Abstract
B-cell lymphoma-2 (Bcl-2) proteins mediate intrinsic-, or mitochondrial-, initiated apoptosis. We have investigated the structure and function of the least characterized Bcl-2 family member, Bcl-B, solving the crystal structure of a Bcl-B:Bim complex to 1.9 Å resolution. Bcl-B is distinguished from other Bcl-2 family members through an insertion of an unstructured loop between helices α5 and α6. Probing Bcl-B interactions with Bcl-2 homology (BH)3 motifs using a combination of biophysical- and cell-based assays revealed a unique BH3-only protein binding profile. Bcl-B has high-affinity interactions with Bim and Bik only. Our results not only delineate the mode of action of Bcl-B but also complete our understanding of the specific interactions between BH3-only proteins and their prosurvival Bcl-2 counterparts. Notably, we conclude that Bim is the universal prosurvival antagonist as no other BH3-only protein binds all six prosurvival proteins and that Mcl-1 and Bcl-xL form a distinct prosurvival dyad.
Similar content being viewed by others
Main
The network of protein–protein interactions among B-cell lymphoma-2 (Bcl-2) family proteins is critical in regulating ‘intrinsic’ or mitochondrial programmed cell death.1 Bcl-2 proteins are activated in response to intracellular stress signals to kill cells and are integral in regulating this process. The presence of several conserved sequence motifs known as Bcl-2 homology (BH) motifs (or domains) characterize the Bcl-2 family. Structural and functional analysis has demonstrated that the BH motifs form the molecular basis of the interaction surfaces between the pro- and antisurvival Bcl-2 proteins,2 with the balance between these opposing fractions determining cell fate.3 Resolving the interactions and interconnectivity in the Bcl-2 family is pivotal to understanding apoptosis.
Bcl-B (also known as Bcl-2L10 or Nrh and Boo or Diva in the mouse) is a human Bcl-2 protein identified in database searches.4, 5, 6 Sequence and phylogenetic analysis suggest that Bcl-B has diverged from other prosurvival Bcl-2 proteins both in structure and function.7 Bcl-B bears BH1 and BH2 motifs4, 5, 6 and a hydrophobic transmembrane (TM) region,4 but does not have a recognizable BH3 motif8 necessary for proapoptotic activity.9 The bcl-b gene structure, including promoter regions and intron–exon boundaries, is also conserved with other Bcl-2 members.10
Bcl-B confers resistance to ‘intrinsic’ apoptosis induced by cytotoxic insult4, 5 but not by cell surface death receptor (‘extrinsic’) activation,5 and regulates apoptosis through its ability to control Bax.11 Like other prosurvival proteins, Bcl-B colocalizes through its TM motif with intracellular membranes.6 The mouse ortholog of Bcl-B, Boo (Diva),12, 13 although sharing significant sequence identity with Bcl-B, has a distinct expression pattern12, 13, 14 and lacks functional equivalence.8
Defects in apoptosis regulation have been linked to many diseases and are a hallmark of cancer.15 Altered apoptosis regulation is associated with tumorigenesis, metastasis16 and drug resistance.17, 18 Elevated Bcl-B expression has been observed in ovarian and prostate cancers, multiple myeloma and lymphoma/leukemia cell lines19, 20, 21 and induced overexpression of human Bcl-B in the mouse accelerates Eμ-Myc-driven leukemogenesis similar to other prosurvival Bcl-2 proteins.22 Gene amplifications leading to increased Bcl-B levels have been linked to drug resistance in cancer cell lines.23 Overexpression of Bcl-B does not provide as profound protection against cell death as that of some other prosurvival Bcl-2 members, probably reflecting its more selective role in apoptosis signaling.4
Each of the Bcl-2 proteins is unique, having distinct molecular properties, biological activities, localization and ligand binding specificities that reflect their specific biological roles. Here we report the structure of Bcl-B in complex with the BH3 motif of Bim and an investigation into the molecular mechanisms of action underlying Bcl-B prosurvival activity.
Results
Sequence alignment and phylogeny
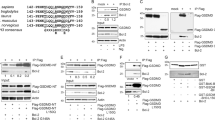
Multiple alignment of the non-redundant Bcl-B sequences (including Bcl-B, Boo, Diva, NR13, vNR13 and Bcl-2L10) against the other multimotif Bcl-2 family members shows they cluster as a separate lineage (this work and Aouacheria et al.6), indicating that Bcl-B derives from a common Bcl-2 ancestor. In addition to their presence in metazoans, a viral Bcl-B (vNR13) is known.24 The shared sequence identity among the Bcl-2 family is generally low. Bcl-B shares about 25% identity with the other human prosurvival proteins. The ‘NR13-like’ sequences from amphibians and fish form a separate clade from the mammalian Bcl-B sequences that are discernible by the presence of multiple sequence insertions in the interhelical loops (Figure 1 and Supplementary Figure S4). The α5–α6 insertion differentiates the mammalian sequences both from other Bcl-B sequences and other Bcl-2 proteins. Phylogenetic analysis of human Bcl-2 sequences shows that Bcl-B has diverged from other human Bcl-2 proteins and forms a distinct lineage (Supplementary Figure S5). A feature conserved in Bcl-B sequences, in addition to the BH1 and BH2 motifs, are an α1 aspartate (D15 human sequence numbering; Figure 1 and Supplementary Figure S4) and an α2 arginine (R40).25 These two residues occur at the mid-points of helices α1 and α2, respectively, in 42 of the 48 Bcl-B sequences (Supplementary Figure S1), but only Bcl-w and Bcl-xL of the other multimotif Bcl-2 proteins retain these residues.
Sequence and structure alignment of Bcl-2 proteins. Sequences were aligned using MEGA5,51 followed by structural alignment with TOP,53 to identify equivalent structural positions and the sequence alignment adjusted accordingly. The sequence numbering is that for Bcl-B. Helices are indicated with colored bars and named α1–α9 below the sequence. C-terminal residues deleted in structure determinations are underlined and the N-terminal residues of Mcl-1 are not shown. The position of BH motifs is indicated by bars above the alignment. Accession numbers for the sequences and structures are given in the Supplementary Information
Protein expression
Sequence analysis of the bcl-b gene indicates that there are two potential start codons (ATG) present separated by 27 nucleotides; however, a predicted hairpin loop five nucleotides from the second ATG suggests that it is the preferred start codon.6 Despite many optimization trials of the published expression and purification protocols11, 20, 21, 26, 27 in our hands, all resulted in the production of insoluble protein. To obtain soluble stable recombinant protein from Escherichia coli fermentations, we found it necessary to both mutate the cysteines 20 and 128 to serine and delete 27 C-terminal residues in the predicted TM region. The C terminally truncated version of Bcl-B (residues A2–A167) was linked to human Bim-BH3 peptide (D51-R76) via a (GS)9 linker to have the Bcl-B hydrophobic groove occupied with its native ligand. Notably, this linker is of sufficient length to enable engagement of a Bcl-B equivalent of the recently identified non-canonical ‘α1-site’ on Bax,28 and is removed together with the affinity tag during the purification via tobacco etch virus (TEV) protease cleavage. This construct yielded well-behaved soluble protein. Dynamic light scattering (DLS) and size-exclusion chromatography showed that the protein was monodisperse and monomeric (Supplementary Figure S1 and S2). The 1H, 15N-HSQC spectrum of Bcl-B gave a set of well-dispersed resonances consistent with a folded and monomeric species (Supplementary Figure S3). In contrast, attempts to express Bcl-B with ligands more weakly binding than Bim failed.
X-ray structure of Bcl-B:Bim complex
We determined the crystal structure of the Bcl-B:Bim complex (Figure 2). Residues 107–121 are absent from the electron density for Bcl-B, indicating their flexibility. The Bim binding site of Bcl-B consists of a wide, shallow hydrophobic groove approximately 25 × 10 Å2 formed by the α2–α5 and α8 helices of Bcl-B (Figure 2a). This region spans the BH1, BH2 motifs and the region on α2 equivalent to the BH3 motif in other multimotif Bcl-2 proteins (Figure 1). The hydrophobic face of the Bim-BH3 amphipathic helix is packed in the Bcl-B interface burying an accessible surface area of 1060 Å2 on Bcl-B and 930 Å2 on Bim. The surface on Bcl-B contacting Bim is shown in Figure 2b and is formed from a combination of hydrophobic and polar interactions. Side chains of the four conserved hydrophobic residues on Bim-BH3 (I58, L62, I65 and F69) are buried in hydrophobic pockets created, in part, by residues conserved across the prosurvival Bcl-2 proteins.29 The highly conserved BH3 leucine, L62, projects into a hydrophobic pocket provided by residues F53, M71, T89 and F93 of Bcl-B (Figure 3). The residues that make up the pocket for I58 are F53, Y57, M71 and F93; the third hydrophobic residue, I65, resides in a pocket generated by L46, H50, F53 and T89, whereas the fourth, that is, F69 of Bim, is within 4 Å of atoms from A42, R45, L46 and F160. Significant polar interactions are also observed between Bcl-B and Bim-BH3. The Bim N70 side-chain carbonyl is hydrogen bonded to Bcl-B via the G85 amide and Bcl-B R86 forms a salt bridge with Bim D67. The BH1 domain is characterized by the presence of a XZGR motif at the N terminus of α5, where X and Z are usually N and W, respectively (Figure 1). This sequence motif forms part of the BH3 binding site in the prosurvival Bcl-2 family and an N-terminal helix-capping motif at the initiation site of helix α5 that is highly conserved in the multimotif Bcl-2 family (including the proapoptotic proteins Bax, Bak and Bok).30 In Bcl-B this sequence is TWGR and the side-chain hydroxyl oxygen of T83 provides the acceptor atom for a hydrogen bond with the donor amide of R86. The TWGR motif also forms an intermolecular hydrogen bond between Bim N70 Oδ1 and the G85 amide, an interaction conserved in BH3:prosurvival complexes.30
Ribbon diagram and BH3 binding surface of Bcl-B. (a) Ribbon diagram of the Bcl-B:Bim complex. Bcl-B helices are labeled α1–α8, the N terminus labeled N and C terminus labeled C. The Bim-BH3 helix (yellow) with N-terminus labeled N′ and C-terminus labeled C′ is bound in a groove formed by helices α2–α5 and α8 of Bcl-B. Helices are colored as indicated in Figure 1. (b) Binding site and interactions of Bcl-B. Surface residues on Bcl-B within 4 Å of an atom on Bim are colored (yellow hydrophobic, red acidic, blue basic and cyan other) and residues on Bim in close contact (≤4 Å) with Bcl-B are shown as sticks and labeled with their residue type and sequence position. The molecular orientation is identical to the ribbon in A
Conserved interactions and structural motifs in the Bcl-B:Bim interface. (a) Molecular details of two highly conserved polar interactions in the BH1 motif of Bcl-2 proteins. A helix capping motif is formed between the acceptor atom of the side chain of the first residue preceding helix α5 (T83) and the donor amide proton of R86.30 Shown is also the ionic interaction between R86 of Bcl-B and D67 of Bim (represented yellow) and the α4–α5 loop residue D78 of Bcl-B. (b) The hydrophobic pocket of Bcl-B enclosing the conserved L62 of Bim. Residues within 4 Å of Bim L62 are labeled. (c) Bcl-B sequences bear a conserved ionic interaction between Asp15 and Arg40 located on α1 and α2 of Bcl-B, respectively
Comparison with other Bcl-2 family complexes
The topology of the Bcl-B:Bim complex is very similar to those of other multimotif Bcl-2 homologs (Supplementary Figure S7). When pairwise superimposed over the common secondary structure elements (Figure 1), the r.m.s.d. from Boo (pdb code 2KUA), Bcl-2 (1G5M), Bcl-xL (1PQ1), Bcl-w (1O0L), Mcl-1 (2NL9) and A1 (2VM6) are 1.3, 1.7, 1.4, 1.7, 1.1 and 1.9 Å, respectively, indicating the close structural similarities of the core residues of these complexes. Although the topology with other multimotif Bcl-2 proteins is well maintained, significant local differences are observed between Bcl-B:Bim and other Bcl-2 family complexes and are shown in the structure–sequence alignment of Figure 1. A point of similarity between Bcl-B and Boo, which differentiates them both from other Bcl-2 proteins, is the presence of the unstructured α5–α6 loop. However, there is little sequence conservation between Bcl-B and Boo residues in these loops (Figure 1).
Structures have been solved for Bcl-2 protein complexes with Bim for Mcl-1, Bcl-xL, A1 and the viral Bcl-2 homolog BHRF1 from Epstein–Bar virus. These complexes are all structurally highly homologous with that of Bcl-B:Bim (Supplementary Figure S7). Details of conserved interactions in Bcl-B:Bim are shown in Figure 3, including a salt bridge between Bim D67 and Bcl-B R86 of the BH1 motif; α-helix N-capping motif in the BH1 motif (Figure 3a); conserved BH3 leucine, L62, buried in the hydrophobic pocket (Figure 3b); and a salt bridge between D15 and R40 (Figure 3c). The non-conserved residues located on α2, equivalent to the BH3 region (A42, R45, L46, I49), provide 100 Å2 to the binding interface contributing only 10% of the total buried surface.
In addition to prosurvival protein:Bim complexes solved, a low-affinity Bim binding site has been detected on Bax located on the opposite face (the α1-site) to that of the BH3-motif binding site of prosurvival Bcl-2 proteins.28 In Bax, the normal BH3-binding groove is blocked by the presence of the C-terminal helical residues and the Bim binding interface is formed from residues located on α1, the α1–α2 loop, α4 and α6 (details given in Supplementary Figure S6).28 Residues equivalent to the Bax α1-site interface on Bcl-B are not conserved with those of Bax (Supplementary Figure S6), making it unlikely that Bim binds this region of Bcl-B.
Binding profile of Bcl-B
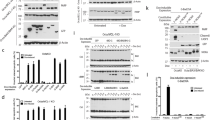
We confirmed that Bcl-B regulates Bax-mediated apoptosis through co-immunoprecipitation experiments (Figure 4) and cell-based assays using MEFs genetically lacking Bax or Bak (Figure 5). Competing prosurvival proteins were selectively inhibited with ABT-737 (an inhibitor of Bcl-2, Bcl-xL and Bcl-w)31 and overexpression of the selective Mcl-1 inhibitor, BimS2A.32 In this context, Bak-dependent apoptosis does not require additional stimulus and results in an absence of colony formation in long-term survival assays (Figure 5).32, 33 Expression of an inactivated form of BimS, BimS4E (a non-binding BH3 where the four hydrophobic residues of the Bim-BH3 motif are replaced by Glu), does not result in cell death in the presence of ABT-737 (Figure 5g) as Mcl-1 neutralization is required for death to occur in this context.33 Overexpression of Bcl-B has a significant effect on cell survival in Bak−/− but not Bax−/− MEFs (Figure 5c), implying Bcl-B inhibits exclusively Bax-dependent cell death. Conversely, Bcl-B overexpression in wild-type and Bax−/− MEF cells did not increase survival in response to ABT-737 and BimS2A (Figure 5d).
Binding profile of Bcl-B and members of the Bcl-2 family. FLAG-tagged Bcl-B was coexpressed with hemaglutinin (HA)-tagged Bcl-2 proteins as indicated. Cell lysates were divided into three equivalent fractions and co-immunoprecipitated with anti-FLAG, anti-HA or control (anti-Glu-Glu) antibodies and immunoblotted for anti-FLAG and anti-HA antibodies
Bcl-B inhibits Bax- but not Bak-initiated apoptosis. (a) Schematic diagram of experiments directed towards determining the Bax or Bak specificity of Bcl-B in the presence of other prosurvival proteins. Neutralization of Bcl-xL, Bcl-2 and Bcl-w is achieved with the drug ABT-737, whereas Mcl-1 is neutralized using a specific inhibitor, BimS2A. Bcl-B binds neither of these inhibitors and its ability to inhibit either Bax- or Bak-initiated apoptosis in the presence of other prosurvival proteins can be determined. (b) Expression of Bcl-B in wild-type (WT) MEF cells does not prevent cell death upon inactivation of endogenous prosurvival proteins Bcl-xL, Bcl-2, Bcl-w and Mcl-1. WT MEFs stably expressing Bcl-B were infected with BimS2A or the inactive BimS4E retrovirus, and then treated for 24 h with 1 μM ABT-737. Colonies were counted after 7 days and expressed as a percentage of control (BimS4E) infected cells. (c) and (d) Bcl-B is able to restrain Bax but not Bak activation. Bak−/− and Bax−/− MEFs stably expressing Bcl-B were infected with BimS2A or the inactive BimS4E retrovirus, and then treated for 24 h with 1 μM ABT-737. Viability was measured by propidium iodide (PI) exclusion (e–g) and expressed as the percentage of control (BimS4E). Long-term survival was measured by following colony formation 7 days after ABT-737 treatment. BimS4E- and BimS2A-infected cells are shown in dark and light bars, respectively
The specificity of Bcl-B for BH3-only binding was determined using co-immunoprecipitation and verified with surface plasmon resonance (SPR) experiments (Figures 4 and 6). The IC50 values determined by SPR for Bik- and Bax-BH3 peptides to dissociate our Bcl-B:Bim complex were 113 and 6.6 μM, respectively. Colony-based assays confirmed Bcl-B co-immunoprecipitation with Bax, but neither Bak nor Bok. Furthermore, ABT-737 failed to displace Bim from the Bcl-B:Bim complex, confirming previous observations34 that ABT-737 is not an antagonist of Bcl-B. Although reported to interact with Bcl-xS,6 a short splice variant of Bcl-xL retaining both the BH3 and hydrophobic C-terminal residues but not the BH1 or BH2 motifs,35 Bcl-B does not co-immunoprecipitate with this protein in our hands (Figure 4). Taken together, these findings show that Bcl-B is neutralized by the BH3-only proteins Bim and Bik and specifically protects cells from Bax-dependent apoptotic pathways.
Peptides spanning the Bax- and Bik-BH3 motifs displace Bim-BH3 from Bcl-B:Bim. Competition assay SPR sensorgrams demonstrate that peptides spanning the BH3 regions of Bax and Bik can displace Bim from the Bcl-B:Bim complex under increasing concentrations of BH3 peptide. Relative responses of samples between 1.25 and 40 μM BH3 peptides are shown. (a) Bax-BH3; (b) Bik-BH3. Other BH3 peptides had no effect
Discussion
Signaling in intrinsic apoptosis in mammals is dependent on heterodimerization between the proapoptotic and prosurvival Bcl-2 family members to ‘activate’ Bax and Bak36 and initiate the caspase cascade. Structural and functional studies have defined the roles of many members of the Bcl-2 family and a structural dichotomy divides the Bcl-2 family: the multi-BH proteins have helical bundle structures and show either prosurvival or proapoptotic activity, whereas the BH3-only proteins are singularly apoptosis inducing9 and are intrinsically disordered proteins, or become them on processing.37 Our current understanding of the network of interactions between the multi-BH motif and BH3-only proteins is incomplete; here we examined the structure and functional characteristics of the last identified and least characterized human Bcl-2 family pro-survival protein, Bcl-B.
The structure of the Bcl-B:Bim complex revealed the anticipated Bcl-2-fold architecture for Bcl-B; a buried central hydrophobic helix (α5) enclosed by seven amphipathic helices (α1–α4 and α6–α8), with α2–α5 and α8 forming the binding groove (Figure 2). The amphipathic Bim-BH3 helix buries its hydrophobic face in the Bcl-B groove provided by Bcl-B and employs a combination of hydrophobic and electrostatic interactions (Figures 2 and 3). The defining features of the BH3 motif are a conserved leucine five residues N terminal to an absolutely conserved aspartate.9, 30, 38 In Bcl-B:Bim, these two conserved interactions are illustrated by the L62 of Bim buried in a hydrophobic pocket and an ionic interaction between Bim D67 and R86 in the BH1 of Bcl-B (Figure 3). Two residues conserved in Bcl-B sequences D15 and R40 form a salt bridge between helices α1 and α2 (Figure 3c). The similarity of Bcl-B:Bim to other prosurvival proteins is illustrated in Supplementary Figure S7 and additionally there is no significant similarity with the α1-site described for Bim binding Bax.28
The apparent human and mouse orthologs, Bcl-B and Boo, share 45.5% sequence identity (60.7% similarity), and although structurally homologous with Bcl-B, Boo bears an altered BH1 sequence (Figure 1) distinguished by the residues SWSQ that substitute TWGR of Bcl-B and result in Boo being unable to bind BH3 motifs from any apoptotic Bcl-2 protein.8 The G to S and R to Q double mutation in the Boo BH1 framework removes a principal ionic interaction with BH3-only proteins required for binding30 and introduces the polar hydroxyl group of serine into the hydrophobic groove.8 Weak BH3 binding affinity can be reconstituted to Boo if these residues are mutated to Gly and Arg, respectively.8 Biochemical data indicate some common features between Bcl-B and Boo, such as their intracellular localization, function in oocyte maturation and early embryonic development,39, 40 but there are also major differences in the protein expression patterns. In adult tissues, Boo is highly expressed in the ovary, but only weakly in other tissues,13, 14 whereas Bcl-B is widely expressed at low levels in most adult tissues4, 5 and highly expressed in the brain, liver, lung and pancreas,4, 6 oocytes and early embryos.39 Immunostaining detected the strongest Bcl-B expression in plasma cells21 and in the terminal stages of B-cell differentiation.26 Combined, these data confirm that Boo and Bcl-B are not orthologous.4 The boo knockout mouse has no overt phenotype;14 however, the knockout in other organisms, where the key BH1 residues are maintained (such as the rat), and that potentially interact with BH3-only proteins, may provide a more appropriate genetic model for investigating the role of Bcl-B in apoptosis.
Insertions and deletions have played an important part in the divergence of sequence and function among the Bcl-2 paralogs.7 The most striking difference between Bcl-B and Boo and the other multi-BH motif Bcl-2 proteins is the presence of an unstructured loop connecting helices α5 and α6, and other Bcl-2 family members have a tight turn at this position. These extra residues do not significantly alter the structure of Bcl-B compared with other multimotif Bcl-2 proteins (Figures 1 and 2) and the lack of observed electron density for these residues indicates that they do not make any significant long-lived contact with the folded core of Bcl-B. A similar situation was found with Boo.8 The presence of intrinsically disordered regions (IDRs) is a feature of the Bcl-2 family37 and the location of these regions varies. Bcl-2 and Bcl-xL have an extended IDR of ∼50 residues inserted between α1 and α2, which in other family members is only ∼13 residues, whereas Mcl-1 has a low complexity sequence of about 160 residues N terminal to its structured region.29 In BHRF1, this loop is unstructured in solution,41 but contains a helix in crystals.42 IDRs in the Bcl-2 proteins frequently bear multiple regulation sites, such as those for phosphorylation, deamidation and ubiquitination,37 but appear absent from viral Bcl-2 homologs.43 The α5–α6 IDR of Bcl-B is formed by a shortening of almost two turns of α5 and a sequence insertion of ∼20 residues (Figure 1) compared with other multi-BH proteins. While Bcl-B and Boo bear a longer α5–α6 loop, the sequence is not well conserved between them, further indicating potential differences between these proteins.
Prosurvival Bcl-2 proteins differ in several facets, including their sequence, subcellular localization and ability to bind proapoptotic proteins. The high-affinity binding of Bcl-B for Bim and Bik-BH3 motifs we observe establishes a narrow BH3-only binding repertoire for Bcl-B compared with other prosurvival Bcl-2 members and is reminiscent of viral Bcl-2 proteins, such as F1L, which only binds Bim, Bak and Bax.44 Our finding that Puma does not bind Bcl-B with substantial affinity leaves only Bim to bind tightly to all six prosurvival proteins, hence making it the only universal apoptotic initiator among the BH3-only proteins and most potent prosurvival antagonist (Figure 7). Bim is activated in response to many apoptotic stimuli in multiple cell types. Other BH3-only proteins have more limited scope for neutralizing prosurvival proteins. For example, Noxa binds only Mcl-1 and A1, whereas Puma binds all prosurvival proteins, with the exception of Bcl-B. No prosurvival protein binds all eight BH3 motifs with high affinity; however, Mcl-1 and Bcl-xL form a dyad that in combination bind all eight BH3-only proteins.
Summary of Bcl-2-regulated apoptotic pathways. (a) Bcl-B, Bcl-w and Bcl-2 selectively inhibit Bax-initiated apoptosis and A1 Bak-initiated apoptosis. Bcl-xL and Mcl-1 inhibit both Bax- and Bax-initiated apoptosis. (b) Bim binds all prosurvival proteins, whereas other BH3-only proteins are more selective. Bcl-B binds only Bim and Bik tightly. Prosurvival proteins are displayed in unfilled boxes, BH3-only proteins in gray boxes and proapoptotic multi-BH proteins in black boxes. Interactions are indicated where binding data suggest a KD<100 nM58, 60
The multimotif proapoptotic proteins, Bax and Bak, are indispensable for intrinsic apoptosis; deletion of both genes renders cells highly resistant to apoptotic stimuli,36 implying they integrate most, if not all, Bcl-2-mediated proapoptotic signals. Our functional studies confirm that Bcl-B acts through Bax and not through Bak or Bok (Figure 4). This finding is consistent with that for chicken NR13, where Bax interaction was demonstrated by co-immunoprecipitation45 and a yeast two-hybrid screen25 and with previous biochemical observations for Bcl-B.20 Thus, Bax is inhibited by Bcl-2, Bcl-w, Bcl-xL, Mcl-1, A1 and Bcl-B, whereas Bak is neutralized by Bcl-xL, Mcl-1 and A1 (Figure 7a). In combination, therefore, Bcl-xL and Mcl-1 neutralize all proapoptotic Bcl-2 proteins.
Perturbed Bcl-B expression has been associated with disease and high levels of Bcl-B have been detected in colorectal tumors, small-cell lung cancer tumors,26 acute myelodysplastic syndromes and acute myeloid leukemia,46 where it has been associated with poor prognosis and drug resistance. Knockdown of Bcl-B can reinstate drug sensitivity to cells responsible for the etiology of these diseases.46 BH3-mimicking inhibitors of prosurvival Bcl-2 proteins are proving efficacious as new cancer therapeutics targeting prosurvival Bcl-2 proteins.31, 47 The BH3-mimetic ABT-737 and its clinical analog, Navitoclax,48, 49, 50 target Bcl-2, Bcl-xL and Bcl-w, and have activity against a range of tumors.31 ABT-737 does not bind Bcl-B34 nor displaces Bim from the Bcl-B:Bim complex, and thus potentially creates Bcl-B-dependent cell survival as a chemoresistance mechanism to ABT-737 and its analogs. Emergence of resistance may ultimately limit the efficacy of treatments as survival factors other than those targeted by ABT-737 enable cancer cells to evade apoptosis. Understanding the structure and function of Bcl-B is a key step towards developing its potential as a therapeutic target.
The combination of genetic, cell biological, biochemical and structural data now available on the BH3-only interactions with prosurvival proteins places them among the most well-understood interactions in apoptosis. Compared with other prosurvival Bcl-2 proteins, Bcl-B has a narrow capacity to bind proapoptotic proteins, defining a more specialized role for it in apoptosis signaling. These data allow us to conclude that Bim is the only BH3-only protein that binds all six of the known mammalian prosurvival proteins, making it the most potent death signal. Solving the structure and defining the selectivity of Bcl-B delimits the role of one of the few remaining Bcl-2 family members without these data, both setting Bcl-B in the context of the Bcl-2 family and declaring Bim as the singular universal prosurvival protein antagonist.
Materials and Methods
Sequence alignment
Sequence and phylogenetic analyses were performed using MEGA5.51 A search for structural homologs of Bcl-B in the Protein Data Bank was performed using secondary structure matching as described.52 Structure alignment was performed using TOP.53 A more comprehensive alignment is presented in the Supplementary Information and Guillemin et al.54
Protein production
To obtain soluble stable recombinant human Bcl-B (Uniprot: Q9HD36) protein from E. coli fermentations for our structural studies, we mutated the cysteines 20 and 128 to serine and deleted 27 C-terminal residues in the predicted TM region. The C-terminally truncated version of Bcl-B (residues A2–A167) was linked to human Bims (Uniprot: O43521-3) BH3 peptide (D51-R76) via a (GS) linker to have the Bcl-B hydrophobic groove occupied with its native ligand. This construct yielded well-behaved soluble protein (Supplementary Figures S1–3). The final expression construct was 6His-MBP-x-Bcl-BΔC27-x-(GS)9-x-Bim-BH3 (where -x- represents a TEV cleavage site ENLYFQGS) in E. coli BL21(DE3). The protein was purified by affinity chromatography on a 5 ml Ni-NTA column before TEV protease cleavage liberation of 6His-MBP and the linker between Bcl-B and Bim-BH3. The final purification step was size-exclusion chromatography on a Superdex S75 16/60 column (GE Healthcare, Rydalmere, Australia). Protein purity was confirmed by SDS-PAGE and electrospray mass spectrometry. DLS and nuclear magnetic resonance (NMR) showed that the protein was monodisperse and monomeric and thermal stability monitored by circular dichroism (CD) showed an irreversible melting point of 58 °C (Supplementary Figure S2). The 1H, 15N-HSQC spectrum of Bcl-B:Bim (Supplementary Figure S3) gave a set of well-dispersed resonances consistent with a folded species and CD spectroscopy confirmed the helical nature of the protein (Supplementary Figure S2). Attempts to express Bcl-B with ligands more weakly binding than Bim were unsuccessful.
The protocol for isotope labeling followed the published method.55 Briefly, E. coli BL21(DE3) were grown at 30 °C in 2 l of Super Broth medium and upon reaching optical density 600∼0.7, cells were pelleted at 5000 × g, and then washed with 500 ml of M9 salt solution that excluded all nitrogen and carbon sources and re-centrifuged. The cell pellet was re-suspended in 500 ml of isotopically labeled M9 medium with 13C6-glucose and/or 15NH4Cl (Cambridge Isotope Laboratories, Andover, MA, USA), and incubated to allow the recovery of growth and the clearance of unlabeled metabolites. After 1 h, protein expression was induced by the addition of IPTG to a final concentration of 1 mM. Cells were harvested after a 3-h incubation period at 30 °C by centrifugation at 4000 × g for 5 min. Protein purification was performed as described above. NMR samples contained 0.3 mM protein in 50 mM sodium acetate (pH 5.0) and 0.05% sodium azide in H2O : 2H2O of 95 : 5. The protocol for selenomethionine labeling followed was based on methionine pathway inhibition as described,56 using identical conditions for the production of isotope-labeled Bcl-B:Bim-BH3 complex.
Crystallization and structure determination
Bcl-B:Bim complex was purified as described above and crystals were grown in sitting drops at 20 °C in 0.1 M MgCl2, 20% PEG 400, 20% PEG 3350 and 0.1 M Tris (pH 8.0). Diffraction data were collected from crystals flash frozen in mother liquor supplemented with 20% PEG 400 at 100 K at the Australian Synchrotron (Melbourne, VIC, Australia; beamline MX2) and processed with HKL2000 and XDS. The crystals belong to the space group C2221, with a=39.843 Å, b=94.845, c=96.372 Å and α=β=δ=90°. The asymmetric units contain 1 Bcl-B:Bim complex with a solvent content of 38% and two Se sites were identified. Clear and continuous electron density was obtained for residues 2–106, and 122–164 of Bcl-B and 53–76 of Bim, whereas the residues constituting the α5–α6 loop (residues 107–121) lack clear electron density and were presumed disordered. The final model refined to a resolution of 1.9 Å and a final R-factor of 0.196 (R-free 0.220). In all, 96.5% of the residues are in the core regions of the Ramachandran plot and none in disallowed regions. Data collection and refinement statistics are summarized in Supplementary Table S1 and Supplementary Information. The coordinates have been deposited in the Protein Data Bank (accession code: 4B4S).
Surface plasmon resonance
Relative affinities of BH3 peptides for Bcl-B were assessed by comparing their abilities to separate the Bcl-B:6His-Bim complex. Measurements were performed at room temperature on a Biacore S51 instrument (GE Healthcare) with 1% DMSO of HBS-P (0.01 M HEPES (pH 7.4), 0.15 M NaCl, 0.005% Surfactant P20) as the running buffer. Taking the response for complex alone as the maximal response (100%), the relative binding response in the presence of competitor peptide at the end of injection was calculated. Serial dilutions of BH3 peptides were performed between 40 and 0 μM (40, 20, 10, 5, 2.5 1.25 and 0 μM). IC50 values (peptide concentration that gives 50% binding) were obtained by nonlinear least-squares fitting of the data to a four parameter sigmoidal equation (XLfit3, IDBS).
Cell biological studies
Tissue culture, transfection and immunoprecipitation
Mouse embryonic fibroblasts (MEFs) were cultured in the FMA media: Dulbecco’s modified Eagle’s (DME) medium, 10% fetal calf serum (FCS), 250 μM L-asparagine and 50 μM 2-mercaptoethanol. All MEFs were generated from E13 to 14.5 embryos and immortalized (at passages 2–4) with SV40 large T antigen. HEK293T and Phoenix Ecotropic packaging cells57 were cultured in DME medium with 10% FCS. Phoenix and HEK293T cells were transfected with Fugene 6 transfection reagent (Roche, Castle Hill, Australia) according to the manufacturer’s protocol.
Neutralization of prosurvival proteins Bcl-xL, Bcl-2 and Bcl-w in MEFs was achieved by treating cells with 1 μm ABT-737 (Abbott Laboratories, Abbott Park, IL, USA) in combination with neutralization of Mcl-1 by BimS2A32 retroviral infection. pMIG retroviral constructs encoding BH3-only protein BimS2A, or the inactive variant BimS4E, were transiently transfected into Phoenix Ecotropic packaging cells and viral supernatants used to infect MEFs as described.58 Short-term cell viability was assayed by PI exclusion. For long-term survival assays, MEFs were infected with pMIG BimS4E or BimS2A vectors in the presence of caspase inhibitor. At 24 h post-infection, 150 GFP-positive cells were sorted unto six-well cell culture plates (in triplicates) and colony numbers counted 7 days later.
Immunoprecipitations were performed using HEK293T cells transiently transfected with Fugene 6 according to the manufacturer’s instructions. Cells were lysed in Onyx lysis buffer with 1% Triton X-100, supplemented with protease inhibitors (Roche; complete). Lysates were divided into three equivalent fractions and immunoprecipitations performed using mouse monoclonal anti-FLAG (M2; Sigma, Castle Hill, Australia) or anti-HA (3F10; Roche) antibodies. Control immunoprecipitations were performed using an anti-mouse Glu-Glu (MMS-115R; Convance Research Product (CRP), Princeton, NJ, USA) antibody. Proteins were resolved by SDS-PAGE (Novex gels; Invitrogen, Mulgrave, Australia), transferred onto nitrocellulose membranes and proteins detected by immunoblotting using mouse FLAG (9H1; Wilson-Annan et al.59); and anti-HA (16B12; CRP). The proteins were detected using enhanced chemiluminescence (GE Healthcare).
Accession codes
Abbreviations
- Bcl-2:
-
B-cell lymphoma-2
- BH:
-
Bcl-2 homology
- CD:
-
circular dichroism
- DLS:
-
dynamic light scattering
- IDR:
-
intrinsically disordered region
- NMR:
-
nuclear magnetic resonance
- SPR:
-
surface plasmon resonance
- TM:
-
transmembrane
References
Youle RJ, Strasser A . The BCL-2 protein family: opposing activities that mediate cell death. Nat Rev Mol Cell Biol 2008; 9: 47–59.
Hinds MG, Day CL . Regulation of apoptosis: uncovering the binding determinants. Curr Opin Struct Biol 2005; 15: 690–699.
Willis SN, Adams JM . Life in the balance: how BH3-only proteins induce apoptosis. Curr Opin Cell Biol 2005; 17: 617–625.
Ke N, Godzik A, Reed JC . Bcl-B, a novel Bcl-2 family member that differentially binds and regulates Bax and Bak. J Biol Chem 2001; 276: 12481–12484.
Zhang H, Holzgreve W, De Geyter C . Bcl2-L-10, a novel anti-apoptotic member of the Bcl-2 family, blocks apoptosis in the mitochondria death pathway but not in the death receptor pathway. Hum Mol Genet 2001; 10: 2329–2339.
Aouacheria A, Arnaud E, Venet S, Lalle P, Gouy M, Rigal D et al. Nrh, a human homologue of Nr-13 associates with Bcl-Xs and is an inhibitor of apoptosis. Oncogene 2001; 20: 5846–5855.
Aouacheria A, Brunet F, Gouy M . Phylogenomics of life-or-death switches in multicellular animals: Bcl-2, BH3-Only, and BNip families of apoptotic regulators. Mol Biol Evol 2005; 22: 2395–2416.
Rautureau GJ, Day CL, Hinds MG . The structure of Boo/Diva reveals a divergent Bcl-2 protein. Proteins 2010; 78: 2181–2186.
Huang DC, Strasser A . BH3-only proteins-essential initiators of apoptotic cell death. Cell 2000; 103: 839–842.
Feng CY, Rise ML . Characterization and expression analyses of anti-apoptotic Bcl-2-like genes NR-13, Mcl-1, Bcl-X1, and Bcl-X2 in Atlantic cod (Gadus morhua). Mol Immunol 2010; 47: 763–784.
Zhai D, Jin C, Huang Z, Satterthwait AC, Reed JC . Differential regulation of Bax and Bak by anti-apoptotic Bcl-2 family proteins Bcl-B and Mcl-1. J Biol Chem 2008; 283: 9580–9586.
Inohara N, Gourley TS, Carrio R, Muniz M, Merino J, Garcia I et al. Diva, a Bcl-2 homologue that binds directly to Apaf-1 and induces BH3-independent cell death. J Biol Chem 1998; 273: 32479–32486.
Song Q, Kuang Y, Dixit VM, Vincenz C . Boo, a novel negative regulator of cell death, interacts with Apaf-1. EMBO J 1999; 18: 167–178.
Russell HR, Lee Y, Miller HL, Zhao J, McKinnon PJ . Murine ovarian development is not affected by inactivation of the bcl-2 family member diva. Mol Cell Biol 2002; 22: 6866–6870.
Hanahan D, Weinberg RA . Hallmarks of cancer: the next generation. Cell 2011; 144: 646–674.
Mehlen P, Puisieux A . Metastasis: a question of life or death. Nat Rev Cancer 2006; 6: 449–458.
Amundson SA, Myers TG, Scudiero D, Kitada S, Reed JC, Fornace AJ . An informatics approach identifying markers of chemosensitivity in human cancer cell lines. Cancer Res 2000; 60: 6101–6110.
Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J et al. The landscape of somatic copy-number alteration across human cancers. Nature 2010; 463: 899–905.
Lee R, Chen J, Matthews CP, McDougall JK, Neiman PE . Characterization of NR13-related human cell death regulator, Boo/Diva, in normal and cancer tissues. Biochim Biophys Acta 2001; 1520: 187–194.
Zhai D, Ke N, Zhang H, Ladror U, Joseph M, Eichinger A et al. Characterization of the anti-apoptotic mechanism of Bcl-B. Biochem J 2003; 376: 229–236.
Luciano F, Krajewska M, Ortiz-Rubio P, Krajewski S, Zhai D, Faustin B et al. Nur77 converts phenotype of Bcl-B, an antiapoptotic protein expressed in plasma cells and myeloma. Blood 2007; 109: 3849–3855.
Beverly LJ, Varmus HE . MYC-induced myeloid leukemogenesis is accelerated by all six members of the antiapoptotic BCL family. Oncogene 2009; 28: 1274–1279.
Yasui K, Mihara S, Zhao C, Okamoto H, Saito-Ohara F, Tomida A et al. Alteration in copy numbers of genes as a mechanism for acquired drug resistance. Cancer Res 2004; 64: 1403–1410.
Aouacheria A, Banyai M, Rigal D, Schmidt CJ, Gillet G . Characterization of vnr-13, the first alphaherpesvirus gene of the bcl-2 family. Virology 2003; 316: 256–266.
Lalle P, Aouacheria A, Dumont-Miscopein A, Jambon M, Venet S, Bobichon H et al. Evidence for crucial electrostatic interactions between Bcl-2 homology domains BH3 and BH4 in the anti-apoptotic Nr-13 protein. Biochem J 2002; 368: 213–221.
Krajewska M, Kitada S, Winter JN, Variakojis D, Lichtenstein A, Zhai D et al. Bcl-B expression in human epithelial and nonepithelial malignancies. Clin Cancer Res 2008; 14: 3011–3021.
Yip KW, Godoi PH, Zhai D, Garcia X, Cellitti JF, Cuddy M et al. A TR3/Nur77 peptide-based high-throughput fluorescence polarization screen for small molecule Bcl-B inhibitors. J Biomol Screen 2008; 13: 665–673.
Gavathiotis E, Suzuki M, Davis ML, Pitter K, Bird GH, Katz SG et al. BAX activation is initiated at a novel interaction site. Nature 2008; 455: 1076–1081.
Day CL, Chen L, Richardson SJ, Harrison PJ, Huang DC, Hinds MG . Solution structure of prosurvival Mcl-1 and characterization of its binding by proapoptotic BH3-only ligands. J Biol Chem 2005; 280: 4738–4744.
Day CL, Smits C, Fan FC, Lee EF, Fairlie WD, Hinds MG . Structure of the BH3 domains from the p53-inducible BH3-only proteins Noxa and Puma in complex with Mcl-1. J Mol Biol 2008; 380: 958–971.
Oltersdorf T, Elmore SW, Shoemaker AR, Armstrong RC, Augeri DJ, Belli BA et al. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature 2005; 435: 677–681.
Lee EF, Czabotar PE, van Delft MF, Michalak EM, Boyle MJ, Willis SN et al. A novel BH3 ligand that selectively targets Mcl-1 reveals that apoptosis can proceed without Mcl-1 degradation. J Cell Biol 2008; 180: 341–355.
van Delft MF, Wei AH, Mason KD, Vandenberg CJ, Chen L, Czabotar PE et al. The BH3 mimetic ABT-737 targets selective Bcl-2 proteins and efficiently induces apoptosis via Bak/Bax if Mcl-1 is neutralized. Cancer Cell 2006; 10: 389–399.
Zhai D, Jin C, Satterthwait AC, Reed JC . Comparison of chemical inhibitors of antiapoptotic Bcl-2-family proteins. Cell Death Differ 2006; 13: 1419–1421.
Boise LH, Gonzalez-Garcia M, Postema CE, Ding L, Lindsten T, Turka LA et al. Bcl-x, a bcl-2-related gene that functions as a dominant regulator of apoptotic cell death. Cell 1993; 74: 597–608.
Zong WX, Lindsten T, Ross AJ, MacGregor GR, Thompson CB . BH3-only proteins that bind pro-survival Bcl-2 family members fail to induce apoptosis in the absence of Bax and Bak. Genes Dev 2001; 15: 1481–1486.
Rautureau GJP, Day CL, Hinds MG . Intrinsically disordered proteins in Bcl-2 regulated apoptosis. Int J Mol Sci 2010; 11: 1808–1824.
Lanave C, Santamaria M, Saccone C . Comparative genomics: the evolutionary history of the Bcl-2 family. Gene 2004; 333: 71–79.
Guillemin Y, Lalle P, Gillet G, Guerin JF, Hamamah S, Aouacheria A . Oocytes and early embryos selectively express the survival factor BCL2L10. J Mol Med 2009; 87: 923–940.
Yoon SJ, Kim EY, Kim YS, Lee HS, Kim KH, Bae J et al. Role of Bcl2-like 10 (Bcl2l10) in regulating mouse oocyte maturation. Biol Reprod 2009; 81: 497–506.
Huang Q, Petros AM, Virgin HW, Fesik SW, Olejniczak ET . Solution structure of the BHRF1 protein from Epstein–Barr virus, a homolog of human Bcl-2. J Mol Biol 2003; 332: 1123–1130.
Kvansakul M, Wei AH, Fletcher JI, Willis SN, Chen L, Roberts AW et al. Structural basis for apoptosis inhibition by Epstein–Barr virus BHRF1. PLoS Pathogen 2010; 6: e1001236.
Kvansakul M, van Delft MF, Lee EF, Gulbis JM, Fairlie WD, Huang DC et al. A structural viral mimic of prosurvival Bcl-2: a pivotal role for sequestering proapoptotic Bax and Bak. Mol Cell 2007; 25: 933–942.
Kvansakul M, Yang H, Fairlie WD, Czabotar PE, Fischer SF, Perugini MA et al. Vaccinia virus anti-apoptotic F1L is a novel Bcl-2-like domain-swapped dimer that binds a highly selective subset of BH3-containing death ligands. Cell Death Differ 2008; 15: 1564–1571.
Lee RM, Gillet G, Burnside J, Thomas SJ, Neiman P . Role of Nr13 in regulation of programmed cell death in the bursa of Fabricius. Genes Dev 1999; 13: 718–728.
Cluzeau T, Robert G, Mounier N, Karsenti JM, Dufies M, Puissant A et al. BCL2L10 is a predictive factor for resistance to azacitidine in MDS and AML patients. Oncotarget 2012; 3: 490–501.
Fesik SW . Promoting apoptosis as a strategy for cancer drug discovery. Nat Rev Cancer 2005; 5: 876–885.
Rudin CM, Hann CL, Garon EB, Ribeiro de Oliveira M, Bonomi PD, Camidge DR et al. Phase 2 Study of single agent navitoclax (ABT-263) and biomarker correlates in patients with relapsed small cell lung cancer. Clin Cancer Res 2012; 18: 3163–3169.
Gandhi L, Camidge DR, Ribeiro de Oliveira M, Bonomi P, Gandara D, Khaira D et al. Phase I study of Navitoclax (ABT-263), a novel Bcl-2 family inhibitor, in patients with small-cell lung cancer and other solid tumors. J Clin Oncol 2011; 29: 909–916.
Wilson WH, O’Connor OA, Czuczman MS, LaCasce AS, Gerecitano JF, Leonard JP et al. Navitoclax, a targeted high-affinity inhibitor of BCL-2, in lymphoid malignancies: a phase 1 dose-escalation study of safety, pharmacokinetics, pharmacodynamics, and antitumour activity. Lancet Oncol 2010; 11: 1149–1159.
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S . MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 2011; 28: 2731–2739.
Krissinel E, Henrick K . Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D 2004; 60: 2256–2268.
Lu GG . TOP: a new method for protein structure comparisons and similarity searches. J Appl Crystallogr 2000; 33: 176–183.
Guillemin Y, Cornut-Thibaut A, Gillet G, Penin F, Aouacheria A . Characterization of unique signature sequences in the divergent maternal protein Bcl2l10. Mol Biol Evol 2011; 28: 3271–3283.
Marley J, Lu M, Bracken C . A method for efficient isotopic labeling of recombinant proteins. J Biomol NMR 2001; 20: 71–75.
Berne PF, Doublie S, Carter CW . Molecular biology for structural biology In: Ducruix A, Giege R (eds.) Crystallization of Nucleic Acids and Proteins. A Practical Approach. Oxford University Press: Oxford, 1999 pp 45–73.
Kinsella TM, Nolan GP . Episomal vectors rapidly and stably produce high-titer recombinant retrovirus. Hum Gene Ther 1996; 7: 1405–1413.
Chen L, Willis SN, Wei A, Smith BJ, Fletcher JI, Hinds MG et al. Differential targeting of prosurvival Bcl-2 proteins by their BH3-only ligands allows complementary apoptotic function. Mol Cell 2005; 17: 393–403.
Wilson-Annan J, O’Reilly LA, Crawford SA, Hausmann G, Beaumont JG, Parma LP et al. Proapoptotic BH3-only proteins trigger membrane integration of prosurvival Bcl-w and neutralize its activity. J Cell Biol 2003; 162: 877–887.
Kuwana T, Bouchier-Hayes L, Chipuk JE, Bonzon C, Sullivan BA, Green DR et al. BH3 domains of BH3-only proteins differentially regulate Bax-mediated mitochondrial membrane permeabilization both directly and indirectly. Mol Cell 2005; 17: 525–535.
Acknowledgements
We thank Catherine Day, Otago University, for reagents. NMR spectra were acquired at the Bio21 Institute NMR Facility, University of Melbourne and crystal screening performed at Bio21 C3. We thank staff at the Australian Synchrotron for assistance. Our research is supported by the Cancer Council Victoria (Grant 575549; MGH), the Leukemia and Lymphoma Society (SCOR 7015-02), National Health and Medical Research Council of Australia (fellowship 637372, MK), Victorian State Government Operational Infrastructure Support and NHMRC IRIISS.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by G Raschellá
Supplementary Information accompanies the paper on Cell Death and Disease website
Supplementary information
Rights and permissions
This work is licensed under the Creative Commons Attribution-NonCommercial-No Derivative Works 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Rautureau, G., Yabal, M., Yang, H. et al. The restricted binding repertoire of Bcl-B leaves Bim as the universal BH3-only prosurvival Bcl-2 protein antagonist. Cell Death Dis 3, e443 (2012). https://doi.org/10.1038/cddis.2012.178
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cddis.2012.178
Keywords
This article is cited by
-
Bcl-B: an “unknown” protein of the Bcl-2 family
Biology Direct (2023)
-
Potent hepatoprotective activity of common rattan (Calamus rotang L.) leaf extract and its molecular mechanism
BMC Complementary Medicine and Therapies (2023)
-
Membrane recruitment of the polarity protein Scribble by the cell adhesion receptor TMIGD1
Communications Biology (2023)
-
A structural investigation of NRZ mediated apoptosis regulation in zebrafish
Cell Death & Disease (2018)
-
Tannin-rich extracts from Lannea stuhlmannii and Lannea humilis (Anacardiaceae) exhibit hepatoprotective activities in vivo via enhancement of the anti-apoptotic protein Bcl-2
Scientific Reports (2018)