice expressing cre-recombinase (Cre) under the control of the tamoxifen-inducible ERT2 promoter (ROSA26-CreERT2) (a kind gift from Pierre Chambon, Illkirch, France, and Anton Berns, Amsterdam, The Netherlands)39 were cross bred to obtain animals that carry the THOC5 (flox/flox) allele and CreERT2 gene (CreERT2 THOC5 (flox/flox)). Genotyping was performed from tail-derived sample DNA extracted with DirectPCR-Tail Lysis Reagent (Peqlab, Erlangen, Germany) by PCR using THOC5 primers, 5′-CCCTCGGCCCCTTTTGAG-3′ and 5′-CAGCACTGGAGCGGGAGATGT-3′ or Cre primers Cre1 5′-CCGGGCTGCCACGACCAA-3′ and Cre2 5′-GGCGCGGCAACACCATTTT-3′.
Macrophage isolation, cell culture and virus infection
Bone marrow cells were isolated from CreERT2 THOC5 (flox/flox) and ROSA26-CreERT2 (control) mice. To remove mature macrophages and bone marrow stroma cells, the cells were incubated for 24 h with L929-conditioned medium, which contained M-CSF. Non-adherent cells were then transferred to another dish and incubated for an additional 48 h in the presence of M-CSF. Macrophages were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% (v/v) FCS and 10% L929 cell supernatant or 10 ng/ml human recombinant M-CSF (Pepro Tech GmbH, Hamburg, Germany). M-CSFR THOC5 (flox/flox) MEF cells were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% (v/v) FCS. Adenovirus carrying GFP and Cre-recombinase (Ad-Cre-GFP) or GFP (Ad-GFP) genes were obtained from Vector Biolab (Philadelphia, PA, USA). Cells were infected with Ad-Cre-GFP or Ad-GFP at multiplicity of infection (MOI) 200.
RT-PCR analysis
RNA was isolated from mouse bone marrow macrophage culture, or M-CSFR THOC5 (flox/flox) MEF cells with the High Pure RNA Isolation kit (Roche Diagnostics, Mannheim, Germany) according to the manufacturer’s instructions. Reverse transcription was carried out using oligo dT primers or random Hexamer primer (Thermo Scientific, Schwerte, Germany) and the Omniscript reverse transcriptase kit (Qiagen, Hilden, Germany) following the instructions provided. Cytoplasmic RNA was isolated from cells as previously described.32 Primer pairs for each PCR are described in Supplementary Information (Supplementary Table S1). Signal intensity was quantified using TINA 2.0 software (raytest Isotopenmessgeraete GmbH, Straubenhardt, Germany). Quantitative (TaqMan) RT-PCR was performed as previously described.40 TaqMan probes (Applied Biosystems (Carlsbad, CA, USA), assay-IDs as follows: Mm99999915-g1 (GAPDH), Mm01175819-m1 (Ets1)) were used with TaqMan Fast Universal PCR Master Mix (2 × ) (Applied Biosystems).
Flow cytometry analysis
Bone marrow-isolated macrophages treated with carrier or tamoxifen were stained with CD206 Alexa Fluor 488 (1 : 10 final dilution, Bio-Rad AbD Serotec Limited, Puchheim, Germany), CD45 Brilliant Violet 421 (1 : 20 final dilution, Cambridge Bioscience Limited, Cambridge, UK), CD11c Alexa Fluor 647 (1:10 final dilution; Bio-Rad AbD Serotec Limited), CD163 PE (1 : 10 final dilution, eBiosciences, San Diego, CA, USA) and cell-surface fluorescence intensity was assessed using a FACS LSRII (BD Biosciences, San Jose, CA, USA) and analyzed using FlowJo 7.6.3 software (Tree Star, Inc. OR, USA).
Immunoblot procedures
Details of immunoblotting have been described previously.41 Monoclonal antibodies against GAPDH and Aly, and polyclonal antibodies against actin, and c-Fms (M-CSFR) antibodies were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA, USA), polyclonal antibodies against cleaved caspase 3, caspase 3, Histone H3, Akt, phosphoAkt, Erk1/2 and phospho Erk1/2 were from Cell Signaling (Cell Signaling Technology, Beverly, MA, USA) and a monoclonal antibody against THOC5 was generated as described previously.42
Immunostaining
Cells were washed with ice-cold PBS, fixed in cold methanol or in 3% paraformaldehyde and permeabilized with 0.1% Triton X100. After washing with PBS, cells were incubated with blocking buffer (2 mg/ml Octagam-PBS) and stained with THOC5-specific antibody or Aly (11G5) (Santa Cruz), followed by incubation with or without anti-mouse IgG goat IgG and then with FITC-conjugated anti-goat or mouse IgG. Cells were concurrently stained with 4,6-diamidino 2-phenylindole (Sigma, München, Germany).
Microarray-based mRNA expression analysis (dual color mode)
Detailed microarray analysis procedure and data processing are described under www.mh-hannover.de/rcut-downloads.html?L=1. Detailed protocol is shown in Supplementary Information (Materials and Methods). The functional classification of transcripts was performed with annotations provided by Ingenuity Pathway Analysis, IPA Content Version 11904312 (2012-01-17) (Ingenuity Systems, Mountain View, CA, USA).
THOC5–mRNA complex isolation
Two sister cultures of 4 × 106 MEFs expressing M-CSF receptor (M-CSFR THOC5 (flox/flox) MEF cells) were transfected with a pCTAP vector (Stratagene, La Jolla, CA, USA) and the same vector carrying THOC5 cDNA using the Polyfect reagent as described by the manufacturer (Qiagen), and the cells were allowed to grow for 24 h. Cells were then incubated without serum for 24 h and were then stimulated with human M-CSF (100 ng/ml) for 4 h.
Details of mRNP complex isolation have been described previously.32, 35 Bound RNAs were used for RT-PCR analysis.
Chromatin precipitation
TAP-THOC5 or TAP control was expressed in M-CSFR THOC5 (flox/flox) MEF cells, and after 24 h serum starvation cells were stimulated with or without M-CSF for 2 h. Cells were cross-linked by adding formaldehyde to a final concentration of 1% for 10 min at 37 °C in the presence of 5% CO2. The reaction was stopped by adding glycine to a final concentration of 125 mM. Nuclear fractions were isolated using lysis buffer (10 mM TrisHCl, pH 8.0, 150 mM NaCl, 0.5% NP40, 1 mM PMSF, 8 mM Na3VO4, protease inhibitor cocktail (Sigma)), and chromatin was then sheared to fragments of 200–500 bp by sonication. Aliquots of extracts were precipitated by Streptavidin beads or pre-coated Protein G Agarose-PLUS (Santa Cruz) with anti-serine 5 phosphorylated RNA polymerase II CTD (CTD Ser 5P, 3E8) antibody or control IgG, and precipitates were then washed three times with washing buffer (10 mM TrisHCl, pH 8.0, 150 mM NaCl, 0.5% NP40, 10 mM beta-mercaptoethanol, 1 mM PMSF, 0.5 mM EDTA). After reverse cross-linking with 200 mM NaCl at 65 °C overnight and proteinase K digestion at 55 °C for 2 h, the bound DNA was isolated using NucleoSpin Extract II (Macherey-Nagel, Düren, Germany). PCR was then performed using Ets1, Egr1 or HoxA1 gene-specific primers as shown in Supplementary Table S1 (ChIP).
Accession codes
Abbreviations
- THOC5:
-
suppressors of the transcriptional defects of hpr1 delta by overexpression complex 5
- M-CSF:
-
macrophage colony-stimulating factor
- CML:
-
chronic myeloid leukemia
- Id:
-
inhibitor of DNA binding
- Hox:
-
Homeobox
- Ets:
-
v-ets erythroblastosis virus E26 oncogene homolog
- Egr1:
-
early growth response 1
- TREX:
-
transcription/export
- MEF:
-
mouse embryonic fibroblast
- Ad-GFP:
-
adenovirus carrying the GFP gene
- Ad-GFP-Cre:
-
adenovirus carrying the GFP and Cre-recombinase genes
- GAPDH:
-
glyceraldehyde-3 phosphate dehydrogenase
References
Metcalf D . Hematopoietic cytokines. Blood 2008; 111: 485–491.
Mossadegh-Keller N, Sarrazin S, Kandalla PK, Espinosa L, Stanley ER, Nutt SL et al. M-CSF instructs myeloid lineage fate in single haematopoietic stem cells. Nature 2013; 497: 239–243.
Wei S, Dai XM, Stanley ER . Transgenic expression of CSF-1 in CSF-1 receptor-expressing cells leads to macrophage activation, osteoporosis, and early death. J Leukoc Biol 2006; 80: 1445–1453.
Yu W, Chen J, Xiong Y, Pixley FJ, Dai XM, Yeung YG et al. CSF-1 receptor structure/function in MacCsf1r−/− macrophages: regulation of proliferation, differentiation, and morphology. J Leukoc Biol 2008; 84: 852–863.
Sherr CJ . Mitogenic response to colony-stimulating factor 1. Trends Genet 1991; 7: 398–402.
Chavez S, Aguilera A . The yeast HPR1 gene has a functional role in transcriptional elongation that uncovers a novel source of genome instability. Genes Dev 1997; 11: 3459–3470.
Chavez S, Garcia-Rubio M, Prado F, Aguilera A . Hpr1 is preferentially required for transcription of either long or G+C-rich DNA sequences in Saccharomyces cerevisiae. Mol Cell Biol 2001; 21: 7054–7064.
Jimeno S, Rondon AG, Luna R, Aguilera A . The yeast THO complex and mRNA export factors link RNA metabolism with transcription and genome instability. EMBO J 2002; 21: 3526–3535.
Piruat JI, Aguilera A . A novel yeast gene, THO2, is involved in RNA pol II transcription and provides new evidence for transcriptional elongation-associated recombination. EMBO J 1998; 17: 4859–4872.
Strasser K, Masuda S, Mason P, Pfannstiel J, Oppizzi M, Rodriguez-Navarro S et al. TREX is a conserved complex coupling transcription with messenger RNA export. Nature 2002; 417: 304–308.
Reed R, Cheng H . TREX SR proteins and export of mRNA. Curr Opin Cell Biol 2005; 17: 269–273.
Pena A, Gewartowski K, Mroczek S, Cuellar J, Szykowska A, Prokop A et al. Architecture and nucleic acids recognition mechanism of the THO complex, an mRNP assembly factor. EMBO J 2012; 31: 1605–1616.
Rehwinkel J, Herold A, Gari K, Kocher T, Rode M, Ciccarelli FL et al. Genome-wide analysis of mRNAs regulated by the THO complex in Drosophila melanogaster. Nat Struct Mol Biol 2004; 11: 558–566.
Masuda S, Das R, Cheng H, Hurt E, Dorman N, Reed R . Recruitment of the human TREX complex to mRNA during splicing. Genes Dev 2005; 19: 1512–1517.
Tamura T, Mancini A, Joos H, Koch A, Hakim C, Dumanski J et al. FMIP, a novel Fms-interacting protein, affects granulocyte/macrophage differentiation. Oncogene 1999; 18: 6488–6495.
Li Y, Lin AW, Zhang X, Wang Y, Wang X, Goodrich DW . Cancer cells and normal cells differ in their requirements for Thoc1. Cancer Res 2007; 67: 6657–6664.
Katahira J, Inoue H, Hurt E, Yoneda Y . Adaptor Aly and co-adaptor Thoc5 function in the Tap-p15-mediated nuclear export of HSP70 mRNA. EMBO J 2009; 28: 556–567.
Merz C, Urlaub H, Will CL, Luhrmann R . Protein composition of human mRNPs spliced in vitro and differential requirements for mRNP protein recruitment. RNA 2007; 13: 116–128.
Pierce A, Carney L, Hamza HG, Griffiths JR, Zhang L, Whetton BA et al. THOC5 spliceosome protein: a target for leukaemogenic tyrosine kinases that affects inositol lipid turnover. Br J Haematol 2008; 141: 641–650.
Griaud F, Pierce A, Gonzalez Sanchez MB, Scott M, Abraham SA, Holyoake TL et al. A pathway from leukemogenic oncogenes and stem cell chemokines to RNA processing via THOC5. Leukemia 2012.
Mancini A, Niemann-Seyde SC, Pankow R, El Bounkari O, Klebba-Farber S, Koch A et al. THOC5/FMIP, an mRNA export TREX complex protein, is essential for hematopoietic primitive cell survival in vivo. BMC Biol 2010; 8: 1.
Essers MA, Offner S, Blanco-Bose WE, Waibler Z, Kalinke U, Duchosal MA et al. IFNalpha activates dormant haematopoietic stem cells in vivo. Nature 2009; 458: 904–908.
Gordon S, Taylor PR . Monocyte and macrophage heterogeneity. Nat Rev Immunol 2005; 5: 953–964.
Dias AP, Dufu K, Lei H, Reed R . A role for TREX components in the release of spliced mRNA from nuclear speckle domains. Nat Commun 2010; 1: 97.
Mancini A, Koch A, Whetton AD, Tamura T . The M-CSF receptor substrate and interacting protein FMIP is governed in its subcellular localization by protein kinase C-mediated phosphorylation, and thereby potentiates M-CSF-mediated differentiation. Oncogene 2004; 23: 6581–6589.
Wang J, Muntean AG, Hess JL . ECSASB2 mediates MLL degradation during hematopoietic differentiation. Blood 2012; 119: 1151–1161.
Bach C, Buhl S, Mueller D, Garcia-Cuellar MP, Maethner E, Slany RK . Leukemogenic transformation by HOXA cluster genes. Blood 2010; 115: 2910–2918.
Hess JL, Bittner CB, Zeisig DT, Bach C, Fuchs U, Borkhardt A et al. c-Myb is an essential downstream target for homeobox-mediated transformation of hematopoietic cells. Blood 2006; 108: 297–304.
Skokowa J, Welte K . Dysregulation of myeloid-specific transcription factors in congenital neutropenia. Ann NY Acad Sci 2009; 1176: 94–100.
Glesne D, Huberman E . Smad6 is a protein kinase X phosphorylation substrate and is required for HL-60 cell differentiation. Oncogene 2006; 25: 4086–4098.
Wheadon H, Ramsey JM, Dobbin E, Dickson GJ, Corrigan PM, Freeburn RW et al. Differential Hox expression in murine embryonic stem cell models of normal and malignant hematopoiesis. Stem Cells Dev 2011; 20: 1465–1476.
Guria A, Tran DD, Ramachandran S, Koch A, El Bounkari O, Dutta P et al. Identification of mRNAs that are spliced but not exported to the cytoplasm in the absence of THOC5 in mouse embryo fibroblasts. RNA 2011; 17: 1048–1056.
Kopytova DV, Orlova AV, Krasnov AN, Gurskiy DY, Nikolenko JV, Nabirochkina EN et al. Multifunctional factor ENY2 is associated with the THO complex and promotes its recruitment onto nascent mRNA. Genes Dev 2010; 24: 86–96.
Katahira J, Okuzaki D, Inoue H, Yoneda Y, Maehara K, Ohkawa Y . Human TREX component Thoc5 affects alternative polyadenylation site choice by recruiting mammalian cleavage factor I. Nucleic Acids Res 2013; 41: 7060–7072.
Ramachandran S, Tran DD, Klebba-Faerber S, Kardinal C, Whetton AD, Tamura T . An ataxia-telangiectasia-mutated (ATM) kinase mediated response to DNA damage down-regulates the mRNA-binding potential of THOC5. Rna 2011; 17: 1957–1966.
Bhatt DM, Pandya-Jones A, Tong AJ, Barozzi I, Lissner MM, Natoli G et al. Transcript dynamics of proinflammatory genes revealed by sequence analysis of subcellular RNA fractions. Cell 2012; 150: 279–290.
Rollenhagen C, Hodge CA, Cole CN . Following temperature stress, export of heat shock mRNA occurs efficiently in cells with mutations in genes normally important for mRNA export. Eukaryot Cell 2007; 6: 505–513.
Elkon R, Ugalde AP, Agami R . Alternative cleavage and polyadenylation: extent, regulation and function. Nat Rev Genet 2013; 14: 496–506.
Vooijs M, Jonkers J, Berns A . A highly efficient ligand-regulated Cre recombinase mouse line shows that LoxP recombination is position dependent. EMBO Rep 2001; 2: 292–297.
Dhamija S, Doerrie A, Winzen R, Dittrich-Breiholz O, Taghipour A, Kuehne N et al. IL-1-induced post-transcriptional mechanisms target overlapping translational silencing and destabilizing elements in IkappaBzeta mRNA. J Biol Chem 2010; 285: 29165–29178.
Koch A, Scherr M, Breyer B, Mancini A, Kardinal C, Battmer K et al. Inhibition of Abl tyrosine kinase enhances nerve growth factor-mediated signaling in Bcr-Abl transformed cells via the alteration of signaling complex and the receptor turnover. Oncogene 2008; 27: 4678–4689.
Mancini A, El Bounkari O, Norrenbrock AF, Scherr M, Schaefer D, Eder M et al. FMIP controls the adipocyte lineage commitment of C2C12 cells by downmodulation of C/EBP alpha. Oncogene 2007; 26: 1020–1027.
Zhao F, Xuan Z, Liu L, Zhang MQ . TRED: a Transcriptional Regulatory Element Database and a platform for in silico gene regulation studies. Nucleic Acids Res 2005; 33: D103–D107.
Perier RC, Praz V, Junier T, Bonnard C, Bucher P . The eukaryotic promoter database (EPD). Nucleic Acids Res 2000; 28: 302–303.
Acknowledgements
We would like to thank M Roussel for providing human M-CSF receptor cDNA, Dirk Eick for providing CTD Ser 5P, 3E8 antibody and P Chambon and A Berns for providing ROSA26-CreERT2 mice. This research was supported by DFG Ta-111/13-1 and PhD program Molecular Medicine and Leistungsorientierte Mittelvergabe with Frauenfaktor from MHH. AJKW and ADW are supported by Leukaemia Lymphoma Research UK.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by A Stephanou
Supplementary Information accompanies this paper on Cell Death and Disease website
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Tran, D., Saran, S., Dittrich-Breiholz, O. et al. Transcriptional regulation of immediate-early gene response by THOC5, a member of mRNA export complex, contributes to the M-CSF-induced macrophage differentiation. Cell Death Dis 4, e879 (2013). https://doi.org/10.1038/cddis.2013.409
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cddis.2013.409
Keywords
This article is cited by
-
RNA editing enzyme APOBEC3A promotes pro-inflammatory M1 macrophage polarization
Communications Biology (2021)
-
Long first exons and epigenetic marks distinguish conserved pachytene piRNA clusters from other mammalian genes
Nature Communications (2021)
-
Differential immune response modulation in early Leishmania amazonensis infection of BALB/c and C57BL/6 macrophages based on transcriptome profiles
Scientific Reports (2019)
-
Myc/Max dependent intronic long antisense noncoding RNA, EVA1A-AS, suppresses the expression of Myc/Max dependent anti-proliferating gene EVA1A in a U2 dependent manner
Scientific Reports (2019)
-
Depletion of three combined THOC5 mRNA export protein target genes synergistically induces human hepatocellular carcinoma cell death
Oncogene (2016)