Abstract
Apoptosis research has been significantly aided by the generation of antibodies against caspase-cleaved peptide neo-epitopes. However, most of these antibodies recognize the N-terminal fragment and are specific for the protein in question. The aim of this project was to create antibodies, which could identify caspase-cleaved proteins without a priori knowledge of the cleavage sites or even the proteins themselves. We hypothesized that many caspase-cleavage products might have a common antigenic shape, given that they must all fit into the same active site of caspases. Rabbits were immunized with the eight most prevalent exposed C-terminal tetrapeptide sequences following caspase cleavage. After purification of the antibodies we demonstrated (1) their specificity for exposed C-terminal (but not internal) peptides, (2) their ability to detect known caspase-cleaved proteins from apoptotic cell lysates or supernatants from apoptotic cell culture and (3) their ability to detect a caspase-cleaved protein whose tetrapeptide sequence differs from the eight tetrapeptides used to generate the antibodies. These antibodies have the potential to identify novel neo-epitopes produced by caspase cleavage and so can be used to identify pathway-specific caspase cleavage events in a specific cell type. Additionally this methodology may be applied to generate antibodies against products of other proteases, which have a well-defined and non-promiscuous cleavage activity.
Similar content being viewed by others
Main
Clinically translatable biomarkers of apoptosis would be beneficial in multiple disease areas, not the least of which is cancer therapeutics. Many chemotherapeutics or biologics ultimately induce apoptosis as their mode of action.1, 2 Thus, the ability to monitor this increased apoptosis in patients would enable an earlier determination of the success (or failure) of a particular treatment. Likewise, from a drug development perspective, earlier readouts of efficacy will improve costs and enable more nimble decision making.3, 4
There are several desired characteristics of an apoptotic biomarker, especially for those with clinical translatability.5 The constraint of non-invasiveness is a high bar over which many biomarkers do not pass; tissues or organs that may be obtained in preclinical rodent models of disease are simply not accessible in patients in the clinic. This immediately narrows down the possibilities for biomarkers into two realms: imaging modalities or those measurable from easy-to-collect biological specimens such as blood, urine, saliva, skin or hair. Another concern is the stability of the analyte; the utility of a biomarker is severely hampered if it is so short lived that special measures must be employed for collection: it must be robust enough to be performed by multiple users, as in a multi-center study. A low background level of signal is another highly desirable characteristic of biomarkers. Although some caspase activity occurs in healthy organisms, the overall levels of apoptosis in a healthy organism compared with the levels induced by chemotherapy should be far above this background, leading to a satisfactory window of signal to noise. Lastly, the very nature of apoptosis affords it an advantage not conferred to all biomarkers: a level of ‘universality’; in that the core mechanism of apoptosis is largely cell-type independent. This allows an apoptotic biomarker to have utility in a wide range of disease types.
Caspase-cleavage products (CCPs) embody many of the characteristics of a desired biomarker: increases in apoptosis should cause elevations in caspase activity leading to increased levels of CCPs; moreover if a CCP is serum- or urine-stable it could be clinically translatable. An advantage of detecting CCPs as a biomarker of apoptosis is specificity: caspase cleavage of a protein creates a neo-epitope that would primarily be present only under apoptotic conditions, as executioner caspases generally are inactive under non-apoptotic conditions. Any proteins detected by such neo-epitope antibodies (NEAs) can reasonably be assumed to have only been formed during apoptosis and thus the levels of such CCPs would correlate with the amount of apoptosis. One such example of a CCP that is currently being utilized as a biomarker of apoptosis is caspase-cleaved cytokeratin 18, marketed as the Apoptosense ELISA from Peviva (Bromma, Sweden) (see, for example, Brandt et al.6). The aim of the present study was to develop antibodies that could identify CCPs without a priori knowledge of the cleavage sites or even the proteins themselves. We describe the successful development and validation of such antibodies.
Results
Immunization strategy
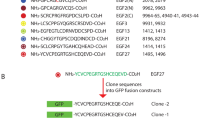
We hypothesized that many CCPs might have a common antigenic shape, given that they must all fit into the same active site of execution-phase caspases. To obtain the broadest coverage of neo-epitope sites, we utilized an immunization scheme wherein rabbits were injected with a cocktail of peptides containing the eight most prevalent c-terminally exposed (after caspase cleavage) tetrapeptide sequences, collectively called ‘DXXD’ (Figure 1a). Each of the eight peptides was coupled to two different linker sequences (designated Scramble 1 and Scramble 2), chosen to closely resemble native rabbit peptides in order to minimize generation of antibodies to these irrelevant parts of the immunogen. The cohort of rabbits immunized with the mixture of eight Scramble 1 peptides were boosted once with the Scramble 2 peptides, and vise versa, such that the only commonality between immunization and boost was the c-terminal DXXD sequences (Figure 1b).
Strategy of immunization. (a) In all, 262 caspase-cleavage sites were analyzed to determine the most prevalent C-terminally exposed (after caspase cleavage) tetrapeptide sequences. Based on this analysis, the top eight were chosen. The tetrapeptides were joined to two different eight amino-acids ‘linker'sequences (Scramble 1 and Scramble 2), chosen to closely resemble native rabbit peptides in order to minimize generation of antibodies to these irrelevant parts of the peptide. (b) The cohort of rabbits immunized with the mixture of eight Scramble 1 peptides were boosted once with the Scramble 2 peptides, and vise versa, such that the only commonality between immunization and boost was the C-terminal DXXD sequences. Scramble 3 was utilized during purification to decrease the isolation of irrelevant antibodies to the other linker sequences
Purified antibodies are specific to the antigen peptides
The antibody purification strategy, outlined in Figure 2a, employed both conventional and affinity chromatography. Following ammonium sulfate precipitation of serum and DEAE ion-exchange column, the flow-through from the DEAE column was applied to the sulfolink affinity resin conjugated with the eight peptides (Scramble 3-DXXD). Each of these eight peptides contains 12 amino acids, comprising a ‘DXXD’ tetrapeptide used for the immunization, and an octapeptide (Scramble 3) never seen by the host animals. Thus the antibodies purified would be those specific to the caspase substrate tetrapeptides, while the antibodies recognizing linkers Scramble 1 or 2, or the carrier protein OMPC, would not bind to the resin. The purification progress of the antibodies was visualized with SDS-PAGE and western blot (Figure 2b). To verify that the purified antibodies specifically recognized ‘DXXD’, direct ELISAs were performed. As shown in Figure 2c, the purified antibodies had high binding affinity to all the eight Scramble 3-DXXD peptides. Much lower binding affinity was observed when tested with similar peptides ending with ‘DXXDZZ’ (in this study ZZ represents two random amino acids except C, D, E and M), indicating that these antibodies specifically recognize the ‘DXXD’ at the end rather than in the middle of the peptides. In other words, these antibodies are potentially selective for cleaved caspase substrates but will not recognize DXXD motifs in the intact (uncleaved) protein. Additionally, it was confirmed by ELISA that the purified antibodies showed little or no binding to the sequences found in Scramble 1 or 2, or OMPC (Figure 2d).
Purification of antibodies. (a) Purification scheme. The serum was fractionated with 50% ammonium sulfate and subsequent DEAE column as described in ‘Materials and Methods’. The flow-through from the DEAE column was further purified with eight peptides (Scramble 3-DXXD) conjugated to sulfolink affinity resin. The eluate was collected and the titer of antibodies was tested by ELISA. (b) SDS-PAGE and western blot showed the purification progress of the antibodies. (c) ELISA showed the titer of antibodies purified from the serum of rabbit 2 against eight specific peptides (open circles). Scramble 1-DXXDZZ peptides were used as negative control to show the titer of antibodies against uncleaved substrate sites (filled circles). (d) The high O.D. values of Scramble 3-DEVD-coated wells compared with Scramble 1-, Scramble 2-, Scramble 3-, and OMPC-coated wells are an example of the specificity of the purified antibodies, in all six rabbits tested
Purified antibodies specifically recognize cleaved caspase substrates with a pattern of ‘DXXD’
Next, we tested whether the purified antibodies were capable of binding cleaved caspase substrates generated in apoptotic cells. HCT116, a human colorectal carcinoma cell line, was treated with a combination of 5-fluorouracil (5-FU) and tumor necrosis-related apoptosis-inducing ligand (TRAIL) to induce apoptosis. The induction of apoptosis was confirmed by examining the positive annexin A5 binding of externalized phosphatidylserine (PS), a hallmark of apoptosis (data not shown). Cell lysates prepared from apoptotic HCT116 cells were immune-precipitated with the purified antibodies, applied to SDS-PAGE gels and western blotted for the detection of cleaved caspase substrates. Caspase 6 and Poly (ADP-ribose) polymerase (PARP) are two well-known caspase substrates, whose caspase cleavage sites are DVVD and DEVD, respectively. These two caspase substrate sequences were included in the eight tetrapeptides used in the immunization. When the western blot was probed with commercially available antibodies which recognize both uncleaved and cleaved forms of caspase 6 (Figure 3a) or PARP (Figure 3b), only the cleaved substrates were detected in the pull-down fraction, demonstrating that our purified antibodies specifically recognize cleaved caspase 6 and PARP. However, when the apoptosis was attenuated by adding QVD-OPH, a pan-caspase inhibitor, to the 5FU/TRAIL-treated HCT116 cells, no specific bands could be detected in the co-immuno-precipitated gel (Figures 3a and b), nor were any specific bands detected in the rabbit IgG immuno-precipitated cell lysate. These results further confirm that the purified antibodies are neo-epitope antibodies (NEAs): they specifically recognize epitopes exposed or generated during apoptosis and can be used to immunoprecipitate such proteins. It should be noted that the rabbit IgG bands were detected by the secondary anti-rabbit antibodies on the blots given that the NEAs and the detection antibodies (i.e. anti-PARP and anti-caspase 6) were raised in the same species (rabbit). To avoid precipitation, the samples were not boiled before loading onto the gels. Therefore IgG oligmers were seen on the blots.
Purified antibodies specifically recognize cleaved caspase substrates with a pattern of ‘DXXD’. Apoptosis was induced by 5-FU and TRAIL in HCT116 cells as described in ‘Materials and Methods’. QVD-OPH was added to block caspase activities as indicated. Cell lysate from 107 cells was immuno-precipitated with 20 μg of purified antibodies, then detected with antibodies against PARP (a), caspase 6 (b) or cleaved CK-18 (c), separately. The purified antibodies specifically recognize not only the cleaved PARP (DEVD) and caspase 6 (DVVD) whose caspase cleavage sites are one of the eight peptides used for immunization, but also the cleaved CK-18 (DALD), whose tetrapeptide cleavage site was not one of the eight peptides, but has aspartic acid at the P1 and P4 position. However, the purified antibodies do not recognize the cleaved caspase 7 (IQAD) or lamin A (VEID) whose tetrapeptide cleavage sites has a ‘D’ at P1 but not P4 position (data not shown). Rabbit IgGs in the immune precipitate were detected as artifacts in western blots (as labeled) by the secondary antirabbit antibodies. This was due to the fact that both the NEAs and the detection antibodies (i.e. anti-PARP and anti-caspase 6) were raised in the same species (rabbit). Furthermore, to avoid precipitation, samples were not boiled before loading onto gels, therefore IgG oligmers were seen on the blots. (d) Purified antibodies specifically pull down cleaved recombinant CK-18. A total of 10 μg of recombinant CK-18 was cleaved with caspase 3 in vitro. QVD-OPH was added to block caspase activities as indicated. The reaction was precipitated with purified antibodies and then detected with antibody against cleaved CK-18. (e) Peptides with a ‘D’ at P1, or P1 and P4 (IETD, LQTD, SECD, DCRD, FRHD and FAED) were coated on the ELISA plates, and probed with purified antibodies. Scramble 3-DETD and Scramble 3 peptides were coated as positive and negative control, respectively. The antibodies only recognize the peptides with aspartic acids at both the P1 and P4 position (i.e. DCRD and DETD), but not the ones with a ‘D’ only at P1 position
Cytokeratin 18 (CK-18) is another well-known caspase substrate. During apoptosis, CK-18 is cleaved sequentially by caspase 3 and caspase 6. A monoclonal antibody (M30) developed by Peviva can specifically detect the caspase-3-cleaved form of CK-18. The sequence of this cleavage site is DALD, which was not one of eight tetrapeptides for immunization. Nonetheless, it conforms to the same ‘DXXD’ pattern and should have a similar three-dimensional structure as other ‘DXXD’ tetrapeptides. If the specificity of the purified NEAs was based solely on the peptide sequence, no cleaved caspase substrates whose cleavage sites are different from the eight tetrapeptides should be precipitated by the antibodies. However, if the specificity of the purified antibodies is based on the structure or shape of the peptides, substrates with any ‘DXXD’ pattern may be recognized by the NEAs. To test whether the purified NEAs can recognize cleaved CK-18, the apoptotic HCT116 cell lysates were immuno-precipitated with our NEAs and probed with M30. Intriguingly, the cleaved CK-18 was pulled down by the purified antibodies (Figure 3c). This was further confirmed by the in vitro pull-down assay using recombinant caspase 3 and CK-18 proteins. In this series of experiments, recombinant CK-18 was incubated with recombinant caspase 3 in the presence or absence of QVD-OPH, followed by the precipitation with the purified NEAs and subsequent detection with M30. As shown in Figure 3d, cleaved CK-18 was only captured by the purified NEAs in the absence of QVD-OPH, indicating that the specificity of the NEAs is based on the peptide structure of caspase-cleaved ‘ends’ of proteins. Moreover, the purified antibodies failed to pull down other cleaved caspase substrates, such as caspase 7 or lamin A, whose cleavage sites do not have Ds at P4 position (data not shown). To further confirm that the specificity of the purified NEAs is structure-based, we coated the ELISA plates with peptides terminating with amino-acid sequences DXXD, XXXD or XXXX, then probed the plates with the purified NEAs to measure the binding affinity. None of the peptides tested had the same DXXD sequences as the eight used for immunization. Nonetheless, as expected, the purified antibodies bound to peptide sequences ending in DXXD with high affinity, but little or no binding was observed to those not having aspartates at both P1 and P4 position (Figure 3e). Aspartates at both positions seem to be critical for the binding of the antibodies, as the peptides ending with XXXD did not show binding affinity to the purified antibodies.
In order to test the ability of the purified NEAs to identify multiple caspase substrates, we performed an immuno-precipitation experiment as follows: lysates from healthy or apoptotic HCT-116 cells (±QVD-OPH) were incubated with NEA. Captured proteins were then run on SDS-PAGE gels and the proteins were identified by mass spectrometric analysis of each lane from the gel. The arrows in Figure 4 indicate (as yet unidentified) proteins present more abundantly in the lysates from apoptotic cells (middle lane) than in lysates from healthy or QVD-OPH-treated cells. We identified at least four proteins, which were present in the lysates from apoptotic cells, but not in the healthy or QVD-OPH-treated lysates. Significantly, CK-18 was one of these proteins, confirming the results obtained in Figure 3c. In addition, DNA protein kinase (catalytic subunit) was also present exclusively in immunoprecipitations from the apoptotic HCT-116 cell lysates; this protein has been previously identified as a caspase 3 substrate.7 Elongation factor 1-B (EF1-B) was similarly identified by NEA precipitation in apoptotic lysates. The caspase substrate sequence DETD is present at amino acid 151–155 of human EF1-B. Moreover, Zebrafish EF1-B is cleaved by the zebrafish ortholog to caspase 3.8 Finally, the 60S ribosomal protein L21 was identified only in the immunoprecipitation of lysates from apoptotic cells. Although there are no literature reports of this protein being a caspase substrate, the amino-acid sequence contains a putative substrate-recognition sequence (DIVD) at positions 38–41. These data indicate that both known and (potentially) novel caspase substrates can be identified using the NEA reagents.
Purified antibodies can immunoprecipitate different proteins from apoptotic cell lysates compared with lysates of healthy or caspase-inhibited cells. Immunoprecipitated cell lysates from cells as in Figure 3 were also resolved by SDS-PAGE and protein bands were visualized by Coomassie blue staining. Clear differences were observed; arrows indicate for example, at least five (as yet unidentified) bands distinctly more visible in the apoptotic cell lysates compared with lysates from healthy or caspase-inhibited cells. Sections of the gel were cut and subjected to trypsin digestion and mass spectrometry identification. Three known caspase substrates (CK-18, DNA PK and EF1-B) and a novel one (60S ribosomal protein L21) were identified in apoptotic cell lysates
The purified neo-epitope antibodies can immunoprecipitate secreted caspase-cleavage products
As a pre-requisite step for validation as plasma biomarkers for apoptosis, it was important to determine whether purified NEAs could bind caspase-cleavage products, such as cleaved CK-18, secreted into the medium from cultures of apoptotic cells. To this end, culture medium from apoptotic HCT116 cells was immuno-precipitated with purified NEAs, and probed with the cleaved CK-18 antibody, M30, or with an antibody recognizing cleaved PARP. As shown in Figure 5, both cleavage products were precipitated from apoptotic cell culture supernatants and were detectable by their respective antibodies. The fact that more than one caspase-cleavage product could be precipitated from the same medium preparation by the same NEA preparation, demonstrates the potential of using purified NEAs to identify novel neo-epitopes generated during apoptosis, providing additional targets for validation as circulating biomarkers of tumor response.
Purified antibodies specifically recognize cleaved PARP and CK-18 in apoptotic media. Apoptosis was induced by 5-FU and TRAIL in HCT116 cells as described in ‘Materials and Methods’. QVD-OPH was added to block caspase activities as indicated. Cell media from 107 cells was concentrated and immuno-precipitated with 20 μg of purified antibodies, then detected with antibodies against PARP (a), or cleaved CK-18 (b), separately. The purified antibodies specifically recognize the cleaved PARP and CK-18 released to the media. The rabbit IgG bands from the antibodies were labeled as well
Discussion
Increased caspase activity is one of the hallmarks of apoptotic cell death. Thus, products cleaved by this class of proteases may serve as effective biomarkers of apoptosis. One example of such a protein is caspase-cleaved cytokeratin 18 (CK-18). Both cleaved and uncleaved CK-18 are serum stable; indeed, commercially available kits (Apoptosense M30 and M65) have been used to measure CK-18 processing as a biomarker of apoptosis or necrosis in a variety of different settings including human clinical trials.9 In this regard, Dix et al.10 reported many of the caspase-cleaved products (CCPs) appear to be stable (relative to the half life of the uncleaved ‘parent’ protein), making a strong case for the identification of other CCPs as potential clinically relevant biomarkers of apoptosis. Such biomarkers could be measured in accessible tissue compartments such as plasma, urine and cerebrospinal fluid.
The aim of the present study was to develop antibody-based tools to enable the discovery of novel CCPs. One advantage of an antibody-based (rather than a proteomics-based) approach to the identification of new apoptosis biomarkers,3 is that the discovery tool (antibody) may itself be further optimized for biomarker detection/validation. Antibodies produced through immunization with peptide cocktails have been applied extensively for the generation of reagents specific for distinct epitopes of the same protein. We applied a similar technique for the first time to create antibodies that would recognize multiple proteins whose commonality was a caspase-cleaved end region. In order to create such neo-epitope antibodies (NEAs), we analyzed the CASBAH database and rank ordered the caspase-cleavage sites within the database. The eight most represented cleavage sites were chosen to use in our immunization scheme. All eight sites had an aspartic acid at positions P1 and P4, leading us to collectively call these eight sequences ‘DXXD’. In fact, DXXD is a very prevalent caspase-cleavage motif, accounting for 33% of the 724 caspase-cleavage sites recently collated (manuscript in preparation). We hypothesized that the purified NEAs would recognize the caspase-cleaved motif based on its ‘shape’ (i.e. based on tertiary structure) rather than the sequence. The fact that the antibodies bind to all eight peptides with similar affinity regardless of whether the antibodies were affinity purified from any of a single-peptide-conjugated column or from eight-peptide-conjugated column, further confirms this hypothesis (data not shown).
In addition to immunizing rabbits with a pool of peptides rather than a single amino-acid sequence, the antigenic boost injections contained an alternate set of eight peptides that differed only in their linker region (i.e. remaining constant in their terminal, ‘caspase cleaved’ sites). This increased the chances of antibody production to the desired DXXD region of the immunogen. Success of this crossover-boosting strategy was validated; even before purification, the titers of the NEA serum to the two linker regions (Scramble 1 or Scramble 2) were considerably lower than to the DXXD region.
As a proof of concept for their utility as broad-specificity caspase-cleaved substrate detectors, the NEAs were used to precipitate proteins from lysates of cells induced to undergo apoptosis in the presence or absence of caspase inhibitors. Using this approach, both known (PARP, caspase 6, CK-18, DNA PK and EF1-B) and novel (60S ribosomal protein L21) caspase substrates were identified in apoptotic cell lysates. Importantly, the non-cleaved forms of PARP, caspase 6 and CK-18 (present in healthy cells) were not detected by the same antisera. It is noteworthy that although the cleavage site within CK-18 maintains the DXXD motif, DALD, this peptide was not one of the eight sequences used in immunization, further supporting our hypothesis that the NEAs can detect many DXXD motifs based on their conformation and not their sequence.
Moreover, in additional ELISA-based validation experiments, the second most highly recognized peptide had the DXXD motif, DCRD (the caspase-cleavage site of NFkB11), but again was not one of the eight original peptide sequences used for immunization. Conversely, the NEAs failed to immunoprecipitate cleaved caspase 7 or Lamin A (data not shown), both of which have caspase-cleavage sites consisting of XXXD; that is, where the P4 amino acid was not aspartic acid. A terminal DXXD sequence is much more likely to represent a caspase cleavage event compared with an internal DXXD sequence. Thus, to fulfill its utility as a tool to find caspase-cleaved proteins, it is imperative that the NEAs recognize DXXD only in the context of the end of a peptide, as would be found after caspase cleavage. This was the case, as the titers of the NEAs were generally much higher for the DXXD peptides compared with peptides constructed to mimic an uncleaved (internal) state, DXXDZZ (where the ZZ represents random amino-acid pairs). Additionally, full-length caspase 6 or PARP are not immunoprecipitated by the NEAs, further confirming the utility of the NEAs as a tool to detect caspase-cleaved proteins by virtue of their newly exposed caspase-cleaved terminus. More intriguing, the purified NEAs were able to recognize not only cleaved CK-18 but also cleaved PARP peptides in media conditioned by apoptotic cells. To our knowledge, this is the first time that cleaved PARP has been shown to be released from apoptotic cells, suggesting a new clinical application: using cleaved PARP as a plasma biomarker for the apoptotic response of solid tumors.
There exist at least two previous examples of antibodies that recognize caspase-cleaved ‘ends’ of more than one protein, although this is by serendipity rather than design. Ab127 was produced by immunizing with a KLH-conjugated peptide corresponding to six amino acids of the PARP cleavage site (KGDEVD).12 Under apoptotic conditions, this antibody recognized a protein of 47 kD, rather than the predicted ∼30 or ∼85 kD fragments given upon caspase cleavage of PARP. The second example is mentioned in the same publication, though the authors were unaware of its ability to recognize more than one caspase-cleavage epitope. sc45, an antibody that is commercially available (Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA) was first reported to detect c-Jun (amino acids 73–87 of rat c-Jun13). The terminal four amino acids of this epitope are NVTD. Later, it was demonstrated that sc45 cross-reacted with other proteins, following caspase 3 cleavage, at sites that did not contain the c-Jun sequence.14 Seryl-tRNA is one such protein,15 although the caspase-cleavage site of seryl-tRNA is EVTD rather than NVTD. What is especially noteworthy about this caspase-cleavage site is that the P1 aspartate is the penultimate amino acid of seryl-tRNA. In other words, caspase cleavage appears to liberate only a single amino acid (the C-terminal alanine). The NEAs generated in the present study also fail to detect a DXXD that is only one amino acid from the end of a peptide; when tested against the peptide ending in DEVD, titers in excess of 1 : 50 000 were observed compared with a titer of only 1 : 3000 when the identical peptide ending in DEVDG was tested. Caspase cleavage events occurring so close to the C-terminal region of proteins are undoubtedly under-represented in both CASBAH and even in non-biased screens for caspase activity.16 In contrast, the NEAs outlined in this study, could potentially identify both internal peptides and terminal/penultimate amino-acid cleavage events.
Mass spectrometric methods of detection, while sensitive enough to detect amino-acid differences in mass or sequence, suffer from the limited ability to capture such events. In this regard, two previous studies have greatly extended the known pool of caspase substrates. In the first, the Cravatt laboratory10 developed a high-content proteomic platform to profile proteolytic events occurring in biological systems termed PROTOMAP (for PROtein TOpography and Migration Analysis Platform). Using this approach, they identified more than 150 proteins not previously known to be cleaved during Jurkat T-cell apoptosis. In the second study, Wells and co-workers16 used an engineered enzyme to selectively biotinylate free-protein N termini for positive enrichment of corresponding N-terminal peptides to study the proteolytic events of apoptosis. They sequenced more than 290 substrates not predicted by in vitro caspase-substrate specificity. However, neither of these studies provides practical tools to follow up on the validation of novel substrates as biomarkers of apoptosis. In contrast, the utility of NEAs described in the present study will expand the prospective pool of proteins that serve as biomarkers of apoptosis.
Materials and Methods
Peptides and conjugates
The CASBAH database17 was examined and the eight most frequent caspase-cleavage sites were determined (DEVD, DETD, DELD, DEPD, DEAD, DSVD, DVVD and DEQD, hereafter collectively referred to as ‘DXXD’). Three different octapeptide linker sequences (Scramble 1, 2 and 3) were chosen to closely resemble native rabbit peptides in order to minimize generation of antibodies to these irrelevant parts of the peptide. (Scramble 1 and 2 were used for immunization, Scramble 3 was utilized during purification to decrease the isolation of irrelevant antibodies to these linker sequences.) All peptides were synthesized by JPT Peptide Technologies (Acton, MA, USA) with an N-terminal cysteine followed by a 6-aminohexanoic acid reside (ahx) in order to aid in the conjugation and quantification of conjugation, respectively.18 Finally, the C-terminal aspartate was synthesized as a free acid to more closely resemble the end of a caspase-cleaved protein. The peptides synthesized are shown in Figure 1a.
The outer membrane protein complex (OMPC) of Neisseria meningitidis is an excellent carrier protein19 to which peptides may be conjugated in order to increase immunogenicity.20 All 16 peptides (eight C-ahx-Scramble 1-DXXD and eight C-ahx-Scramble 2-DXXD) were individually conjugated to OMPC via thiol-based conjugation chemistry essentially as previously described,21 except that the duration of maleimide activation of OMPC was 4 h rather than 1 h, and the peptide thiol/maleimide molar ratio was 1.5 rather than 3. Protein concentrations of the 16 peptide-OMPC conjugates were determined by the Lowry assay. Eight Scramble 1-DXXD (1.25 mg each) or the eight Scramble 2-DXXD conjugates (1.25 mg each) were combined to create antigen set A (the pool of all eight OMPC-Scramble 1-DXXD conjugates) and antigen set B (combination of 1.25 mg of each of the OMPC-Scramble 2-DXXD conjugates), respectively.
Antibody production
Polyclonal antibodies were produced by SDIX (Newark, DE, USA) according to their established 99-day protocol, and in facilities that are registered and accredited with AAALAC, USDA, PHS/NIH/OLAW, DEA and APHIS. Rabbits were pre-bled, and the six whose plasma showed the least signal when immunoblotted with antirabbit secondary abs were chosen. A group of three rabbits was initially immunized and boosted (on days 13, 26 and 41) with antigen set A, while another group of three rabbits was immunized and boosted on the same days with antigen set B. On day 70, the final boost was switched such that the rabbits immunized with antigen set A were given their final boost with antigen set B and vise versa, such that the only commonality between immunization and final boost was the C-terminal DXXD sequences. Serum containing polyclonal antibodies from all six rabbits was collected at day 99.
Column conjugation and antibody purification
SulfoLink Immobilization kit (Pierce Biotechnology, Rockford, IL, USA) was used to conjugate the peptide and purify the antibodies. The manufacturer's instruction was followed. In brief, every 2 mg of Scramble 3-DXXD peptides (either single peptide or the mix of 0.25 mg of each of the eight peptides) were dissolved in 2 ml of coupling buffer (50 mM Tris, 5 mM EDTA-Na, pH 8.5), reduced with 25 mM TCEP and incubated for 30 min. The sulfhydryl-containing peptides were then coupled to 2-ml Sulfolink columns by mixing with the SulfoLink resin for 15 min followed by a 30-min incubation. Non-specific binding sites were blocked with 50 mM cysteine by mixing for 15 min and subsequent incubation for 30 min. The columns were then equilibrated with binding/wash buffer (TBS) before the affinity purification step. All steps were performed at room temperature.
The antibody purification scheme is summarized in Figure 2a. The fractions from each step were monitored by SDS-PAGE and western blot to confirm the purification of antibodies; the specificity of the antibodies was verified by ELISA. The serum from each of the six rabbits was first fractionated by ammonium sulfate precipitation. The serum was centrifuged at 10 000 × g for 30 min at 4°C to remove any large aggregates. The supernatant was collected, and an equal volume of saturated ammonium sulfate solution (Pierce Biotechnology) was added to the supernatant drop by drop on ice with gentle stirring. The mixed solution was rotated for at least 3 h (up to over night) at 4°C followed by centrifugation at 10 000 × g for 30 min at 4°C. The pellet was resuspended with a minimal volume of 20 mM Tris-HCl (pH 7.5), and dialyzed against 20 mM Tris-HCl+50 mM NaCl (pH 7.5) before being applied to a HiPrep 16/10 DEAE_FF ion-exchange column (GE Healthcare Biosciences, Piscataway, NJ, USA). A 200 ml of linear gradient from 50 mM to 500 mM of NaCl was applied to the column to elute the bound protein. The flow-through was pooled and applied to the Scramble 3-DXXD peptides-conjugated SulfoLink column. The column was incubated at room temperature with rocking for 30–60 min to allow binding to occur. Following the wash step, the antibodies that specifically recognize the eight DXXD peptides were eluted with 0.1 M glycine (pH 3.0), and neutralized with 1 M Tris (pH 8.5).
ELISA assays
Titers of pre-bleed, post-bleed sera and final-purified aliquots of antibodies were determined via colorimetric ELISA. Plates (96-well) were coated with 10 μg/ml of peptide for a minimum of 2 h (up to overnight at 4°C), then blocked (without removing coating solution) with TBS-T/BSA (150 mM NaCl, 50 mM Tris, 0.05% Tween-20 and 1% BSA) for an additional 2 h. The wells were rinsed three times with TBS-T, aspirated and 95 μl of serially diluted sera/antibodies (1 : 103 up to 1 : 106) were added and allowed to incubate at room temperature for 1–2 h on an orbital shaker. The wells were again rinsed three times with TBS-T, aspirated and 60 μl of 1 : 5000 donkey anti-rabbit-HRP antibody (GE Healthcare Biosciences) was added and incubated for 1–2 h on an orbital shaker. Wells were aspirated and rinsed three times with TBS-T after which 100 μl of TMB substrate (Pierce Biotechnology) was added and incubated in the dark until appropriate coloration was observed, at which point 50 μl of 1 M H2SO4 was added to stop the reaction and the absorbance (450 nm wavelength) was measured on a spectramax M2e spectrophotometer (Molecular Devices, Sunnyvale, CA, USA).
Apoptosis induction, immunoprecipitation and immunoblotting
The conditions and time points of apoptosis induction for HCT-116 cells (ATCC) have been optimized to give maximal induction. Briefly, an ∼80% confluent 10-cm plate of cells was incubated with 400 μM 5-Fluorouracil (5-FU, in DMSO, Sigma-Aldrich, St. Louis, MO, USA) for 17 h, then 200 ng/ml recombinant TRAIL (MP Biomedical, Solon, OH, USA) was added to the media and cells were incubated for an additional 3 h. This dosing paradigm reproducibly induced apoptosis (>50% annexin A5 positive) with minimal amounts of membrane permeability as measured by propidium iodide uptake. That this apoptotic induction was caspase-dependent, was confirmed in that concomitant addition of the caspase inhibitor QVD-OPH (MP Biomedical) reduced peak Annexin A5 positivity by 50% or more (data not shown).
Apoptotic cells (107) (or non-induced and 5-FU/TRAIL/QVD-OPH treated) were harvested and lysed with RIPA lysis buffer (Pierce Biotechnology) containing protease inhibitor cocktail (Pierce Biotechnology) at 4°C. Cell lysates were collected with QIAshredder column (Qiagen, Valencia, CA, USA) at 10 000 × g for 2 min at 4°C, then the protein concentration was determined with a BCA Protein Assay kit (Pierce Technology). About 500 μg of cell lysates were pre-cleared by mixing with 50 μl of GammaBind Plus Sepharose (GE Healthcare Biosciences) for 1 h to overnight at 4°C to decrease the background. Pre-cleared cell lysates were incubated with 10 μg of purified neo-epitope antibodies (NEAs) for 1 h at 4°C followed by mixing with 50 μl of GammaBind Plus Sepharose for another 1 h at 4°C. After the incubation, the resin was washed with RIPA buffer, and antibody-bound proteins were eluted with 2 × SDS buffer (Boston Bioproducts, Ashland, MA, USA). Rabbit IgG (Invitrogen, Carlsbad, CA, USA) was mixed with cell lysates and GammaBind Plus Sepharose in parallel as a negative control.
The following primary antibodies were used in immunoblotting: anti-PARP (Cell Signaling Technologies, Danvers, MA, USA), anti-Caspase 6 (Cell Signaling Technologies), M30 (Peviva through Diapharma, West Chester, OH, USA).
Immunoprecipitated cell lysates were loaded onto Bis-Tris gradient gels (Invitrogen) and run according to the manufacturer's directions. Gels were transferred to nitrocellulose using a wet-transfer protocol and blocked for 1 h in Odyssey blocking buffer (LI-COR Biosciences, Lincoln, NE, USA). Blots were incubated overnight at 4°C with primary antibodies in blocking buffer supplemented with 0.1% Tween-20 (Sigma-Aldrich). Blots were rinsed with TBS-T, and IR-dye-labeled secondary antibodies (LI-COR) were applied at 1 : 15 000 in blocking buffer supplemented with 0.1% Tween-20 and 0.01% SDS for 1 h. All blots were imaged using the Odyssey infrared scanner (LI-COR). To avoid precipitation, the samples were not boiled before loading onto the gels. Therefore IgG oligmers were seen on the gels.
Immunoprecipitated cell lysates from apoptotic cells (or non-induced and 5-FU/TRAIL/QVD-OPH treated) were also resolved by SDS-PAGE and protein bands were visualized by Coomassie blue staining. The protein bands were cut and subjected to trypsin digestion and mass spectrometry identification.
Recombinant caspase 3 cleavage
The in vitro caspase 3 cleavage assay was performed in buffer composed of 50 mM HEPES (pH 7.3), 100 mM NaCl, 5% (w/v) glycerol, 0.5 mM EDTA, 0.05% (w/v) CHAPs and 5 mM dithiothreitol. Recombinant CK-18 (10 μg) (MP Biomedical) was mixed with 0.2 μg of recombinant caspase 3 (Sigma-Aldrich) in the presence or absence of QVD-OPH (MP Biomedical). The reaction was incubated at 37°C for 1 h and terminated with SDS buffer.
Abbreviations
- CASBAH:
-
CAspase Substrate dataBAse Homepage
- CCPs:
-
caspase-cleavage products
- CK-18:
-
cytokeratin 18
- DEAE:
-
diethylaminoethyl
- DMSO:
-
Dimethyl sulfoxide
- DNA PK:
-
DNA-dependent protein kinase
- EF1-B:
-
elongation factor 1-B
- ELISA:
-
enzyme-linked immunosorbent assay
- 5-FU:
-
5-fluorouracil
- HRP:
-
horseradish peroxidase
- KLH:
-
keyhole limpet hemocyanin
- NEAs:
-
neo-epitope antibodies
- NFkB:
-
nuclear factor kappa-light-chain-enhancer of activated B cells
- OMPC:
-
outer membrane protein complex
- PARP:
-
poly (ADP-ribose) polymerase
- PROTOMAP:
-
PROtein TOpography and Migration Analysis Platform
- PS:
-
phosphatidylserine
- QVD-OPH:
-
quinolyl-valyl-O-methylaspartyl-(-2, 6-difluorophenoxy)-methyl ketone
- RIPA:
-
radio-immunoprecipitation assay
- SDS-PAGE:
-
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- TRAIL:
-
tumor necrosis-related apoptosis-inducing ligand
- TBS:
-
tris-buffered saline
- TCEP:
-
tris(2-carboxyethyl)phosphine
- TMB, 3,3′,5:
-
5′-tetramethylbenzidine
References
Fulda S, Debatin KM . Extrinsic versus intrinsic apoptosis pathways in anticancer chemotherapy. Oncogene 2006; 25: 4798–4811.
Fulda S . Exploiting apoptosis pathways for the treatment of pediatric cancers. Pediatr Blood Cancer 2009; 53: 533–536.
Sarker D, Workman P . Pharmacodynamic biomarkers for molecular cancer therapeutics. Adv Cancer Res 2007; 96: 213–268.
Amur S, Frueh FW, Lesko LJ, Huang SM . Integration and use of biomarkers in drug development, regulation and clinical practice: a US regulatory perspective. Biomark Med 2008; 2: 305–311.
Ward T, Cummings J, Dean E, Greystoke A, Hou JM, Backen A et al. Biomarkers of apoptosis. Br J Cancer 2008; 99: 841–846.
Brandt D, Volkmann X, Anstatt M, Langer F, Manns MP, Schulze-Osthoff K et al. Serum biomarkers of cell death for monitoring therapy response of gastrointestinal carcinomas. Eur J Cancer 2010; 46: 1464–1473.
Teraoka H, Yumoto Y, Watanabe F, Tsukada K, Suwa A, Enari M et al. CPP32/Yama/apopain cleaves the catalytic component of DNA-dependent protein kinase in the holoenzyme. FEBS Lett 1996; 393: 1–6.
Valencia CA, Bailey C, Liu R . Novel zebrafish caspase-3 substrates. Biochem Biophys Res Commun 2007; 361: 311–316.
Linder S, Alaiya A . Serum efficacy biomarkers for oncology. Biomark Med 2009; 3: 47–54.
Dix MM, Simon GM, Cravatt BF . Global mapping of the topography and magnitude of proteolytic events in apoptosis. Cell 2008; 134: 679–691.
Ravi R, Bedi A, Fuchs EJ, Bedi A . CD95 (Fas)-induced caspase-mediated proteolysis of NF-kappa B. Cancer Res 1998; 58: 882–886.
Siman R, Bozyczko-Coyne D, Meyer SL, Bhat RV . Immunolocalization of caspase proteolysis in situ: evidence for widespread caspase-mediated apoptosis of neurons and glia in the postnatal rat brain. Neuroscience 1999; 92: 1425–1442.
Terwel D, Van de Berg W . c-Jun/AP-1 (N) directed antibodies cross-react with ‘apoptosis-specific protein’ which marks an autophagic process during neuronal apoptosis. Neuroscience 2000; 96: 445–446.
Ribera J, Ayala V, Esquerda JE . c-Jun-like immunoreactivity in apoptosis is the result of a crossreaction with neoantigenic sites exposed by caspase-3-mediated proteolysis. J Histochem Cytochem 2002; 50: 961–972.
Casas C, Ribera J, Esquerda JE . Antibodies against c-Jun N-terminal peptide cross-react with neo-epitopes emerging after caspase-mediated proteolysis during apoptosis. J Neurochem 2001; 77: 904–915.
Mahrus S, Trinidad JC, Barkan DT, Sali A, Burlingame AL, Wells JA . Global sequencing of proteolytic cleavage sites in apoptosis by specific labeling of protein N termini. Cell 2008; 134: 866–876.
Luthi AU, Martin SJ . The CASBAH: a searchable database of caspase substrates. Cell Death Differ 2007; 14: 641–650.
Nahas DD, Palladino JS, Joyce JG, Hepler RW . Amino acid analysis of peptide loading ratios in conjugate vaccines: a comparison of direct electrochemical detection and 6-aminoquinolyl-N-hydroxysuccinimidyl carbamate pre-column derivatization methods. Bioconjug Chem 2008; 19: 322–326.
Donnelly JJ, Deck RR, Liu MA . Immunogenicity of a Haemophilus influenzae polysaccharide-Neisseria meningitidis outer membrane protein complex conjugate vaccine. J Immunol 1990; 145: 3071–3079.
Bianchi E, Liang XP, Ingallinella P, Finotto M, Chastain MA, Fan J et al. Universal influenza B vaccine based on the maturational cleavage site of the hemagglutinin precursor. J Virol 2005; 79: 7380–7388.
Fan JA, Liang XP, Horton MS, Perry HC, Citron MP, Heidecker GJ et al. Preclinical study of influenza virus A M2 peptide conjugate vaccines in mice, ferrets, and rhesus monkeys. Vaccine 2004; 22: 2993–3003.
Acknowledgements
We thank D Nahas for her technical assistance in conjugation of the peptides to OMPC and the quantitation thereof; A Woods for her expert advice on ELISA procedures; H Cardasis and the mass spectrometry group for their support; S Roy for advice on peptide antigen design; D Roush and Y Wang for their expert advice on antibody purification; and M Hoek, L Kong, L Hernandez and C Houde for helpful and critical discussion.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
All the authors are employees of Merck and Co.
Additional information
Edited by G Melino
Rights and permissions
This work is licensed under the Creative Commons Attribution-NonCommercial-No Derivative Works 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Ai, X., Butts, B., Vora, K. et al. Generation and characterization of antibodies specific for caspase-cleaved neo-epitopes: a novel approach. Cell Death Dis 2, e205 (2011). https://doi.org/10.1038/cddis.2011.91
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cddis.2011.91
Keywords
This article is cited by
-
Preferential rabbit antibody responses to C-termini of NOTCH3 peptide immunogens
Scientific Reports (2023)
-
Impact of conjugation strategies for targeting of antibodies in gold nanoparticles for ultrasensitive detection of 17β-estradiol
Scientific Reports (2019)
-
Analysis of apoptosis methods recently used in Cancer Research and Cell Death & Disease publications
Cell Death & Disease (2012)