Abstract
The BH3-only protein BAD binds to Bcl-2 family proteins through its BH3 domain. Recent studies suggest that BAD binds to both Bcl-2 and Bcl-XL, however mediates its pro-apoptotic functions through inhibition of Bcl-XL, but not Bcl-2. In this paper we addressed this issue using a BAD mutant within the BH3 domain, by substitution of Asp 119 with Gly (BADD119G), which selectively abrogates an ability to interact with Bcl-2. Confocal microscopy revealed that mutation of BAD at D119 does not affect BAD targeting to the mitochondrial membrane in serum-starved COS-7 cells. However, co-precipitation assays indicated that, whereas wild-type BAD (BADwt) directly interacts with Bcl-2 and Bcl-XL, BADD119G interacts only with Bcl-XL. Nevertheless both BADwt and BADD119G could introduce apoptosis and diminish the anti-apoptotic effect of Bcl-2 and Bcl-XL in a similar manner in a co-transfection assay. These data thus suggest that Asp119 is a crucial site within the BH3 domain of BAD for interaction of BAD with Bcl-2, but is dispensable for the interaction of BAD with Bcl-XL, for its targeting to mitochondria, and most importantly, for its pro-apoptotic functions. Thus, we confirm that neutralization of Bcl-2 function is marginal for BAD-mediated apoptosis.
Similar content being viewed by others
Introduction
Bcl-2 family proteins can function to either positively or negatively regulate apoptosis. Anti-apoptotic members of this family include Bcl-2, Bcl-XL, Bcl-w, BHRF1, and E1B-19K, while pro-apoptotic family members include Bax, Bak, and Bok. These molecules share homologous domains BH1 to BH4 (reviewed in1,2). Bcl-2 proteins interact with other molecules which express an α-helical structure termed the BH3-domain and this interaction is believed to be important in the regulation of apoptosis (reviewed in3). A large number of BH3-only proteins have been identified including BAD, Bid, Bik, Bim, Bmf,4 BNIP3, Hrk, and Noxa.5 All of these BH3-only proteins are pro-apoptotic molecules and their pro-apoptotic activity is believed to be mediated via interaction with Bcl-2 family proteins. Thus the BH3 domain of these proteins is believed to be crucial for their pro-apoptotic functions.
BH3-only proteins can be divided into two groups. The first group of BH3-only proteins, which includes BAD and Bim, can bind anti-apoptotic molecules selectively, and inactivate their anti-apoptotic functions. For example, BAD is phosphorylated by survival signals, bound to 14-3-3 scaffold proteins and localized in the cytoplasm in an inactive form.6,7,8 Once survival signals are abrogated BAD is dephosphorylated, dissociated from 14-3-3 and localized to the mitochondria. Free, mitochondrial BAD molecules are then believed to interact with either Bcl-2 or Bcl-XL and neutralize their anti-apoptotic functions. Similarly, Bim is normally complexed with the microtubule-associated dynein light chain LC8. However once death signals are activated Bim is released from the complex. Free Bim then binds to Bcl-2 or Bcl-XL and inactivates their anti-apoptotic functions.9 Thus inhibition of the anti-apoptotic functions of Bcl-2 family members appears to be a pivotal role of this group of pro-apoptotic BH3-only molecules. The second group of BH3-only proteins, which is exemplified by Bid, can bind to both anti-apoptotic and pro-apoptotic Bcl-2 family members. Bid is localised in the cytoplasm in non-apoptotic cells. However activated caspase-8 cleaves p22 Bid to generate a p15 active tBid fragment which is then targeted to the mitochondria where it binds to the pro-apoptotic Bcl-2 family members Bax or Bak as well as to the anti-apoptotic Bcl-2.10,11,12 Formation of tBid-Bax or tBid-Bak oligomers induces apoptotic signals. Importantly, neutralization of anti-apoptotic Bcl-2 family members is believed to play only a marginal role in induction of apoptosis by these proteins.
Recently, it has been found that BAD and Bim, which belong to the first group of BH3-only proteins described above, require expression of Bax or Bak for their pro-apoptotic functions.13 However since neither BAD nor Bim can directly interact with Bax or Bak it is unlikely that they can directly activate Bax or Bak. Indirect activation of Bax or Bak via inactivation of Bcl-2 or Bcl-XL has been proposed.13 As described above the pro-apoptotic functions of BAD appear to be directly mediated through interaction of BAD with Bcl-2 or Bcl-XL via the BH3 domain of BAD and subsequent inactivation of the pro-apoptotic activity of Bcl-2 or Bcl-XL. In fact, elimination of the entire BH3 domain from BAD abrogates its ability to interact with Bcl-2 family members and simultaneously abrogates its pro-apoptotic activity.8 Although BAD can interact with both Bcl-2 and Bcl-XL, BAD binds more strongly to Bcl-XL than Bcl-2 and BAD can reverse the death repressor activity of Bcl-XL, but not that of Bcl-2.6 In addition, survival signals induce the phosphorylation of BAD, which results in its dissociation from Bcl-XL, but not from Bcl-2.14 The accumulated data therefore suggests that the pro-apoptotic function of BAD is primarily mediated via its neutralization of Bcl-XL, and that the neutralization of Bcl-2 by BAD plays a marginal role in this function of BAD. However, these studies are all indirect observations and it is also conceivable that the death repressor activity of Bcl-2 is much stronger than that of Bcl-XL, and thus can overcome the death activity of BAD.
Since BAD interacts with both Bcl-2 and Bcl-XL via the BH3 domain, but interacts more strongly with Bcl-XL,6 it was a possibility the BAD might interact with these two molecules via different regions of the BH3 domain. To date, at least three reports have indicated that replacement of the conserved leucine (L) residue within the BH3 domain of BAD, Noxa or Bmf with alanine (A) extensively reduced their ability to bind BCl-2 or Bcl-XL and abolished their pro-apoptotic activity.4,5,8 However, to our knowledge, there has been no study to investigate whether a second residue, aspartic acid (D) at position 119, which is also well conserved among BH3-domains of Bcl-2 family proteins, could contribute to the binding and pro-apoptotic function of BH3-only proteins. We therefore generated a mutant form of BAD with an amino acid substitution of Gly for Asp119 within the BH3 domain (BADD119G) and tested the effect of this mutation on BAD protein–protein interactions and BAD function. We found that this mutation abolished the ability of BAD to interact with Bcl-2, but had no effect on its binding to Bcl-XL. Thus different regions within the BH3 domain of BAD appear to differentially contribute to the interaction of BAD with Bcl-2 and Bcl-XL. Importantly, BADD119G was still targeted to the mitochondria and could induce apoptosis in COS-7 cells as effectively as BADwt. In addition, they can reverse the death repressor activity of Bcl-2 and Bcl-XL in a similar level. This study is the first conclusive demonstration that BAD can induce apoptotic signals independently of its interaction with the anti-apoptotic Bcl-2.
Results
BAD overexpression augments apoptosis in COS-7 cells
The BH3-only protein BAD is abundantly expressed in many epithelial cells and functions to induce apoptotic signals.6,7,8,15 This suggests that BAD may play pivotal roles in the maintenance of epithelial structure by execution of excess or abnormal cells. To analyze the function of BAD in epithelial cell death, and the role of the BH3 domain of BAD in this function, we expressed GFP-tagged human full-length BAD (BADwt) or its BH3 domain deletion mutants in kidney epithelial COS-7 cells. When transfectants were deprived of FCS after transfection, overexpression of BADwt strongly induced cell death, but a BAD mutant, in which the entire BH3 domain has been deleted, could not induce cell death signals (Figure 1A). This indicates that BAD overexpression is able to augment apoptosis in COS-7 cells, and confirms previous observations indicating that the BH3 domain of BAD is essential for induction of apoptosis and that deletion of the BH3 domain totally abrogates pro-apoptotic functions of BAD.7
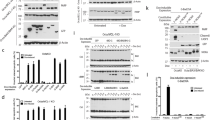
The BADD119G mutant has similar apoptotic activity to BADwt. (A) BAD overexpression augments cell death. COS-7 cells were transfected with the indicated expression vectors, and cultured in the absence of FCS for 24 h to induce the apoptotic process. Cell death was evaluated by trypan blue exclusion assay. The columns display the mean±standard deviation (S.D.) of data from three separate experiments. (B) Sequence homology of BH3 domains. Alignment of amino acid sequences of BH3 domains of Bcl-2 family members including BH3-only proteins. Highly conserved sequences (L and D) are indicated in lines. (C) Overexpression of GFP-tagged BAD proteins. COS-7 cells were transiently transfected with control pEGFP vector alone (MOCK), GFP-human BADwt (BAD), or the GFP-BADD119G mutant (BADD119G). Transfectants were harvested at 24 h following transfection and subjected to immunoblot analysis with the anti-GFP antibody. The anti-Hsc70 antibody was used as a control to show the same amount of protein loaded in each lane. (D) BAD overexpression augments cell death. COS-7 cells were transfected with the indicated expression vectors and cultured in the absence of FCS for 12 (open bars) or 24 h (closed bars). Cell death was evaluated by trypan blue exclusion assay. The columns display the mean±S.D. of data from three separate experiments. (E) The BADD119G mutant enhances caspase-3 activity to a similar extent as BADwt. COS-7 cells were transfected with the indicated vectors, and cultured in the absence of FCS for 9 (open bars) or 12 h (closed bars). Transfectants were washed with ice-cold PBS and caspase-3 activity was measured with a Caspase-3 Colorimetric Protease Assay Kit. The numbers represent the fold increase in absorbance at 400 nm relative to the mock transfectants. Each column displays the mean±S.D. of data from three separate experiments
BADD119G mutant has proapoptotic activity
The BH3 domains are well conserved among the BH3-only protein family and other Bcl-2 family members. Alignment of amino acid sequences of these BH3 domains clearly reveals two highly conserved residues, leucine (L) and aspartic acid (D) as shown in Figure 1B. Although the conserved leucine has been reported to be critical for pro-apoptotic activity of BAD there have been no studies to date on the role of the conserved aspartic acid. We therefore determined the effect of a point mutation at this conserved aspartate on the pro-apoptotic activity of BAD. For this purpose we generated a mutant BAD (BADD119G) in which the aspartate at aa 119 was mutated to glycine (G) and investigated its pro-apoptotic activity in COS-7 cells. Transfection of a BADD119G cDNA expression vector resulted in a lower level of BAD expression compared to transfection of wild-type BAD (BADwt) (Figure 1C). However BADD119G displayed a similar pro-apoptotic activity to that of BADwt, as assessed by cell death and activation of caspase-3 (Figure 1D, E). Thus the BADD119G mutant may even have a higher pro-apoptotic activity than BADwt. Furthermore the well-conserved Asp among Bcl-2 family members appears to be dispensable for the pro-apoptotic activity of BAD.
Mutation of BAD at aa119 in the BH3 domain abrogates its interaction with Bcl-2
The BH3 domain of BAD is believed to be crucial for the interaction of BAD with Bcl-2 family members and subsequent inactivation of Bcl-2 pro-apoptotic function. We therefore determined whether mutation of BAD at the conserved D119 position could alter the interaction of BAD with Bcl-2 family proteins. For this study we expressed both BADwt and BADD119G as GST fusion proteins and determined their interaction with Bcl-2 proteins in lysates of Bcl-2-transfected HEK 293 cells. The purified GST, GST-BADwt and GST-BADD119G proteins used for the experiment are shown in Figure 2A. The fusion proteins were immobilized on glutathione beads and incubated with a total cell lysate of HEK 293 cells transfected with full-length, GFP-labeled Bcl-2 proteins. Binding of Bcl-2 to the fusion proteins was detected following Western blotting with an anti-GFP antibody. As shown in Figure 2B only GST-BAD but not GST-BADD119G or GST alone could specifically bind to Bcl-2. Thus mutation of BAD at D119 can abolish its ability to bind to Bcl-2. To confirm this finding we also carried out the reciprocal experiment to determine if GST-Bcl-2 proteins could co-precipitate GFP-BADwt or GFP-BADD119G proteins expressed in HEK 293 cells. The purified human full-length GST-Bcl-2 fusion protein used for this experiment is shown in Figure 2C. When GST-Bcl-2, immobilized on glutathione beads, was incubated with lysates of HEK 293 cells expressing GFP-BADwt or GFP-BADD119G only the BADwt and not the BADD119G, was specifically co-precipitated by GST-Bcl-2 (Figure 2D). We thus conclude that mutation of BAD at D119 abrogates the binding of BAD to Bcl-2. Since BADD119G can still induce apoptosis (Figure 1) this data indicates that the pro-apoptotic activity of BAD is not necessarily mediated via inhibition of the anti-apoptotic function of Bcl-2.
Interaction of human BAD and BADD119G mutant with Bcl-2. (A) Expression of GST-BAD and GST-BADD119G fusion proteins. 10 μl of glutathione-bead-bound GST, GST-BAD or GST-BADD119G were loaded onto 12% SDS–PAGE and stained with Coomassie Blue G-250. Molecular size markers are shown in kDa. (B) Specific binding of Bcl-2 proteins to the GST-BAD fusion protein. Cell lysates from HEK 293 cells transfected with GFP-tagged-Bcl-2 or -BAX (INPUT) were incubated with the indicated GST-fusion proteins. The proteins bound to the washed beads (PULL-DOWN) were resolved by 12% SDS–PAGE and identified by Western blotting using an anti-GFP antibody. (C) Expression of the GST-Bcl-2 protein. Glutathione beads bound by GST or GST-Bcl-2 were resolved on 12% SDS–PAGE and stained with Coomassie Blue G-250. Molecular size markers are shown in kDa. (D) Specific binding of BADwt but not BADD119G to GST-Bcl-2. Cell lysates from HEK 293 cells transfected with GFP-tagged-BADwt or BADD119G (INPUT) were incubated with GST- or GST-Bcl-2-bound beads. The bound proteins were resolved by 12% SDS–PAGE and identified by Western blotting using an anti-BAD antibody. (BAD)
The BADD119G mutant is able to bind to Bcl-XL. and has similar anti-apoptotic activity in the presence of Bcl-2 or Bcl-XL
Previous data, discussed above, suggests that the pro-apoptotic function of BAD may be mediated via its interaction with Bcl-XL rather than Bcl-2.6 We thus determined whether mutation of BAD at D119 could also abrogate the binding of BAD to Bcl-XL. Incubation of GST-BAD fusion proteins with GFP-Bcl-XL clearly revealed that BADD119G is able to bind to Bcl-XL (Figure 3A). The reciprocal experiment confirmed that BADD119G can bind to Bcl-XL (Figure 3B). This suggests that the observed pro-apoptotic activity of BADD119G (Figure 1) might be mediated by interaction of BADD119G with Bcl-XL and further suggests the hypothesis that Bcl-2 may be irrelevant for the pro-apoptotic activity of BAD. As a further test of the importance of Bcl-2 for BAD pro-apoptotic activity we determined whether the pro-apoptotic functions of BADwt and the BADD119G mutant were affected differently following co-transfection with Bcl-2 or Bcl-XL. Co-transfection of BADwt or BADD119G with Bcl-2 or Bcl-XL resulted in similar levels of expressed proteins in COS-7 cells (Figure 4A). Augmentation of caspase-3 activity following serum deprivation of transfected cells for 18 h was taken as a measure of apoptotic activity. Although a previous report demonstrated that BAD can reverse the anti-apoptotic activity of Bcl-XL but not that of Bcl-2 in hematopoietic transfectants,6 the augmented caspase-3 activity induced by BADwt or BADD119G (see Figure 1) was similarly inhibited following co-transfection of either of the BAD proteins with Bcl-2 or Bcl-XL (Figure 4B). Thus the pro-apoptotic function of BAD can be diminished by both Bcl-2 and Bcl-XL. More importantly, since the anti-apoptotic activity of Bcl-2 was similar in the presence of co-transfected BADwt or the BADD119G mutant with which it cannot interact, this observation further suggests that interaction between Bcl-2 and BAD is marginal for the pro-apoptotic function of BAD at least in COS-7 cells.
Effect of the D119G mutation on the physical and functional interaction of human BAD with Bcl-XL. (A) Interaction of Bcl-2 and Bcl-XL with GST-BAD fusion proteins. Cell lysates from HEK 293 cells transfected with GFP-tagged Bcl-XL or Bcl-2 (INPUT) were incubated with the indicated GST, GST-BAD or GST-BADD119G-fusion proteins. Proteins bound to the washed beads were identified by Western blots using an anti-GFP antibody. (B) Specific binding of BADwt and BADD119G to GST- Bcl-XL. Cell lysates from HEK 293 cells transfected with GFP-tagged-BADwt or BADD119G (INPUT) were incubated with GST- or GST-Bcl-XL-bound beads. The bound proteins were resolved by 12% SDS–PAGE and identified by Western blotting using an anti-BAD antibody (BAD). The loaded SDS–PAGE gel was stained with Coomassie Blue G-250 and shows specific binding of both BAD and BADD119G to Bcl-XL (lower panel). Molecular size markers are shown in kDa
Effect of BAD and BADD119G overexpression on Bcl-2 or Bcl-XL functions. (A) Expression of co-transfected GFP-tagged-BADwt or BADD119G with Bcl-2 or -Bcl-XL proteins. COS-7 cells were transiently co-transfected with the indicated expression vectors. Transfectants were harvested at 24 h following transfection and subjected to immunoblot analysis using the indicated antibodies. Arrows indicate the bands corresponding to GFP-Bcl-XL (upper) and GFP-Bcl-2 (lower). (B) BAD and BADD119G display a similar inhibitory effect on death repressor activity of Bcl-2 and Bcl-XL. COS-7 cells were co-transfected with the indicated vectors, and cultured in the absence of FCS for 18 h. Transfectants were washed with ice-cold PBS and caspase-3 activity was measured with the Caspase-3 Colorimetric Protease Assay Kit. The numbers represent the fold decrease in absorbance at 400 nm relative to the BAD/Mock or BADD119G/Mock transfectants. Each column displays the mean±S.D. of data from three separate experiments, which consistently produced quite similar results
The BADD119G mutant is targeted to mitochondria
It is well established that BAD is activated and targeted to the mitochondria during the induction of the apoptotic process following exposure of cells to a variety of apoptotic stimuli or following deprivation of survival signals.6,7,16,17,18 We thus used GFP-tagged BADwt and BADD119G constructs to investigate if mutation of BAD at D119 could alter its subcellular localization in COS-7 cells grown under normal or apoptosis-inducing conditions. Transfected COS-7 cells were therefore cultured in the presence (normal conditions) or absence (apoptosis-inducing conditions) of FCS. In FCS-deprived transfectants, confocal microscopical analysis revealed that both GFP-tagged BADwt and the BADD119G mutant are visible in the cytoplasm at 12 h following gene-transfection, especially at the perinuclear region. At 24 h following transfection, extensive overlap of the GFP-BAD signals with the mitochondrial MitoTracker marker dye was observed (Figure 5). When transfectants were cultured in the medium in the presence of FCS, the overlap of the BAD signals with mitochondria was substantially reduced although still observable (data not shown). These results suggested both that overexpression of BAD per se can result in localization of activate BAD to the mitochondria and that the mutant BADD119G maintains its ability to be targeted to the mitochondria both in response to apoptotic stimuli and even independently of them. (Figure 5). In contrast BAD deletion mutants, in which the BH3 domain has been completely removed, did not show any distinct overlap with MitoTracker regardless of the culture conditions. Thus, although the BH3 domain of BAD is required for targeting to mitochondria, mutation of D119 to G within this domain does not alter BAD mitochondrial localization. Since this mutation abrogates interaction of BAD with Bcl-2 this data also suggests that mitochondrial targeting of BAD does not require interaction with Bcl-2 and provides further support for the hypothesis that Bcl-2 interaction is not important for BAD pro-apoptotic function.
Mitochondrial localization of BADwt, BADD119G and dBAD. COS-7 cells were transfected with either pEGFP-BADwt (BAD), pEGFP-BADD119G or pEGFP-dBAD (aa1-72) (green as indicated) and deprived of FCS for 24 h to induce apoptosis. Cellular mitochondria were then stained with MitoTracker (red) for 30 min. The cells were fixed with methanol at −20°C. Images were obtained by confocal laser microscopy. An overlay of the BAD and mitochondrial images is indicated at right (Merge) in which co-localization is indicated by a yellow color. Bars, 10 μm
Discussion
In this paper we demonstrate that the pro-apoptotic activity of BAD is not mediated by interaction with Bcl-2 and subsequent inhibition of Bcl-2 anti-apoptotic activity. Studies carried out to date provided indirect evidence that BAD pro-apoptotic activity selectively neutralizes Bcl-XL, but not Bcl-2.6,8 In this study we demonstrated that BAD can neutralize both Bcl-2 and Bcl-XL at a similar level in COS-7 cells. In addition, a BADD119G mutant that is unable to bind Bcl-2, still retains its pro-apoptotic activity and can neutralize both Bcl-2 and Bcl-XL. These data strongly suggest that previous observations showing selective inhibitory effect of BAD on Bcl-XL is not general. Indeed, a recent report shows that BAD can induce apoptosis in cells overexpressing Bcl-2 or Bcl-XL in COS-7 and leukemic cells.19 Thus, previous data could not exclude the possibility that BAD cannot neutralize Bcl-2 because of its strong anti-apoptotic activity in some cell types. In this regard, we have conclusively shown that neutralization of Bcl-2 functions is totally dispensable for the pro-apoptotic activity of BAD.
Recently, a novel phosphorylation site, S170 has been identified as a regulator of BAD pro-apoptotic activity. Since mutation of this region (Ser170 to Ala) results in an enhanced ability of BAD to induce apoptosis, phosphorylation of BAD at S170 contributes to inactivation of its pro-apoptotic activity.20 Importantly, the inactive mutant (Ser170 to Asp) can still associate with Bcl-XL, although they do not induce apoptosis. Thus, the association of BAD with Bcl-XL is not sufficient to introduce apoptosis. This implies that though retained pro-apoptotic functions of our mutant BADD119G appears to depend upon its ability to interact with Bcl-XL, other or additional mechanism(s) may rather be involved in its pro-apoptotic activity.
As shown in this and previous papers,8 deletion of the entire BH3 domain of BAD abrogates both mitochondrial targeting and the pro-apoptotic functions of BAD. Thus, BH3 is a crucial region for pro-apoptotic functions of BAD. The BH3 domain of BAD contains two conserved residues, an L at position 114 and a D at aa119. A previous report indicated that mutation of the L residue strongly reduces the ability of BAD to interact with Bcl-XL or Bcl-2 and destroys BAD pro-apoptotic functions.8 In this study, we found that a single amino acid substitution of BAD at D119 within the BH3 domain selectively destroyed its ability to interact with Bcl-2, but not with Bcl-XL. In this regard, our study sheds light on the importance of the three-dimensional (3D) conformation of the BH3 domain in BAD function. To date, there have been several studies of the solution structures of Bcl-2 family members. Studies of Bid and Bcl-XL reveal that their BH3 domains contain an α-helical structure in which highly conserved hydrophobic residues (including the conserved L residue) are surface exposed and available for binding to a hydrophobic cleft in another molecule.21,22,23 This hydrophobic cleft is considered to be crucial for the formation of heterodimers with other Bcl-2 family members. In contrast, the side chain of the highly conserved charged residue D95 of Bid extends towards the interior of Bid yet is partially surface exposed and is thus in a position to form a salt bridge with another Bcl-2 family member.22 The predicted structure of BAD indicates that the charged D156 residue in murine BAD (D119 in human BAD) is similarly positioned.8 Thus, it seems likely that the D119 residue, which was mutated in this study, may contribute to the formation of a salt bridge which is crucial for heterodimerization with Bcl-2, but not with Bcl-XL.
Our data also indicate that the interaction of BAD with Bcl-2 is marginal for its recruitment to the mitochondria. Previous reports suggest that BAX can contribute to the recruitment of Bid to the mitochondria.24,25 However, there has been little information regarding the mechanism of mitochondrial targeting of BAD. Since neither we, nor others,26 can detect a distinct interaction between BAX and BAD in co-precipitation assays (Figure 2), it is unlikely that BAX can directly translocate BAD to the mitochondria. It is rather conceivable that the interaction with Bcl-XL is required for the translocation, since our mutant BADD119G can interact only with Bcl-XL and has an ability to translocate to the mitochondria. It must be noted however that tBid expression induces a conformational change in the amino-terminus of Bax, which allows BAK/BAX oligomerization and subsequent pore formation.27,28 Since the interaction between tBid and BAX is transient, it is difficult to detect their interaction and tBid is thus believed to act by a ‘hit-and-run’ mechanism to activate BAX. Since BAD is also cleaved and activated during the apoptotic process,29 and its pro-apoptotic functions absolutely require BAX/BAK expression,13 we cannot exclude the possibility that BAX can transiently interact with the cleaved BAD and transport it to the mitochondria. Whatever the transport mechanism is, mutant BADD119G can be translocated to the mitochondria as BADwt, and thus D residue within the BH3 domain is not necessary for the recruitment. Since mitochondrial targeting of BAD is obviously crucial for its apoptotic process, clarification of the precise molecular mechanism is an important avenue of future research.
Materials and methods
Cell culture and transfection
Green monkey kidney epithelial COS-7 and human embryonal HEK 293 cells were provided from the Japanese Cancer Research Resources Bank (Tokyo, Japan) and grown in Dulbecco-modified Eagles medium (D-MEM) supplemented with 10% fetal calf serum (FCS) at 37°C in 5% CO2. The cDNA (3 μg/cm2) constructs described below were transfected into these cells as indicated in the text using LipofectAMINE 2000 Reagent (GIBCO Life and Technologies, MD, USA) or Effectene (Qiagen, CA, USA).
Plasmids
Mutant BAD, in which Asp119 was replaced with Gly (BADD119G), was generated according to a PCR mutagenesis method.30 PCR was performed with the following oligonucleotide primers: 5′-CCGGAGGATGAGTGGCGAGT TTGTGGAC-3′ and 5′-GTCCACAAACTCGCCACTCATCCTCCGG-3′ (corresponding to nucleotides 342–369 in the human BAD cDNA). Full-length human BAD cDNA, (BADwt, kindly provided by Dr. JC Reed of the Burnham Institute), mutant BAD, full-length human Bcl-231 and Bcl-XL32 (kindly provided by Dr. Y Tsujimoto, Osaka University, Japan) were subcloned into GFP expression vectors pEGFPs (Clontec, CA, USA). BADD119G, BADwt, and Bcl-2 cDNAs were also subcloned into the prokaryote pGEX 4T expression vectors (Pharmacia) for the generation of GST-fusion proteins. BAD C-terminal BH3 domain deletion mutants were generated by PCR using two different sets of oligonucleotide primers: 5′-GCGGATCCGCGGCCCCGAAAGGGGCTGGGC-3′ and 5′-GCGGATCCGCCACCATGTT CCAGATCCCAGAG-3′ (corresponding to amino acids 1–72 in human BAD), and were subcloned into the mammalian expression plasmids pcDNA3 and pEGFP.
Reagents and antibodies
Antibody against BAD and Bcl-XL were purchased from Cell Signaling Technology (Beverly, MA, USA) and Transduction Labs (San Diego, CA, USA), respectively. Antibodies against Hsc70, BAD, Bcl-2, and GFP were from Santa Cruz Biotechnology Inc. (Santa Cruz, CA, USA).
Apoptosis quantification
To evaluate cell viability, cells were mixed with the same volume of 0.4% trypan blue solution, and immediately examined under the light microscope for dye exclusion. The caspase-3 colorimetric protease assay was performed following the manufacturer s protocol (CPP32/Caspase-3 Colorimetric Protease Assay Kit, MBL). Briefly 1×106–107 cells were lysed in 250 μl of Cell Lysis Buffer. Total cell lysates were collected and their protein concentration was evaluated using a Protein Assay (BioRad, NY, USA). These lysates (100 μg) were mixed with the same volume of 2× Reaction Buffer, and incubated with the 4 mM DEVD-pNA caspase-3 substrate (200 μM final concentration) at 37°C for 2 h. The samples were then analysed at 400 nm in a spectrophotometer.
Western blotting
As described previously,33 following washing with ice-cold PBS, cells were lysed with 50 μl of lysis buffer containing 100 mM NaCl, 2 mM EDTA, 1 mM PMSF, 1% NP-40 and 50 mM Tris-HCl (pH 7.2). Total cell lysates were collected, separated in 15% SDS–PAGE gels (80 μg/lane) and then electrophoretically transferred to PVDF membranes (Millipore, MA, USA) at 18 V for 70 min. Membranes were soaked into Block Ace (Dainippon Pharmacia Co., Japan) overnight at 4°C and washed with washing buffer containing 140 mM NaCl, 25 mM Tris-HCl (pH 7.8) and 0.05% Tween 20. The membranes were incubated with primary antibodies overnight at 4°C and thereafter incubated with the corresponding peroxidase-linked secondary antibodies (Amersham Pharmacia Biotech, IL, USA) for 1 h at room temperature. Blots were visualized by an ECL system (Amersham Pharmacia Biotech).
GST fusion protein expression and co-precipitation assays
pGEX plasmids encoding full-length BADwt, BADD119G, and Bcl-2 as GST fusion proteins were generated as described above. Expression of the fusion proteins was induced in BL21-competent bacteria with isopropyl-β-D-thiogalactoside and the fusion proteins were purified. For control experiments GST alone was generated from the pGEX-4T vector. Briefly, the bacteria were lysed with sodium deoxycholate, and the supernatant was collected and incubated with a 50% slurry of glutathione-sepharose 4B (Amersham Pharmacia Biotech). The beads were washed and resuspended in 10 mM Tris, pH 7.4, 150 mM NaCl, 5% glycerol, 1% phenylmethylsulfonyl fluoride and 1 μg/ml leupeptin. The concentration of GST-fusion-protein bound to the beads was estimated visually from Coomassie Blue-stained SDS–PAGE with albumin standards. For co-precipitation assays whole cell lysates (1 mg) of HEK 293 transfected cells described above were incubated with 20 μl of GST-fusion-protein-bound beads for 1–2 h at 4°C. The beads were washed at least five times with ice-cold lysis buffer followed by separation via SDS–PAGE. Proteins were then subjected to Western blot analysis.
Confocal laser microscopy
COS-7 cells were cultured on BSA-coated glass slides and transfected with the indicated expression vectors. Twenty-four hours following transfection, the cells were washed with cold PBS and thereafter fixed with cold methanol for 10 min. To visualize mitochondria, cells were incubated in D-MEM containing 500 nM MitoTracker RED CMXRos (Molecular Probes, OR, USA). Cells were washed three times with PBS and the coverslips were mounted on glass-slides using VECTASHILD (Vector Laboratories, Burlingame, CA, USA). Images were taken with an inverted confocal laser scanning microscopy (LSM 510; Zeiss, Jena, Germany) with a 100× oil objective lens, and processed by Adobe Photoshop version 5.0.
Abbreviations
- GFP:
-
green fluorescent protein
- GST:
-
glutathione S-transferase
References
Adams JM, Cory S . 1998 The Bcl-2 protein family: arbiters of cell survival Science 281: 1322–1326
Gross A, McDonnell JM, Korsmeyer SJ . 1999 BCL-2 family members and the mitochondria in apoptosis Genes Dev. 13: 1899–1911
Kelekar A, Thompson CB . 1998 Bcl-2-family proteins: the role of the BH3 domain in apoptosis Trends Cell Biol. 8: 267–271
Puthalakath H, Villunger A, O'Reilly LA, Beaumont JG, Coultas L, Cheney RE, Huang DCS, Strasser A . 2001 Bmf: A proapoptotic BH3-only protein regulated by interaction with the myosin V actin motor complex, activated by anoikis Science 293: 1829–1832
Oda E, Ohki R, Murasawa H, Nemoto J, Shibue T, Yamashita T, Tokino T, Taniguchi T, Tanaka N . 2000 Noxa, a BH3-only member of the Bcl-2 family and candidate mediator of p53-induced apoptosis Science 288: 1053–1058
Yang E, Zha J, Jockel J, Boise LH, Thompson CB, Korsmeyer SJ . 1995 Bad, a heterodimeric partner for Bcl-XL and Bcl-2, displaces Bax and promotes cell death Cell 80: 285–291
Zha J, Harada H, Yang E, Jockel J, Korsmeyer SJ . 1996 Serine phosphorylation of death agonist BAD in response to survival factor results in binding to 14-3-3 not BCL-XL Cell 87: 619–628
Zha J, Harada H, Osipov K, Jockel J, Waksman G, Korsmeyer S J . 1997 BH3 domain of BAD is required for heterodimerization J. Biol. Chem. 272: 24101–24104
O'Connor L, Strasser A, O'Reilly LA, Hausmann G, Adam JM, Cory S, Huang DC . 1998 Bim: a novel member of the Bcl-2 family that promotes apoptosis EMBO J. 17: 384–395
Li H, Zhu H, Xu CJ, Yuan J . 1998 Cleavage of BID by caspase 8 mediates the mitochondrial damage in the Fas pathway of apoptosis Cell 94: 491–501
Luo X, Budihardjo I, Zou H, Slaughter C, Wang X . 1998 Bid, a Bcl2 interacting protein, mediates cytochrome c release from mitochondria in response to activation of cell surface death receptors Cell 94: 481–490
Wei MC, Lindsten T, Mootha VK, Weiler S, Gross A, Ashiya M, Thompson CB, Korsmeyer SJ . 2000 tBID, a membrane-targeted death ligand, oligomerizes BAK to release cytochrome c Genes Dev. 14: 2060–2071
Zong W-X, Lindsten T, Ross AJ, MacGregor GR, Thompson CB . 2001 BH3-only proteins that bind pro-survival Bcl-2 family members fail to induce apoptosis in the absence of Bax and Bak Genes Dev. 15: 1481–1486
Hirai I, Wang H-G . 2001 Survival-factor-induced phosphorylation of Bad results in its dissociation from Bcl-x(L) but not Bcl-2 Biochem. J. 359: 345–352
Kitada S, Krajewska M, Zhang X, Scudiero D, Zapata JM, Wang HG, Shabaik A, Tudor G, Krajewski S, Myers TG, Johnson GS, Sausville EA, Reed JC . 1998 Expression and location of pro-apoptotic Bcl-2 family protein BAD in normal human tissues and tumor cell lines Am. J. Pathol. 152: 51–61
Nechushtan A, Smith CL, Lamensdorf I, Yoon S-H, Youle RJ . 2001 Bax and Bak coalesce into novel mitochondria-associated clusters during apoptosis J. Cell Biol. 153: 1265–1276
Kelekar A, Chang BS, Harlan JE, Fesik SW, Thompson CB . 1997 Bad is a BH3 domain-containing protein that forms an inactivating dimer with Bcl-XL Mol. Cell. Biol. 17: 7040–7046
Wang HG, Pathan N, Ethell IM, Krajewski S, Yamaguchi Y, Shibasaki F, McKeon F, Bobo T, Franke TF, Reed JC . 1999 Ca2+-induced apoptosis through calcineurin dephosphorylation of BAD Science 284: 339–343
Schimmer AD, Hedley DW, Pham NA, Chow S, Minden MD . 2001 BAD induces apoptosis in cells over-expressing Bcl-2 or Bcl-xL without loss of mitochondrial membrane potential Leuk Lymphoma 42: 429–443
Dramsi S, Scheid MP, Maiti A, Hojabrpour P, Chen X, Schubert K, Goodlett DR, Aebersold R, Duronio V . 2002 Identification of a Novel Phosphorylation Site, Ser-170, as a Regulator of Bad Pro-apoptotic Activity J. Biol. Chem. 277: 6399–6405
Chou JJ, Li H, Salvesen GS, Yuan J, Wagner G . 1999 Solution structure of BID, an intracellular amplifier of apoptotic signaling Cell 96: 615–624
McDonnell JM, Fushman D, Milliman CL, Korsmeyer SJ, Cowburn D . 1999 Solution structure of the proapoptotic molecule BID: a structural basis for apoptotic agonists and antagonists Cell 96: 625–634
Petros AM, Medek A, Nettesheim DG, Kim DH, Yoon HS, Swift K, Matayochi ED, Oltersdorf T, Fesik SW . 2001 Solution structure of the antiapoptotic protein bcl-2 Proc. Natl. Acad. Sci. USA 98: 3012–3017
Wang K, Gross A, Waksman G, Korsmeyer SJ . 1998 Mutagenesis of the BH3 domain of BAX identifies residues critical for dimerization and killing Mol. Cell Biol. 18: 6083–6089
Wang K, Yin X-M, Chao DT, Milliman CL, Korsmeyer SJ . 1996 BID: a novel BH3 domain-only death agonist Genes Dev. 10: 2859–2869
Ottilie S, Diaz JL, Horne W, Chang J, Wang Y, Wilson G, Chang S, Weeks S, Fritz LC, Oltersdorf T . 1997 Structural and functional complementation of an inactive Bcl-2 mutant by Bax truncation J. Biol. Chem. 272: 30866–30872
Wei MC, Lindsten T, Mootha VK, Weiler S, Gross A, Ashiya M, Thompson CB, Korsmeyer SJ . 2000 tBID, a membrane-targeted death ligand, oligomerizes BAK to release cytochrome c Genes Dev. 14: 2060–2071
Wei MC, Zong W-X, Cheng EH-Y, Lindsten T, Panoutsakopoulou V, Ross AJ, Roth KA, MacGregor GR, Thompson CB, Korsmeyer SJ . 2001 Proapoptotic BAX and BAK: a requisite gateway to mitochondrial dysfunction and death Science 292: 727–730
Condorelli F, Salomoni P, Cotteret S, Cesi V, Srinivasula SM, Alnemri ES, Calabretta B . 2001 Caspase cleavage enhances the apoptosis-inducing effects of BAD Mol. Cell Biol. 21: 3025–3036
Vallette F, Mege E, Reiss A, Milton A . 1989 Construction of mutant and chimeric genes using the polymerase chain reaction Nucleic Acid Res. 17: 723–733
Takaoka A, Adachi M, Okuda H, Sato S, Yawata A, Hinoda Y, Takayama S, Reed JC, Imai K . 1997 Anti-cell death activity promotes pulmonary metastasis of melanoma cells Oncogene 14: 2971–2977
Shimizu S, Eguchi Y, Kosaka H, Kamiike W, Matsuda H, Tsujimoto Y . 1995 Prevention of hypoxia-induced cell death by Bcl-2 and Bcl-xL Nature 374: 811–813
Yawata A, Adachi M, Okuda H, Naishiro Y, Takamura T, Hareyama M, Takayama S, Reed JC, Imai K . 1998 Prolonged cell survival enhances peritoneal dissemination of gastric cancer cells Oncogene 16: 2681–2686
Acknowledgements
We thank Drs. JC Reed (Burnham Institute) and Y Tsujimoto for providing cDNAs, and Ms Miyuki Itoh for helping with construction of plasmids and generation of mutant BAD. Supported by Grants-in-Aid for Cancer Research and for Scientific Research from the Ministry of Education, Science, Sports and Culture of Japan (M Aadachi, K Imai).
Author information
Authors and Affiliations
Corresponding author
Additional information
Edited by H Ichijo
Rights and permissions
About this article
Cite this article
Adachi, M., Imai, K. The proapoptotic BH3-only protein BAD transduces cell death signals independently of its interaction with Bcl-2. Cell Death Differ 9, 1240–1247 (2002). https://doi.org/10.1038/sj.cdd.4401097
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.cdd.4401097
Keywords
This article is cited by
-
Curriculum vitae of CUG binding protein 1 (CELF1) in homeostasis and diseases: a systematic review
Cellular & Molecular Biology Letters (2024)
-
Dual role of PID1 in regulating apoptosis induced by distinct anticancer-agents through AKT/Raf-1-dependent pathway in hepatocellular carcinoma
Cell Death Discovery (2023)
-
Combined inhibition of BADSer99 phosphorylation and PARP ablates models of recurrent ovarian carcinoma
Communications Medicine (2022)
-
Inhibition of BAD-Ser99 phosphorylation synergizes with PARP inhibition to ablate PTEN-deficient endometrial carcinoma
Cell Death & Disease (2022)
-
BAD sensitizes breast cancer cells to docetaxel with increased mitotic arrest and necroptosis
Scientific Reports (2020)