Abstract
Changes in mitochondrial genome such as mutation, deletion and depletion are common in cancer and can determine advanced phenotype of cancer; however, detailed mechanisms have not been elucidated. We observed that loss of mitochondrial genome reversibly induced overexpression and activation of proto-oncogenic Ras, especially K-Ras 4A, responsible for the activation of AKT and ERK leading to advanced phenotype of prostate and breast cancer. Ras activation was induced by the overexpression of 3-hydroxy-3-methyl-glutaryl-CoA reductase (HMGR), the rate-limiting enzyme of the mevalonate pathway. Hypoxia is known to induce proteasomal degradation of HMGR. Well differentiated prostate and breast cancer cells with high mitochondrial DNA content consumed a large amount of oxygen and induced hypoxia. Loss of mitochondrial genome reduced oxygen consumption and increased in oxygen concentration in the cells. The hypoxic-to-normoxic shift led to the overexpression of HMGR through inhibiting proteasomal degradation. Therefore, reduction of mitochondrial genome content induced overexpression of HMGR through hypoxic to normoxic shift and subsequently the endogenous induction of the mevalonate pathway activated Ras that mediates advanced phenotype. Reduction of mitochondrial genome content was associated with the aggressive phenotype of prostate cancer in vitro cell line model and tissue specimens in vivo. Our results elucidate a coherent mechanism that directly links the mitochondrial genome with the advanced progression of the disease.
Similar content being viewed by others
Main
Prostate cancer (PCa) is a significant health problem, representing the leading cancer diagnosis and the second leading cause of cancer death in North American men.1 PCa begins as an androgen-dependent cancer that undergoes clinical regression in response to pharmacological or surgical strategies that reduce testosterone concentration. Despite this treatment, the cancer frequently recurs as an androgen-independent or castration-resistant cancer with metastatic phenotype. Therefore, elucidation of the mechanisms involved in the development to an aggressive metastatic phenotype is a critical topic.
Mitochondria are essential organelles that generate ATP through oxidative phosphorylation. This process is accomplished by a series of protein complexes and mitochondrial respiratory chains encoded by nuclear DNA (nDNA) and mitochondrial DNA (mtDNA). Human mtDNA is remarkably small (16 569 bp) compared with nDNA (∼109 bp) and encodes 13 proteins in the mitochondrial respiratory chains (MRC), 22 transfer RNAs, and 2 ribosomal RNAs.2
Several reports showed that mutations of mtDNA had been identified in various types of cancer, including breast, colon, prostate, pancreatic and other cancers.3 The growth advantage of cancer cells with specific mutant mtDNA has also been demonstrated in an in vivo mouse model system using cybrid (trans-mitochondrial hybrid) cells, suggesting that specific mutations of mtDNA give a survival advantage and induce metastasis.4 In PCa specimens, Chen et al.5 reported an extremely high incidence of somatic mutation (90%) in the D-loop region that may cause reduction of mtDNA content.6
The roles of mtDNA and mitochondrial respiratory function in biological functions has been demonstrated using mtDNA-deficient cells.7 We previously showed that TNF and serum starvation could not induce apoptosis in mtDNA-depleted cells, whereas they induced apoptosis in parental cells and cells reconstituted with normal mtDNA.8 These results indicate that mitochondrial respiratory function and mtDNA are critically important in apoptosis. We showed following observations to demonstrate the mechanisms involved in the induction of advanced phenotype of PCa by the reduction of mitochondrial genome content. Reduction of mtDNA content shifted (1) androgen-dependent PCa cells to an androgen-independent phenotype in vitro and in vivo9 and induced (2) epithelial-to-mesenchymal transition changes that may lead to PCa progression,10 (3) hypermethylation of CpG islands of the putative tumor suppressor genes11 and (4) abnormal activation of NF-κB,12 AP-1,12 AKT,13 ERK10 and JNK10 that may lead to aggressive phenotype in PCa progression.
Ras-GTPases are potential candidates that can transduce the constitutive and concurrent activation of a broad array of effectors such as ERK and AKT. As small GTPases, Ras are dynamically activated and inactivated with the association of GTP or GDP, respectively. Human cancers (∼25%) often harbor (oncogenic) mutations in Ras that disable the ability of the GTPases to dynamically be inactivated, leading to the constitutive and concurrent activation of a diverse array of signal pathways, for example, Raf/MEK/ERK and PI3K/Akt.14 Although tissue studies have revealed the constitutive and concurrent activation of ERK and Akt in the progression of PCa, the majority of PCa tumors do not harbor RAS mutation (oncogenes).15 Yet, it is generally acknowledged that the aberrant activation of Ras can shift PCa from androgen-dependent to independent,16 whereas the expression of dominant-negative Ras attenuates androgen-independent phenotype,17 indicating that Ras activation is responsible for the phenotype of PCa. In the current study of PCa, we investigated a pathway of metabolic and proto-oncogenic processes including Ras activation that aberrantly activates ERK and Akt. We denoted this as the mitochondrial-generated progression signal (mitoGPS) that transduces changes in mitochondrial genome in the early-stages of PCa into the advanced progression of the disease.
Results
Reduction of the mitochondrial genome content is associated with PCa progression
We first investigated whether mitochondrial genome content changes associated with aggressive phenotype of PCa in human tissue specimens. Mitochondrial genomic analysis in a single cell revealed that the mitochondrial genome copy-number/cell was significantly reduced in high-grade tumors (Gleason grade 8 and 9) as compared with that in low-grade tumors (Gleason grade 6 and 7) (Figure 1a) (P<0.01). The in vivo observations were verified in human cell lines the early stages androgen-dependent PCa (LNCaP), or an array of PCa (C4-2, PC-3 and DU-145). Classical cell lines for the in vitro evaluation of PCa, include: the castration sensitive LNCaP established from a metastatic (supraclavicular lymph node) lesion of human prostate adenocarcinoma;18 the castration resistant (CR) C4-2 isolated from recurrent tumors that develop after castration of mice inoculated with LNCaP;19 and CR PCa cell lines derived from metastasis in human bone (PC-3) and brain (DU-145).20, 21 In the current study, mitochondrial genomic analysis showed a low mitochondrial genome content in PC-3 and DU-145, in addition to a significant reduction in C4-2, relative to the parental LNCaP (Figure 1b).
Reduction of the mitochondrial genome content in PCa and BCa reversibly induced aberrant activation of ERK and AKT. (a) Mitochondrial genome content per cell in low (6–7) and high (8–9) Gleason grade PCa separated by laser microdissection. Student's t-test compared PCa with a high Gleason grade (*P<0.01) (n=8 and 6, respectively). (b) Mitochondrial genome contents of single cells (n=10) of an early-stage (LNCaP) PCa cell line, and an array of (C4-2, PC-3 and DU-145) PCa cell lines. (c) FISH of the mitochondrial genome. Green: mitochondrial genome. Red: mitochondria. Scale bar represents 20 μm. (d) Mitochondrial genome content in single cells (n=8) of the parental (LNCaP and MCF-7), mtDNA-KO (LNρ0-8 and MCFρ0) and mtDNA-KI (LNCyb and MCFCyb) cells. (e) Phospho-ERK, total ERK, phospho-Akt, total AKT and β-actin in total-protein lysates (30 μg) of the indicated cell lines
Reduction of the mitochondrial genome content reversibly induces the aberrant activation of ERK and Akt
We evaluated the biological impact of the reduction of the mitochondrial genome content in the progression of PCa by the experimental depletion of the genome from LNCaP. We additionally utilized the MCF-7 cell line, a classical breast cancer (BCa) established from the metastasis (pleural effusion) of a human breast adenocarcinoma.22 The knock-out of the mtDNA (mtDNA-KO) from LNCaP and MCF-7 was demonstrated in the creation of the LNρ0-8 and MCFρ0 cell lines, respectively.9, 10 To confirm that experimental observations that follow the loss of the mitochondrial genome content are not of consequence to an incidental mutation to the nuclear genome induced by ethidium bromide treatment, the mitochondrial genome was reconstituted to the mtDNA-KO cell lines (LNρ0-8 and MCFρ0) in the creation of the mtDNA-knock in (mtDNA-KI) cells lines, denoted as LNCyb and MCFCyb.
The mitochondrial genome contents of the indicated cell lines were visually confirmed by fluorescent in situ hybridization (FISH). Mitochondria were labeled with MitoTracker Red and showed that presence of mitochondria in parental, mtDNA-KO and mtDNA-KI cells (Figure 1c). Microscopic (confocal) images also show the abundance of the mtDNA probes localized in the mitochondria of the parental and mtDNA-KI cells; no mtDNA probes were observed in the mtDNA-KO cells (Figure 1c). The results were confirmed by the quantification of the mitochondrial genome content by QRT-PCR (Figure 1d); the mitochondrial genome content in LNCyb and MCFCyb was partially or completely restored, respectively, to that in LNCaP and MCF-7. We observed that the amount of ATP in LNCaP was significantly reduced in LNr0-8 (113.3±2.0 and 73.2±2.1 nM/mg protein, respectively) (average±S.E., P-value=0.0056) and lactate generation was increased (1463±66 and 3416±82 nM, respectively) (average±S.E., P-value=0.000049). The results suggest that loss of mitochondrial genome reduced ATP generation possibly by the reduction of oxidative phosphorylation that might increase in anaerobic glycolysis.
It is well documented that ERK and Akt are constitutively activated in the progression of advanced PCa tumors.15 Our data reveal ERK and Akt to be constitutively activated in the mtDNA-KO of PCa and BCa, relative to the parental and mtDNA-KI cells (Figure 1e). Of the ERKs, that is, ERK1 (p44) and ERK2 (p42), ERK2 was the most dramatically phosphorylated in LNρ0-8 and MCFρ0.
Reduction of the mitochondrial genome content in PCa and BCa constitutively activates Ras
The interaction of activated GTP-bound Ras with the Ras binding domain (RBD) of Raf-1,23 generates GTP-bound Ras–Raf complexes that lead to the activation of ERK and Akt.24, 25 We investigated whether Ras is activated by depletion of mitochondrial genome. No active Ras was pulled-down from the total-protein lysates (160 μg) of parental (LNCaP and MCF-7) cells in comparison with a positive control (Figure 2a). Meanwhile, active Ras from mtDNA-KO was observed even when the quantity of LNρ0-8 and MCFρ0 lysates probed was reduced to 0.5 μg or 5 μg, respectively, active Ras was observed as two bands of intensity similar to the positive control (Figure 2a). The experiment demonstrates that the depletion of the mitochondrial genome in PCa leads to the constitutive activation of Ras. Association of the reduction of mitochondrial genome content with the activation of Ras was verified in an array of PCa C4-2, PC-3 and DU-145 that harbor low mitochondrial genome contents, relative to the early stages of PCa LNCaP (Figures 1b and 2b).
Reduction of the mitochondrial genome content in PCa and BCa constitutively activates Ras. (a) Activated (GTP-bound) Ras was pulled down and determined by western blotting. Negative controls (*GDP) and positive controls (**GTPγS): before pull-down, an aliquot of total protein lysate was pre-loaded with GDP or GTPγS, respectively. (b) Activated Ras in total-protein lysates (160 μg) of the indicated PCa cells lines. (c) DAB2IP in total-protein lysates (90 μg) of normal PrEC and the indicated PCa cell lines
The shift of normal epithelial cells to the early-stages of PCa and BCa is associated with the genetic silencing of RasGAPs, particularly the DOC-2/DAB2 interactive protein (DAB2IP).26 The attenuation of DAB2IP activity in PCa diminishes the cells ability to amplify the GTPase activity of Ras, leading to the prolonged accumulation of active GTP-bound Ras.27 We observed the expression of DAB2IP to be expressed in the prostate epithelial cells (PrEC), but not in all PCa including LNCaP, C4-2, DU-145 and PC-3 (Figure 2c). Collectively the experiments demonstrate that the shift of normal prostate epithelial cells to PCa is associated with a silencing of a RasGAP that is not enough to explain why Ras is not activated in early stages of PCa LNCaP.
Reduction of the mitochondrial genome content in PCa overexpresses proto-oncogenic K-Ras 4A
The human cell contains three nuclear-encoded RAS genes that encode four isoforms of Ras: H-Ras, N-Ras and two splice variations of K-Ras, K-Ras 4A and K-Ras 4B. We observed the protein level of K-Ras 4A to be greatly elevated in the mtDNA-KO (LNρ0-8 and MCFρ0), relative to the parental cells. Reconstitution of the mitochondrial genome to the mtDNA-KO attenuated the aberrant K-Ras 4A expression (Figure 3a). Expression level of K-Ras 4A in mtDNA-KO cells was exceptionally high as compared with that of K-Ras 4B, H-Ras and N-Ras. A similar expression pattern was observed for the transcript levels of K-Ras 4A (Figure 3b). Association of the reduction of mitochondrial genome content with increased expression of K-Ras 4A was verified in an array of PCa C4-2, PC-3 and DU-145 that harbor low mitochondrial genome contents, relative to the early stages of PCa LNCaP (Figures 1b and 3c).
Reduction of the mitochondrial genome content in PCa overexpresses proto-oncogenic K-Ras 4A. (a) Ras isoforms in total protein lysates (30 μg) of the indicated cell lines. Exposure time was 10 s, excluding K-Ras 4A (0.5 s). (b) RT-PCR analysis of Ras (cDNA) transcripts in total-cDNA constructed from total-RNA lysates of the indicated cell lines. (c) K-Ras 4A in total-protein lysates (30 μg) of the indicated cell lines. (d) Phospho-ERK and phospho-Akt in total-protein lysates (30 μg) of LNρ0-8 transfected in the absence (control) or presence of K-RAS siRNA or non-specific siRNA. Protein levels of K-Ras 4A were evaluated in *(0.5 μg) total-protein lysates
We note that the constitutive activation of Ras observed with the depletion of the mitochondrial genome was not a consequence of oncogenic mutation in nucleotide sequences that encode for codons 12, 13 or 61 of K-Ras.28 In a subsequent experiment we demonstrated that silencing of K-RAS in LNρ0-8, by transfection of K-RAS siRNA, reduced the expression of K-Ras 4A and the phosphorylation of ERK and Akt, but not total ERK nor total AKT (Figure 3d). Collectively, the experiments demonstrate that the reduction of the mitochondrial genome leads to the aberrant expression and activation of Ras, predominantly proto-oncogenic K-Ras 4A, and a process that is reversible with the reconstitution of the mitochondrial genome.
HMGR (3-hydroxy-3-methyl-glutaryl-CoA reductase) expression is responsible for the aberrant activation of Ras mediated by the reduction of the mitochondrial genome content
Ras must translocate to cellular membrane by post-translational modifications that enable the (GTP-bound) Ras–Raf complexes to activate effectors at cellular membranes.29 Localization of Ras to the cellular membrane is mediated by the prenylation of Ras, that is, the addition of farnesyl to the first cysteine (CAAX) of the C-terminus.30 Farnesyl moieties for the prenylation of Ras are derived from farnesyl pyrophosphate, an intermediate of the mevalonate pathway that is catalytically transferred by farnesyltransferase (FTase).31
Immunohistochemical analysis confirmed that expression of Ras was greatly increased in LNρ0-8 as compared with LNCaP (Figure 4a). Figures 4a and b also demonstrates that Ras located only in cytosolic fraction in LNCaP and cellular membrane and certain organelle in LNρ0-8 suggesting that the depletion of the mitochondrial genome signals the localization of Ras to the cellular membrane and specific organelle. Farnesylation of Ras can be repressed with lovastatin-mediated inhibition of the rate-limiting step of the mevalonate pathway – the metabolism of HMG-CoA to mevalonate by HMGR.32 Addition of lovastatin to the culture of LNρ0-8 generated the cytosolic accumulation of Ras, in addition to a shift to LNCaP-like cell morphology, both of which were reversed with co-application of exogenous mevalonate (Figure 4b). To directly inhibit the farnesylation of Ras, farnesyltransferase inhibitors (FTIs) have been designed as peptidomimetics of the C-terminus (CAAX) of Ras.32 The supplementation of lovastatin or FTI-276 diminished the phosphorylation of ERK and Akt in LNρ0-8 (Figures 4c and d). The results suggest that the endogenous activity of the mevalonate pathway is necessary for the constitutive activation of Ras in LNρ0-8.
The endogenous mevalonate pathway mediates the maturation of aberrant Ras signals in PCa devoid of the mitochondrial genome. (a) Confocal images (z-slices) of anti-total-Ras (Alexa-488 labeled-anti-mouse IgG) within an intracellular plane of formaldehyde fixed cultures of the indicated cell lines. Matched image sets were mapped to a common linear gray-scale range (*overexposed image of LNCaP). Scale bars represent 20 μm. (b) Confocal images (z-slices) of anti-total-Ras in mtDNA-KO (LNρ0-8) cells cultured 12 h in the absence (control) or presence of lovastatin (5 μM) with or without mevalonate (1 mM). (c and d) Phospho-ERK, total ERK, phospho-Akt, total AKT and β-actin in total-protein lysates (30 μg) of LNρ0-8 cells cultured 24 h in the absence (control) or presence of lovastatin (c), or (d) 48 h in the absence (control) or presence of FTI-276. (e) HMGR in total-protein lysates (30 μg) of the parental, mtDNA-KO and mtDNA-KI of LNCaP. (f) HMGR in total-protein lysates (30 μg) of indicated cells as assessed by western blot with anti-HMGR
Then, we further investigated the potential cause for upregulation of mevalonate pathway. We observed an elevation of the expression of HMGR with the depletion of the mitochondrial genome (LNρ0-8) could be attenuated with the reconstitution of the genome (LNCyb) (Figure 4e). We also observed the expression of HMGR was enhanced associated with the reduction of the mitochondrial genome content in PCa cell lines (Figure 4f). These results indicate that an elevation in the endogenous expression of HMGR is responsible for the activation of Ras when the mitochondrial genome content is reduced.
Inhibition of mitochondrial respiration is responsible for HMGR expression possibly through the inhibition of proteasomal degradation
Mitochondrial genome encodes 13 proteins in MRC and therefore reduction of mitochondrial genome leads to the inhibition of mitochondrial respiration. To determine if the induction of HMGR expression is induced by the inhibition of mitochondrial respiration, LNCaP cells were cultured in the absence or presence of inhibitors of various complexes of the MRC: rotenone inhibits complex I; antimycin A inhibits complex III; oligomycin inhibits Fo-ATPase of complex V. Rotenone, antimycin A and oligomycin at the concentration used inhibited the oxygen consumption rate of LNCaP (32.6%, 35.2% and 34.4%, respectively) and enhanced the expression of HMGR (Figure 5a).
Inhibitors of mitochondrial respiratory chain and proteasomal degradation enhanced expression of HMGR. (a) HMGR expression and ERK phosphorylation were evaluated in total-protein-lysates (30 μg) of LNCaP cultured 6 h in the presence or absence of rotenone (400 nM), antimycin A (160 ng/ml) and oligomycin (8 μg/ml). (b) HMGR in total-protein lysates of LNCaP cultured 6 h in the absence (control) or presence of ALLN at the indicated dosages
HMGR is regulated by multiple feedback mechanisms, including: transcriptional and/or translational regulation, PTMs, and/or proteasomal degradation by both sterol and non-sterol products of the mevalonate pathway.33 We investigated whether proteasomal degradation is associated with high expression of HMGR. Incubation with proteasomal inhibitor N-[N-(N-Acetyl-L-leucyl)-L-leucyl]-L-norleucine (ALLN) in LNCaP increased the protein level of HMGR (Figure 5b), suggesting that early stage PCa maintain low expression level of HMGR by proteasomal degradation.
HMGR expression in PCa is enhanced by the hypoxic-to-normoxic shift
A recent study has demonstrated that hypoxia induces the proteasomal degradation of HMGR.34, 35 As expected, oxygen consumption is well associated with the content of mitochondrial genome in PCa cell lines (Figures 1b and 6a). From this observation, we hypothesized that LNCaP cells induce hypoxia by high consumption rate of oxygen leading to hypoxia-induced HMGR degradation, and that in advanced PCa the hypoxic state is retarded with the inhibition of respiration, that is, by the reduction of mitochondrial genome content, leading to the accumulation of HMGR expression. Interestingly, the oxygen concentration of surrounding cells was greatly reduced with the culture of LNCaP and MCF-7, as observed with the induction of hypoxia (12.3 and 4.2 μM, respectively) in an hour (Figures 6b and c, respectively). The hypoxia-inducing activity of the parental cells was greatly reduced with the depletion of the mitochondrial genome, and recovered with the reconstitution of the genome (Figures 6b and c). To investigate whether the hypoxic-to-normoxic shift induced by the reduction of mitochondrial genome content augments the expression of HMGR, the oxygen condition in the culture of LNCaP cells was increased (30%, 310 μM) to establish an increased intracellular oxygen condition. The expression of HMGR and the phosphorylation of ERK were amplified within 6 h of culture in the enriched oxygen condition (Figure 6d). The study was further refined by the culture of LNρ0-8 in low oxygen environments to exogenously induce a normoxic-to-hypoxic shift in LNρ0-8. The greatly reduced-oxygen condition (0.2%, 2.1 μM), but not 1, 5 or 20%, attenuated the expression of HMGR and the phosphorylation of ERK (Figure 6e), within 6 h of culture in the 0.2% oxygen environments (Figure 6f). The data indicate that hypoxic-to-normoxic or normoxic-to-hypoxic shifts regulate the protein levels of the HMGR enzyme. We observed the hypoxia-inducing activity of advanced PCa cell lines to be low (Figure 6g), an observation that correlates with the oxygen consumption (Figure 6a) and the dysregulation/reduction of the steady state of the mitochondrial genome content in the progression of PCa (Figure 1b).
HMGR expression in PCa is enhanced by the hypoxic-to-normoxic change. (a) Oxygen consumption rates of the indicated cell lines (1 × 106 cells/ml) (mean±S.E.) (n=3). (b and c) Oxygen concentration surrounding the indicated cell lines (2 × 106 cells/ml) (mean±S.E.) (n=3). (d) HMGR and phospho-ERK in total-protein-lysates (30 μg) of LNCaP cultured in 30% oxygen for the indicated time periods. LNCaP were cultured in 20% oxygen as a control. (e) HMGR and phospho-ERK in total-protein-lysates (30 μg) of LNρ0-8 seeded (1 × 106 cells/ml) overnight, then cultured 6 h in the indicated oxygen concentration, or (f) in 0.2% oxygen for the indicated time periods. LNρ0-8 cells were cultured in 20% oxygen as a control. (g) Oxygen concentration surrounding the indicated cell lines (2 × 106 or 1 × 106 cells/ml) (mean±S.E.) (n=3) after 2 h incubation
Endogenous or exogenous induction of the mevalonate pathway in PCa signals the progression of the advanced phenotype in vitro and in vivo
To verify that exogenous androgens amplify the viability of the early-stages PCa, LNCaP were cultured in androgen-deprived media without (Figure 7a) or with synthetic androgen (R1881) (Figure 7b). Meanwhile, the androgen-deprived media was supplemented with exogenous mevalonate to induce the mevalonate pathway in the absence or presence of exogenous androgen (Figures 7c and d, respectively). Figure 7e demonstrates that exogenous androgens amplify the growth and proliferation of LNCaP in a dose-dependent fashion that is diminished in the presence of exogenous mevalonate. Alternatively, mevalonate alone supports growth in the absence of exogenous androgen (Figures 7c and e). The experiment reveals that exogenous mevalonate induces androgen independence.
Endogenous or exogenous induction of the mevalonate pathway in PCa signals the progression of the phenotype. Microscopic images of LNCaP cultured 7 days in androgen-deprived media with 5% charcoal stripped bovine serum (no androgen) (a) supplemented with synthetic androgen R1881 (b), mevalonate (c), or mevalonate and R1881 (d). Scale bar represents 100 μm. (e) Total cell viability was quantified by a calcein assay (arbitrary unit (A.U.)±S.E.) (n=3). Student's t-test compared the control group in the absence or presence of R1881 (*P<0.01, except for 10 pM and 100 pM, P<0.05), and the control and mevalonate groups cultured in the absence of R1881 (**P<0.01). (f) Viability of the parental, mtDNA-KO and mtDNA-KI of LNCaP cultured 4 days in the absence or presence of lovastatin (n=3). Student's t-test compared LNCaP and LNρ0-8 cultured in the same lovastatin concentration (*P<0.01). (g) Addition of mevalonate to the lovastatin-mediated inhibition of the viability of LNρ0-8. Total cell viability was quantified by a crystal violet assay (average AD590±S.E.) (n=3). Student's t-test compared the lovastatin groups with the control (no lovastatin, no mevalonate) (*P<0.001)
In subsequent experiments we evaluated the essentialness of the endogenous mevalonate pathway on the viability of PCa that harbor or are devoid of the mitochondrial genome. The viability of LNρ0-8 was significantly diminished in the presence of lovastatin, relative to the parental and mtDNA-KI cells (Figure 7f); the lovastatin-mediated inhibition of the viability of the mtDNA-KO was prevented by mevalonate (Figure 7g). The experiments demonstrate that a reduction of the mitochondrial genome content in PCa is transduced into the advanced phenotype via an endogenous induction of the mevalonate pathway.
LNCaP cells are known to gradually acquire more aggressive growth within in vitro and in vivo environments.36, 37 In order to investigate, in vivo, the role of mevalonate pathway in the shift of LNCaP to more aggressive growth phenotype, we inoculated LNCaP cells into athymic nude mice. Following the development of tumors with diameters of 9–10 mm, mice were randomly assigned to four experimental groups as follows; (1) control, (2) lovastatin, (3) mevalonate and (4) lovastatin and mevalonate. The mean tumor volume for each group was quantified on Day 0 (pre-treatment): 377±105, 336±106, 454±86 and 383±44 (average±S.D. mm3). Subsequently, pumps were utilized to systemically deliver either mevalonate (group 3 and group 4) or isotonic saline (group 1 and group 2) into mice, whereas lovastatin was consumed in the diet of mice of group 2 and group 4. Figure 8a demonstrates that the progressive growth of LNCaP (group 1) in mice was significantly reduced with the treatment of lovastatin (group 2); the growth inhibitory effect of lovastatin was reversed with exogenous mevalonate (group 4). Meanwhile, the administration of exogenous mevalonate alone (group 3) slightly but significantly enhanced the growth of LNCaP. These results indicate that the mevalonate pathway is responsible for in vivo advanced growth in the xerographic model of PCa.
The effect of lovastatin and mevalonate on tumor formations of LNCaP in athymic nude mice. Athymic BALB/c (nu/nu) mice were subcutaneously inoculated with LNCaP. The effect of lovastatin and mevalonate on tumor growth was investigated as described in Materials and Methods. The tumor volumes were measured weekly and calculated. The relative tumor volume was calculated as percent of control±S.D. by assuming the rate of each tumor volume on day 0 to be 100% (n=6). Student's t-test compared the control and lovastatin group showed significance at day 14, 21 and 28 days (*P<0.01), and the control and mevalonate groups at day 14 and 21 (**P<0.01) and 28 days (***P<0.02)
Discussion
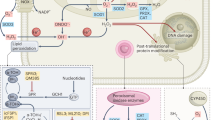
Our collective report demonstrates in PCa that the (1) inhibition of respiration, by reduction of the mitochondrial genome content, shifts cells from a hypoxic-to-normoxic state (2) that attenuates the endogenous proteasomal degradation of the HMGR enzyme; (3) the increase in mevalonate pathway (4) induces the constitutive activation of nuclear-encoded proto-oncogenic Ras (5) that signal for an array of cancer progression signals (Figure 8b). We denote the coherent series of events as the mitoGPS.
The pathological condition of a deprivation in an adequate oxygen supply to the body is referred to as hypoxia, a condition that is transduced at the molecular level into hypoxia-related signals that allow a cell(s) to adjust to the extracellular environment. We are the first to directly establish the mitochondrion as a direct physiological source of hypoxia in an in vitro system. Our results demonstrate that the reduction of the mitochondrial genome content diminishes the hypoxic state of the early stages of PCa and BCa, generating a hypoxic-to-hyperoxic shift in the oxygen content that is transduced into the overexpression of HMGR; subsequently, the induction of the mevalonate pathway activates proto-oncogenic Ras that signals the constitutive and concurrent activation of a diverse array of cancer progression pathways that advance the disease. Reports indicate that hypoxia inhibits lanosterol demethylase, an oxygen requiring enzyme, and accumulates lanosterol (molecules in cholesterol pathway) and that accumulation of lanosterol induces Insig-mediated degradation of HMGR. Hypoxia also activates HIF-1 to induce expression of insig.34, 35 Whether this pathway is activated in our system is now under investigation. The roles of ROS may not be likely as mtDNA-deficient cells generate reduced superoxide.38
With primary PCa composed of a mixture of cells with high or low mitochondrial genome content,39 cells with high mitochondrial content may generate more hypoxia-induced changes that benefit the growth and maintenance of both groups under high cell density conditions, that is, the primary tumor site. In contrast, we speculate in the initiation of metastasis that the progressive induction of hyperoxia with the reduction of cell-density further enhances the mitoGPS, advancing the phenotype of cancer that harbor a reduced copy-number of the mitochondrial genome. It is plausible that the mitoGPS is a ubiquitous (patho)physiological response to the etiology and/or progression of a broad spectrum of human diseases that are attributed to respiratory incompetent mitochondria in various degenerative diseases (e.g., metabolic syndrome and diabetes). For example, as K-Ras 4A is responsible for aldosterone signaling,40 the roles of mtDNA changes on hypertension is implicated.
In conclusion, our report has identified an intricate interaction of prospective (mitochondrial and nuclear encoded) markers for the selective penetrance, progression and prognosis of PCa, and potentially a diverse array of respiratory-deficient (e.g., aging) pathologies.
Materials and Methods
Materials
Uridine, sodium pyruvate, rotenone, antimycin A, oligomycin A, mevalonate and FTase-276 were purchased from Sigma-Aldrich (St. Louis, MO, USA).; Lovastatin from A.G. Scientific (San Diego, CA, USA) and ALLN from Fisher Scientific (Pittsburgh, PA, USA).
Cell culture
LNCaP and C4-2 were purchased from UROCOR (Minnetonka, MN, USA); MCF-7, PC-3 and DU-145 were purchased from ATCC (Manassas, VA, USA); and, PrEC was purchased from Lonza (Allendale, NJ, USA). PC-3, DU-145, LNCyb, MCF-7 and MCFCyb were cultured in Dulbecco's modified Eagle medium with a high level of D-glucose (4.5 g/l) and sodium pyruvate (110 mg/l) (Invitrogen, Allendale, NJ, USA), additionally supplemented with 5% fetal calf serum (FCS, Thermo Scientific, Barrington, IL, USA). PrEC was cultured in prostate epithelial cell basal media (PrEBM, Lonza) supplemented with PrEGM SingleQuots: BPE, insulin, HC, GA-1000, retinoic acid, transferrin, T3, epinephrine and rhEGF (no. CC-4177, Lonza). Media for LNρ0-8 and MCFρ0 was additionally supplemented with uridine (50 μg/ml) and sodium pyruvate (100 μg/ml). Media for BCa cells was additionally supplemented with bovine insulin (0.01 mg/ml) (Sigma-Aldrich).
Microdissection
Samples and matched clinical information were obtained from the Division of Urology at McClellan's Memorial Veteran's Hospital (Little Rock, AR, USA). All patient-derived biological specimens were collected and archived under protocols approved by the Human Subjects Committee of the University of Arkansas for Medical Science Institutional Review Board (UAMS IRB no. 55578). In all, 14 PCa patients participated in the study (age ranging from 49–73 years; average 61.7 years). According to the clinical information available, none of these individuals had any medical condition or had any treatment known to affect the prostate. In all, 14 PCa tissues were collected by radical prostatectomy specimens. The cancers ranged from Gleason grade 3 + 3, 3 + 4, 4 + 3, 4 + 4, 4 + 5 or 5 + 4. Fresh specimens were frozen at −80 °C and sectioned by cryostat to yield 8 μm sections. Each section was fixed in methanol and stained in Mayer's hematoxylin. Before microdissection, cut sections were dehydrated in ethanol. An expert pathologist selected cancer and normal epithelial regions, and then individual cells were collected by laser microdissection using the Leica AS LMD system (Leica Microsystems, Richmond, IL, USA). The dissected cells were transferred by gravity alone into a microcentrifuge tube placed directly underneath the tissue section. The tube was filled with digestion buffer (20 μl) containing SDS (0.0015%), proteinase K (0.2 mg/ml) and EDTA (4.5 mM) to ensure the isolation of DNA. After incubation (55 °C for 3 h), the samples were boiled (95 °C for 10 min) to inactivate proteinase K.
FISH of mitochondrial genome
Alexa-488 dye labeled mtDNA-probes (∼600 bp) were created from six overlapping PCR products (∼3.3 kb in length) that collectively represent the entire human mitochondrial genome were prepared. An equal amount of PCR product from each reaction were combined, and the pooled products were nick-translated and labeled with the Alexa-488 dye as instructed in the ARES Alexa-488 DNA Labeling Kit (Invitrogen).
Cells were seeded (60% confluence) on submerged cover slips. To counter stain the mitochondria, cell cultures were incubated (20 min at 37 °C) in MitoTracker Red dye (300 nM) (Molecular Probes). Cells were then washed in PBS, and fixed (10 min at 37 °C) in 4% formaldehyde (Ted Pella, Redding, CA, USA). Fixed cells were washed in PBS, and permeabilized (5 min at 25 °C) in 0.1% Triton X-100 (Sigma-Aldrich). Cells were then washed in PBS, and treated (90 min at 37 °C) with 0.1 mg/ml DNase-free RNase (Roche Diagnostics, Indianapolis, IN, USA). Cells were then washed with PBS; meanwhile, the mtDNA probes (0.5 ng/ml) were denatured (10 min at 74 °C) in formamide (50%), dextran sulfate (10%) and 2 × saline sodium citrate (SSC) buffer. Cover slips were mounted with denatured probes (15 μl) onto pre-warmed slides (74 °C). Cover slips and the mtDNA probes were sealed to the slides with rubber cement and left overnight in a dark humidified chamber (37 °C) to hybridize. After hybridization the cover slips were removed from the slides and washed in 2 × SSC buffer at 25 °C, followed by sequential washes in 2 × SSC and 0.5 × SSC buffer at 40 °C. Cover slips were then rinsed in PBS and mounted to slides by ProLong antifade reagent with or without 4′, 6-diamidino-2-phenylindole dihydrochloride (DAPI) (Invitrogen).
Wide field fluorescence microscopy
Wide field microscopy imaging was performed with a Zeiss Axioskop 40 microscope (Carl Zeiss, Thornwood, NY, USA) fitted with a × 40/0.75 NA objective. Images were captured with a Zeiss AxioCam MRm camera, and collected using AXIOVISION software (Carl Zeiss). Matched image sets were mapped to a common linear gray-scale range. Images were prepared for publication with ADOBE PHOTOSHOP software (Adobe Systems, San Jose, CA, USA).
Spinning disk fluorescence microscopy
Confocal imaging was performed with a Zeiss Axiovert 200 M microscope (Carl Zeiss) fitted with a × 100/1.40 NA objective and a CARV II spinning disk confocal accessory (BD Bioimaging, RockVille, MA, USA) mounted to the sideport of the microscope. Images were captured with a Retiga EXi camera (Qlmaging, Surey, BC, Canada). The microscope was operated in multitrack configuration with alternative excitation, as controlled with IVISION MAC (BioVision Technologies, Exton, PA, USA). Image stacks were collected through the entire cell depth, and then compressed into a single plane using maximum intensity projection and IVISION (Atlanta, GA, USA) software. When noted, a single z-slice (plane) was captured using a × 100/1.40 NA objective. Matched image sets were mapped to a common linear gray-scale range. Images were prepared for publication with ADOBE PHOTOSHOP software (Adobe Systems).
SDS-PAGE and western blotting
Cells were lysed in radioimmunoprecipitation assay buffer supplemented with DTT (0.5 mM), and protease and phosphatase inhibitors (100 × Halt protease and phosphate inhibitor cocktail, no. 1861281, Thermo Scientific). Cytosolic extracts were isolated by centrifugation (12 000 g for 15 min at 4 °C), and the protein concentration was determined by a protein assay (Bio-Rad Protein Assay, Bio-Rad, Hercules, CA, USA). Total-protein lysates were suspended in reducing SDS gel-loading buffer, denatured (95 °C for 5 min) and subjected to electrophorectic separation in polyacrylamide slab gels in the presence of SDS (i.e., SDS-PAGE), and then transferred to immobilon-P membrane (Millipore, Bellelica, MA, USA). After a wash in Tris Buffered Solution (TBS), the membrane was incubated overnight (4 °C) in primary antibody diluted (1 : 1000) in bovine serum albumin (5% BSA, Sigma-Aldrich). Antibodies utilized in the current study are as follows: rabbit anti-phospho ERK(T202/Y204) (no. 9101), rabbit anti-total ERK (no. 9102), rabbit anti-phospho Akt(Ser473) (no. 9271), rabbit anti-total Akt (no. 9272), rabbit anti-β-actin (no. 4967, Cell Signaling, Beverly, MA, USA), mouse anti-total Ras (clone RAS10, no. 05-516, Millipore), rabbit anti-H-Ras (C-20, no. sc-520), rabbit anti-K-Ras 4A (C-17, no. sc-522), rabbit anti-K-Ras 4B (C-19, no. sc-521), rabbit anti-N-Ras (C-20, no. sc-519, Santa Cruz Biotechnology, Santa Cruz, CA, USA), rabbit anti-total DAB2IP (no. A302-439A, Bethyl Laboratories, Montgomery, TX, USA), rabbit anti-HMGR reductase (no. 07-457, Millipore), and mouse anti-GAPDH (no. MAB374, Millipore). The membrane was then washed in TBS, and incubated (1 h at RT) with HRP-linked anti-mouse IgG or anti-rabbit IgG diluted (1 : 10 000) in 5% BSA-TBS (no. 7074 and no. 7076, respectively, Cell Signaling). The membrane was then washed in TBS, and analyzed with ECL Plus (GE Healthcare Bio-Sciences AB, Uppsala, Sweden) by autoradiography film.
Ras activation (Pull Down) assay
Activated (GTP-bound) Ras was pulled down from total-protein lysates of cell cultures by a Ras Activation Assay kit (Kit no. 17-218, Millipore). Briefly, an agarose-bound GST fusion protein corresponding to the RBD (residues 1–149) of human c-Raf was utilized to bind and precipitate GTP-bound Ras from total-protein lysates. As a negative control, before pull-down an aliquot of total protein lysate was pre-loaded with GDP to deactivate (GTP-bound) Ras to the GDP-bound state. As a positive control, before pull-down an aliquot of total protein lysate was pre-loaded with nonhydrolyzable GTPγS to active (GDP-bound) Ras to the GTP-bound state. Protein levels of pulled down Ras in samples were analyzed by SDS-PAGE and western blot.
Genomic sequencing of K-RAS
A DNA extraction kit (ZR Genomic DNA II Kit, Zymo Research, Irvine, CA, USA) was used to prepare genomic DNA. Primers were custom synthesized (Integrated DNA Technologies, Coralville, IA, USA) to amplify the K-RAS gene in exon 1 (codon 12 and 163 bp) and in exon 2 (codon 61, 133 bp): exon 1: exon 1 (sense 5′-GGCCTGCTGAAAATGACTGAA-3′ and anti-sense 5′-GGTCCTGCACCAGTAATATGC-3′) and exon 2 (sense 5′-CAGGATTCCTACAGGAAGCAAGTA-3′ and anti-sense 5′-CACAAAGAAAGCCCTCCCCA-3′). PCR was carried out with La Taq DNA polymerase (TaKaRa, Shiga, Japan): 94 °C for 1 min; 30 sequential cycles of 94 °C for 15 s, 55 °C (exon 1) or 60 °C (exon 2) for 30 s, and 72 °C for 10 s; with a final elongation at 72 °C for 1 min. PCR products were loaded onto an agarose gel (2%) to confirm a single target band. The target band was cut out and purified using the Wizard PCR Preps DNA Purification System (Promega, Madison, WI, USA), then subjected to direct sequencing with a 3100 Genetic Analyzer (Applied Biosystems, Carlsbad, CA, USA; UAMS DNA Sequencing Core Facility, Little Rock, AR, USA).
Measurement of oxygen consumption
Cells (1 × 106) were suspended in 1 ml of culture medium pre-equilibrated to 20% oxygen and then placed in a sealed respiration chamber equipped with a thermostat control and a micro-stirring device (Oxytherm, Hansatech Instrument, King's Lynn, Norfolk, UK). The oxygen contents in the starting medium were normalized assuming an O2 concentration of 217 μM in air-saturated medium at 37 °C (n=3).
Oxygen concentration measurements by OxoPlate
The OxoPlate OP96U (PreSens) contains oxygen-sensitive, polystyrene particles PSLi-Pt-1 (Opto-Sense). Sulforhodamin is covalently attached to these particles as reference dye, and a platinum porphin is incorporated as indicator dye. The sensor has a thickness of about 10 μm and is fixed at the bottom of each well of a 96-round bottom microtiter plate (Greiner, Monroe, NC, USA). Cells (200 μl) were added to each well at the indicated cell density. Wells containing oxygen-free water (cal 0) and air-saturated water (cal 100) served as standards. Oxygen-free water was prepared by dissolving sodium sulfite (1%) in water; in the wells, the oxygen-free water was overlaid with mineral oil (50 μl) to prevent the diffusion of oxygen into the wells. The oxygen concentration (fluorescence) in each well was quantified for 3 h in 5-min intervals, as measured in a dual kinetic mode (Synergy HT, BIO-TEK, Einooski, VT, USA): filter pair 1 (544/650 nm) detects fluorescence of the indicator dye; the second measures fluorescence (544/590 nm) of the reference dye (n=3).
Calculation of oxygen concentration
The relationship between fluorescence and dissolved oxygen is nonlinear, as described by the Stern–Volmer equation:

where, IR,0 is the fluorescence intensity ratio in the absence of oxygen; IR is the fluorescence intensity ratio at the oxygen concentration [O2], and KSV the Stern–Volmer parameter. Intensity ratios IR are calculated for each individual well by dividing the intensity of the indicator dye by the intensity of the reference dye. A two-point calibration is sufficient: [O2]=0 and at [O2]=[O2]*, where [O2]* is the saturation concentration.
Oxygen concentration as a percentage of air saturation was calculated for each well, as follows:

where the constant K0 is defined as IR of well filled with cal 0. Analogously, K100 is defined as IR of well filled with cal 100.
Culture of cells in high or low oxygen condition
LNCaP (0.25 × 106 cells) and LNρ0-8 (1 × 106 cells) were seeded in a 6-cm culture dish overnight in a humidified atmosphere of 5% CO2 in an incubator. Increased and reduced oxygen incubations were carried out in a C-chamber (no. C-174, BioSpherix, Lacona, NY, USA), regulated by a PRO-OX 110 oxygen controller (Biospherix) to establish the indicated oxygen concentrations. (DMEM +5% FCS, 50 mM HEPES) and Additional Gases (nexAir: Nitrogen Liquid Medical Grade LS180 ltrs 22psi (no. LQ NI-NF-LS180) (Santa Cruiz, CA) and Oxygen Medical Grade 200+cft (no. HP OX-USP-2)).
QRT-PCR of the mitochondrial genome content
The mitochondrial genome content of individual cells was quantified by QRT-PCR as previously described.39
RT-PCR of RAS
Total-RNA was isolated with a Qiagen RNA-Plus Mini Easy kit (no. 74134, Qiagen , Valencia, CA, USA), subjected (2 μg) to a gDNA eliminator spin column, primed with oligo (dT)18−20 (Invitrogen), and reverse transcribed by Superscipt III Reverse Transcriptase (Invitrogen). Primers were custom synthesized (Integrated DNA Technologies) to amplify the cDNA for H-RAS (111 bp), K-RAS 4A (560 bp), K-RAS 4B (711 bp), N-RAS (682 bp), HMGR (280 bp) and GAPDH (89 bp): H-RAS (sense 5′-ATGACGGAATATAAGCTGGT-3′ and anti-sense 5′-CTCTATAGTGGGGTC GTATT-3′); K-RAS 4A (sense 5′-GAGAGAGGCCTGCTGAAAATG-3′ and anti-sense 5′-ACACAGCCAGGAGTCTTTTCTTC-3′); K-RAS 4B (sense 5′-AGCGCGGCGCAGGCACTGAA-3′ and anti-sense 5′-TTAGTGTAATGTACAAAAATTACCA-3′); N-RAS (sense 5′-CTGTGGTCCTAAATCT;GTCC-3′ and anti-sense 5′-CAGTGCAGCTTGAAAGTG -3′); HMGR (sense 5′-TGCTGCTTTGGCTGTATGTC-3′ and anti-sense 5′-TGAGCGTGAACAAGAACCAG-3′); GAPDH (sense 5′-GGTGGTCTCCTCTGACTTCAA-3′ and anti-sense 5′-AGCTTGACAAAAGTGGTCGTTG-3′). The entire coding sequence of GAPDH cDNA was amplified as a control. Ex Taq (TaKaRa) was used for the amplification of Ras cDNA or HMGR cDNA from total-cDNA, 50 ng or 200 ng, respectively. The PCR cycle to amplify target cDNA is as follows: 94 °C for 1 min, followed by 20 (Ras) or 35 (HMGR/GAPDH) sequential cycles of 94 °C for 15 s, the denoted annealing temperature (as follows) for 1 min, and 72 °C for 1 min; with a final elongation at 72 °C for 5 min. Annealing temperatures for primers: H-RAS (56 °C); K-RAS 4A (50 °C); K-RAS 4B (56 °C); N-Ras (58 °C); HMGR (58 °C); GAPDH (58 °C). PCR products were separated in agarose gels (RAS cDNA, except H-RAS 1.2%; HMGR 1.5%; H-RAS and GAPDH 2%) supplemented with ethidium bromide (1 : 20 000), and then visualized by a UV transilluminator (UVP, Upland, CA, USA).
K-RAS siRNA
Stealth siRNA duplex oligonucleotides against K-RAS (KRASHSS180200) were custom synthesized (Invitrogen). The sequences were as follows: (RNA)-UGUGGUAGUUGGAGCUGGUGGCGUA and UACGCCACCAGCUCCAACUACCACA. LNρ0-8, seeded at 30% confluence in a 25 cm2 culture flask overnight, were transfected with 100 ng of K-RAS siRNA or random siRNA (no. 12935-200 (Low GC), Invitrogen), using 10 μg of Lipofectamine 2000 (Invitrogen) as directed by the manufacturer's protocol. Protein lysates were collected 72 h post-transfection.
Immunocytochemistry of anti-total-Ras
Cells were seeded (60% confluence) on submerged cover slips. Cover slips were washed with PBS, then fixed and permeabilized as previously described. Permeabilized cells were blocked (1 h) in 5% normal goat serum (Vector Laboratories, Burlingame, CA, USA). Ras was probed with anti-total-Ras diluted (1 : 1000) in blocking solution overnight at 4 °C, and then washed in PBS. Alexa-488 anti-mouse IgG (Invitrogen) was applied (1 h at RT) to the cover slips (1 : 500) in blocking solution. Cover slips were then washed in PBS and mounted on top of ProLong antifade reagent with or without DAPI (Molecular Probes).
Detection of cell viability by microscopic analysis
Cells were cultured (2000 cells per 200 μl/well) in a 96 well flat-bottomed microtiter plate. Phase-contrast image analysis was conducted by a Fisher Scientific MICROMASTER INVERTED microscope fitted with a × 40/1.5 NA objective. Image were collected and prepared for publication with ADOBE PHOTOSHOP software (Adobe Systems).
Assay of cell viability by calcein
Cells were cultured (2000 cells per 200 μl/well) in a 96-well flat-bottomed microtiter plate (Fisher Scientific) that was centrifuged (250 g for 5 min at 4 °C) to avoid the loss of cells in the staining process. The media was removed (170 μl) and PBS (100 μl) was added to each well, followed by the addition of Calcein-AM working solution (10 μl) (EMD Chemicals, Philadelphia, PA, USA). The plate was then incubated (37 °C for 1 h) in a CO2 incubator. The fluorescence of the samples was measured by a microtiter plate spectrophotometer (BioTek Synergy HT plate reader, BIO-TEK) (excitation 485±10 nm and emission 520±10 nm). All cultures were performed in triplicate (n=3) and the mean was displayed as an arbitrary unit (A.U.)±S.E.
Assay of cell viability by crystal violet
Cells were cultured in a 96-well flat-bottomed microtiter plate (2000 cells per 200 μl/well). Subsequently, the supernatants were discarded and the remaining viable adherent cells were stained (5 min) with crystal violet (0.2%) in 20% ethanol. The microtiter plate was then rinsed with water and solubilization solution (100 μl of H2O : methanol : ethanol=5 : 1 : 4) was added to each well. The absorbance of each well was read at 590 nm with a microtiter plate spectrophotometer. All cultures were performed in triplicate (n=3) and results were expressed by mean±S.E.
Animal studies
Athymic BALB/c (nu/nu) mice (Clea Japan Inc., Tokyo, Japan), 4 weeks of age, were subcutaneously inoculated with LNCaP cells (5.0 × 106 cells per 50 μl of medium). Following the development of tumors with diameters of 7–8 mm, 24 mice were randomly assigned to four experimental groups as follows: group 1, isotonic saline (control) (6 mice); group 2, lovastatin (5 mg/kg/day) (6 mice); group 3, mevalonate (36 μmol/day) (6 mice); and group 4, lovastatin (5 mg/kg/day) and mevalonate (36 μmol/day) (6 mice). 200-μl miniosmotic pumps (flow rate 0.25 μl/h, 28-days pumping life) (Alzet Corp., Palo Alto, CA, USA) were subcutaneously implanted in the intrascapular area of all mice at Day 0. Pumps delivered either mevalonate (group 2 and group 4) or isotonic saline (group 1 and group 3) from day 0 to day 28. The diet contain an additional lovastatin were fed to mice of group 2 and group 4 from day 0 to day 28. The tumor volumes were measured weekly for 4 weeks and calculated by using the following formula: volume (a rotational ellipsoid)=M1 × M22 × 0.5236, where M1=the long axis and M2=the short axis. The relative tumor volume was calculated by assuming the rate of each tumor volume on day 0 to be 100%. Experiments in each group were performed in six plicate and results were expressed by mean±S.D. All the aspects of the experimental design and procedure were reviewed and approved by the institutional ethics and animal welfare committees of The Hyogo University of Health Sciences (2009-19-1).
Abbreviations
- HMGR:
-
3-hydroxy-3-methyl-glutaryl-CoA reductase
- nDNA:
-
nuclear DNA
- mtDNA:
-
mitochondrial DNA
- PCa:
-
prostate cancer
- MRC:
-
mitochondrial respiratory chains
- FISH:
-
fluorescent in situ hybridization
- DAB2IP:
-
DOC-2/DAB2 interactive protein
- PrEC:
-
prostate epithelial cells
- FPP:
-
Farnesyl pyrophosphate
- FTase:
-
farnesyltransferase
- FTI:
-
farnesyltransferase inhibitor
- ALLN:
-
N-[N-(N-Acetyl-L-leucyl)-L-leucyl]-L-norleucine
- mitoGPS:
-
mitochondrial-generated progression signal
References
American Cancer Society. Cancer Facts & Figures. ACS: Atlanta, GA, 2009.
Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J et al. Sequence and organization of the human mitochondrial genome. Nature 1981; 290: 457–465.
Carew JS, Huang P . Mitochondrial defects in cancer. Molecular Cancer 2002; 1: 1–12.
Ishikawa K, Takenaga K, Akimoto M, Koshikawa N, Yamaguchi A, Imanishi H et al. ROS-generating mitochondrial DNA mutations can regulate tumor cell metastasis. Science 2008; 320: 661–664.
Chen JZ, Gokden N, Greene GF, Mukunyadzi P, Kadlubar FF . Extensive somatic mitochondrial mutations in primary prostate cancer using laser capture microdissection. Cancer Res 2002; 62: 6470–6474.
Barthelemy C, de Baulny HO, Lombes A . D-loop mutations in mitochondrial DNA: link with mitochondrial DNA depletion? Human Genetics 2002; 110: 479–487.
Chandel NS, Schumacker PT . Cells depleted of mitochondrial DNA (rho0) yield insight into physiological mechanisms. FEBS Letters 1999; 454: 173–176.
Higuchi M, Aggarwal BB, Yeh ET . Activation of CPP32-like protease in tumor necrosis factor-induced apoptosis is dependent on mitochondrial function. J Clin Invest 1997; 99: 1751–1758.
Higuchi M, Kudo T, Suzuki S, Evans TT, Sasaki R, Wada Y et al. Mitochondrial DNA determines androgen dependence in prostate cancer cell lines. Oncogene 2006; 25: 1437–1445.
Naito A, Cook CC, Mizumachi T, Wang M, Xie CH, Evans TT et al. Progressive tumor features accompany epithelial-mesenchymal transition induced in mitochondrial DNA-depleted cells. Cancer Science 2008; 99: 1584–1588.
Xie CH, Naito A, Mizumachi T, Evans TT, Douglas MG, Cooney CA et al. Mitochondrial regulation of cancer associated nuclear DNA methylation. Biochem Biophys Res Commun 2007; 364: 656–661.
Nishimura G, Proske RJ, Doyama H, Higuchi M . Regulation of apoptosis by respiration: cytochrome c release by respiratory substrates. FEBS Lett 2001; 505: 399–404.
Suzuki S, Naito A, Asano T, Evans T, Reddy S, Higuchi M . Constitutive Activation of AKT Pathway Inhibits TNF-induced Apoptosis in Mitochondrial DNA-Deficient human myelogenous leukemia ML-1a. Cancer Lett 2008; 268: 31–37.
McCubrey JA, Steelman LS, Chappell WH, Abrams SL, Wong EW, Chang F et al. Roles of the Raf/MEK/ERK pathway in cell growth, malignant transformation and drug resistance. Biochem Biophys Acta 2007; 1773: 1263–1284.
Gioeli D, Mandell JW, Petroni GR, Frierson Jr HF, Weber MJ . Activation of mitogen-activated protein kinase associated with prostate cancer progression. Cancer Res 1999; 59: 279–284.
Gao H, Ouyang X, Banach-Petrosky WA, Gerald WL, Shen MM, Abate-Shen C . Combinatorial activities of Akt and B-Raf/Erk signaling in a mouse model of androgen-independent prostate cancer. Proc Natl Acad Sci USA 2006; 103: 14477–14482.
Bakin RE, Gioeli D, Bissonette EA, Weber MJ . Attenuation of Ras signaling restores androgen sensitivity to hormone-refractory C4-2 prostate cancer cells. Cancer Res 2003; 63: 1975–80.
Horoszewicz JS, Leong SS, Chu TM, Wajsman ZL, Friedman M, Papsidero L et al. The LNCaP cell line – a new model for studies on human prostatic carcinoma. Prog Clin Biol Res 1980; 37: 115–132.
Wu HC, Hsieh JT, Gleave ME, Brown NM, Pathak S, Chung LW . Derivation of androgen-independent human LNCaP prostatic cancer cell sublines: role of bone stromal cells. Int J Cancer 1994; 57: 406–412.
Kaighn ME, Narayan KS, Ohnuki Y, Lechner JF, Jones LW . Establishment and characterization of a human prostatic carcinoma cell line (PC-3). Invest Urol 1979; 17: 16–23.
Stone KR, Mickey DD, Wunderli H, Mickey GH, Paulson DF . Isolation of a human prostate carcinoma cell line (DU 145). Int J Cancer 1978; 21: 274–281.
Soule HD, Vazguez J, Long A, Albert S, Brennan M . A human cell line from a pleural effusion derived from a breast carcinoma. J Natl Cancer Inst 1973; 51: 1409–1416.
Zhang XF, Settleman J, Kyriakis JM, Takeuchi-Suzuki E, Elledge SJ, Marshall MS et al. Normal and oncogenic p21ras proteins bind to the amino-terminal regulatory domain of c-Raf-1. Nature 1993; 364: 308–313.
Minden A, Lin A, McMahon M, Lange-Carter C, Derijard B, Davis RJ et al. Differential activation of ERK and JNK mitogen-activated protein kinases by Raf-1 and MEKK. Science 1994; 266: 1719–1723.
Rodriguez-Viciana P, Warne PH, Dhand R, Vanhaesebroeck B, Gout I, Fry MJ et al. Phosphatidylinositol-3-OH kinase as a direct target of Ras. Nature 1994; 370: 527–532.
Dote H, Toyooka S, Tsukuda K, Yano M, Ouchida M, Doihara H et al. Aberrant promoter methylation in human DAB2 interactive protein (hDAB2IP) gene in breast cancer. Clin Cancer Res 2004; 10: 2082–2089.
Min J, Zaslavsky A, Fedele G, McLaughlin SK, Reczek EE, De Raedt T et al. An oncogene-tumor suppressor cascade drives metastatic prostate cancer by coordinately activating Ras and nuclear factor-kappaB. Nat Med 2010; 16: 286–294.
Jancik S, Drabek J, Radzioch D, Hajduch M . Clinical relevance of KRAS in human cancers. J Biomed Biotechnol 2010; 2010: 150960.
Konstantinopoulos PA, Karamouzis MV, AG . Post-translational modifications and regulation of the RAS superfamily of GTPases as anticancer targets. Nature Reviews. Drug Discovery 2007; 6: 541–555.
Willumsen BM, Christensen A, Hubbert NL, Papageorge AG, Lowy DR . The p21 ras C-terminus is required for transformation and membrane association. Nature 1984; 310: 583–586.
Manne V, Roberts D, Tobin A, O’Rourke E, De Virgilio M, Meyers C et al. Identification and preliminary characterization of protein-cysteine farnesyltransferase. Proc Natl Acad Sci USA 1990; 87: 7541–7545.
Reiss Y, Goldstein JL, Seabra MC, Casey PJ, Brown MS . Inhibition of purified p21ras farnesyl:protein transferase by Cys-AAX tetrapeptides. Cell 1990; 62: 81–88.
Chang TY, Chang CC, Ohgami N, Yamauchi Y . Cholesterol sensing, trafficking, and esterification. Annu Rev Cell Dev Biol 2006; 22: 129–157.
Song BL, Javitt NB, DeBose-Boyd RA . Insig-mediated degradation of HMG CoA reductase stimulated by lanosterol, an intermediate in the synthesis of cholesterol. Cell Metabolism 2005; 1: 179–189.
Nguyen AD, McDonald JG, Bruick RK, DeBose-Boyd RA . Hypoxia stimulates degradation of 3-hydroxy-3-methylglutaryl-coenzyme A reductase through accumulation of lanosterol and hypoxia-inducible factor-mediated induction of insigs. J Biol Chem 2007; 282: 27436–27446.
Igawa T, Lin FF, Lee MS, Karan D, Batra SK, Lin MF . Establishment and characterization of androgen-independent human prostate cancer LNCaP cell model. Prostate 2002; 50: 222–235.
Thalmann GN, Sikes RA, Wu TT, Degeorges A, Chang SM, Ozen M et al. LNCaP progression model of human prostate cancer: androgen-independence and osseous metastasis. Prostate 2000; 44: 91–103.
Higuchi M, Manna SK, Sasaki R, Aggarwal BB . Regulation of the activation of nuclear factor kappaB by mitochondrial respiratory function: evidence for the reactive oxygen species-dependent and -independent pathways. Antioxidants Redox Signaling 2002; 4: 945–955.
Mizumachi T, Muskhelishvili L, Naito A, Furusawa J, Fan CY, Siegel ER et al. Increased distributional variance of mitochondrial DNA content associated with prostate cancer cells as compared with normal prostate cells. Prostate 2008; 68: 408–417.
Terada Y, Kobayashi T, Kuwana H, Tanaka H, Inoshita S, Kuwahara M et al. Aldosterone stimulates proliferation of mesangial cells by activating mitogen-activated protein kinase 1/2, cyclin D1, and cyclin A. J Am Soc Nephrol 2005; 16: 2296–2305.
Acknowledgements
This work was supported by Taiho Pharmaceutical Co. Ltd, State of Arkansas Tobacco Settlement, NIH grant CA100846 to MH and DOD PCRP Prostate Cancer Training Award to CCC. We thank Drs. Kiyoshi Kita, Michael Douglas and Yung C Lin for reviewing the manuscript. We also are thankful for the technical assistance of Drs. Takatsugu Mizumachi, Dwight Pierce, Kim Light and Brian Storrie.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by A Finazzi-Agró
Rights and permissions
This work is licensed under the Creative Commons Attribution-NonCommercial-No Derivative Works 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Cook, C., Kim, A., Terao, S. et al. Consumption of oxygen: a mitochondrial-generated progression signal of advanced cancer. Cell Death Dis 3, e258 (2012). https://doi.org/10.1038/cddis.2011.141
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cddis.2011.141
Keywords
This article is cited by
-
Age Structure Can Account for Delayed Logistic Proliferation of Scratch Assays
Bulletin of Mathematical Biology (2019)
-
Reduction–oxidation (redox) system in radiation-induced normal tissue injury: molecular mechanisms and implications in radiation therapeutics
Clinical and Translational Oncology (2018)
-
Molecular and functional heterogeneity of early postnatal porcine satellite cell populations is associated with bioenergetic profile
Scientific Reports (2017)
-
Intracellular oxygen determined by respiration regulates localization of Ras and prenylated proteins
Cell Death & Disease (2015)
-
mtDNA haplogroup and single nucleotide polymorphisms structure human microbiome communities
BMC Genomics (2014)