Abstract
The mammalian target of rapamycin (mTOR)/regulatory-associated protein of mTOR (Raptor) pathway transmits and integrates different signals including growth factors, nutrients, and energy metabolism. Nearly all these signals have been found to play roles in skeletal biology. However, the contribution of mTOR/Raptor to osteoblast biology in vivo remains to be elucidated as the conclusions of recent studies are controversial. Here we report that mice with a deficiency of either mTOR or Raptor in preosteoblasts exhibited clavicular hypoplasia and delayed fontanelle fusion, similar to those found in human patients with cleidocranial dysplasia (CCD) haploinsufficient for the transcription factor runt-related transcription factor 2 (Runx2) or those identified in Runx2+/− mice. Mechanistic analysis revealed that the mTOR-Raptor-S6K1 axis regulates Runx2 expression through phosphorylation of estrogen receptor α, which binds to Distal-less homeobox 5 (DLX5) and augments the activity of Runx2 enhancer. Moreover, heterozygous mutation of raptor in osteoblasts aggravates the bone defects observed in Runx2+/− mice, indicating a genetic interaction between Raptor and Runx2. Collectively, these findings reveal that mTOR/Raptor signaling is essential for bone formation in vivo through the regulation of Runx2 expression. These results also suggest that a selective mTOR/Raptor antagonist, which has been developed for treatment of many diseases, may have the side effect of causing bone loss.
Similar content being viewed by others
Main
Osteoblasts, the bone-forming cells active during bone development and remodeling,1, 2 are derived from bone marrow mesenchymal stem cells (BMSCs). The differentiation of osteoblasts from BMSCs is controlled by transcription factors that are expressed in a defined temporal and spatial sequence. Among them, runt-related transcription factor 2 (Runx2) is considered to be the master transcription factor3, 4 as mice deficient in Runx2 exhibit a lack of mineralization in the skeleton and absence of mature osteoblasts.5 Runx2-heterozygous mice display clavicular hypoplasia and delayed closure of the fontanelles, a phenotype resembling the cleidocranial dysplasia (CCD) syndrome caused by mutations of Runx2 in humans.6 Runx2 expression at appropriate times and sites is essential for bone development and bone remodeling. However, regulation of Runx2 expression, especially the upstream signaling pathways involved, has not been completely clarified.
The mammalian/mechanistic target of rapamycin (mTOR) is an evolutionarily conserved protein kinase. mTOR functions in two structurally and functionally distinct multiprotein complexes, namely mTORC1 and mTORC2,7 which are distinct in their unique components and downstream targets. mTORC1 contains Raptor and is sensitive to rapamycin, while mTORC2 contains Rictor and is resistant to rapamycin. S6 kinase1 (S6K1) is the mTORC1 downstream target and can phosphorylate a series of substrates including estrogen receptor alpha (ERα) and S6 ribosomal protein (S6) to control gene transcription, protein synthesis, and other biological processes.8, 9, 10 mTORC1 signaling is considered as the check-point of several extracellular and intracellular signals including growth factors, nutrients, energy metabolism and stress11 and has been the target for drug development in many diseases,11 which highlights the necessity of studying the role of mTORC1 in osteoblasts and bone development to monitor and avoid possible side effects on bone.
Recently, different studies of the role of mTORC1 signaling in osteoblast differentiation and bone development produced controversial results.12 Previous in vitro studies shown that mTORC1 can activate13, 14 or inhibit15 osteogenesis, and these controversies may be resulted from the differences in the cell types or cell differentiation-stages examined. Furthermore, disturbing of mTOR signaling in osteoblast-lineage cells induced various skeletal disorders. A mouse model with increased mTORC1 activity in neural crest-derived cells due to deletion of tuberous sclerosis 1 (Tsc1) led to increased bone mass through enlargement of the osteoprogenitor pool.16 Interesting, two independent studies indicated that osteoblast-specific inactivation of Tsc complex caused osteoblasts to differentiate poorly and produce disorganized bone.17, 18 On the other hand, Chen et al. reported that enhanced mTORC1 signaling due to heterozygous mutations in the fibrillin-1 gene resulted in osteopenia.19 Furthermore, decreased mTORC1 signaling with deletion of mTOR or Raptor in mesenchyme resulted in death shortly after birth and skeletal discrepancy.20 Raptor deletion in Osterix-expressing preosteoblasts led to osteopenia.21 In contrast, an in vitro study showed that depletion of Raptor promoted osteoblast differentiation of BMSCs.22 Taken together, these data reveal that the role of mTORC1 signaling in osteoblasts is still ambiguous and the underlying mechanisms have not been fully illuminated.
In the present study, we found that loss of mTORC1 signaling in preosteoblasts through the deletion of mTOR or Raptor in mice induced severe skeletal defects secondary to impaired osteogenesis and osteoblast differentiation. Further molecular mechanism studies revealed that the mTOR-Raptor-S6K1 axis could promote osteoblast differentiation through regulating Runx2 expression by augmenting the activity of Runx2 enhancer.
Results
mTOR deficiency in preosteoblasts causes a CCD phenotype with impaired osteoblast differentiation
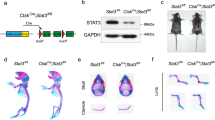
To determine the role of mTOR signaling in preosteoblasts in vivo, we generated conditional mTOR knockout mice (mTORfl/fl;Osx-cre, hereafter mTOROsx) (Figure 1a) by crossing mTORfl/fl mice with Osx-cre mice, a transgenic line in which Cre activation is confined to osteoblast precursors. Western blot assay confirmed the loss of mTOR protein in parietal bones from mTOROsx newborns (Figure 1b). Compared to WT mice and mice heterozygous for the mTOR floxed allele (mTORfl/+;Osx-cre, hereafter mTOROsx/+), mTOROsx mice showed slower growth rate (Figures 1c and d). Alizarin red and Alcian blue staining showed although newborn mTOROsx mice had similar skeletal size to their WT littermates (Figure 1e), mTOROsx mice exhibited the CCD phenotype with clavicular hypoplasia and hypomineralization of the calvarium (Figure 1f), features of mice heterozygous for Runx2.
Preosteoblast-specific knockout of mTOR-induced CCD-like bone defects in mice. (a) Illustration of mTOR deletion in Osx-expressing preosteoblasts. (b) Western blot assay of mTOR in parietal bone lysate of WT and mTOROsx newborns. (c) Representative view of 4-week-old WT, mTOROsx/+ and mTOROsx mice. (d) Body weight of male WT, mTOROsx/+and mTOROsx littermates measured at different age points (n=5 for each group, t-test). Data represent means±s.d. *P<0.05 in comparison with WT mice, #P<0.05 when compared with mTOROsx/+ mice. (e) Representative images of skeleton of WT, mTOROsx/+ and mTOROsx newborn littermates by alcian blue and alizarin red staining. (f) Clavicular hypoplasia and calvarial hypomineralization of mTOROsx newborns. Arrow indicates calvarial lesions in mTOROsx mice
In addition to the clavicular and calvarial phenotypes, mTOROsx mice displayed a substantial reduction in bone mass. As shown in Figures 2a and b, femoral trabecular bone of 4-week-old male mTOROsx mice displayed an approximately 70% reduction in bone volume fraction (BV/TV) compared to WT littermates while mTOROsx/+ showed an intermediate effect (Figure 2b). The significant reduction in bone mass between mTOROsx and mTOROsx/+ confirmed that the decreased bone density in mTOROsx mice is due to the loss of mTOR in osteoblasts, not to the presence of the Cre transgene. mTOROsx mice also displayed a decrease of trabecular number (Tb.N., Figure 2c) and trabecular thickness (Tb.Th., Figure 2d). mTORosx mice also displayed a dramatic reduction of the cortical thickness (Ct.Th., Figure 2e).
mTOR deficiency in osteoblasts resulted in reduced bone mass with impaired osteoblast differentiation. (a) Micro-CT images of femur from 4-week-old WT, mTOROsx/+ and mTOROsx littermates. (b–e) Quantitative parameters of Micro-CT. BV/TV: bone volume/total volume, Tb.N.: trabecular number, Tb.Th.: trabecular thickness, Ct.Th.: cortical thickness. Data represent means±s.d. *P<0.05, n=5. (f) Calvarial osteoblasts were cultured with osteogenic medium from 5-day-old WT and mTOROsx littermates. ALP staining and alizarin red staining were performed to analyze osteoblast differentiation on the 7th and 14th day respectively. (g) Immunohistochemistry for Runx2 of distal femur from 7-day-old WT and mTOROsx mice. (h) Number of Runx2-positive osteoblasts counted in the distal femur from 7-day-old WT and mTOROsx mice. Data represent means±s.d. *P<0.05, n=3. (i) In situ hybridizationfor Col1α1 and Ocn in femur from 7-day-old WT and mTOROsx littermaters. (j) Number of Col1α1-positive osteoblasts counted in the trabecular bone of distal femur from 7-day-old WT and mTOROsx littermaters. Data represent means±s.d. *P<0.05, n=3. (k) Number of Ocn-positive osteoblasts calculated in the cortical bone of distal femur from 7-day-old WT and mTOROsx littermaters. Data represent means±s.d. *P<0.05, n=3
We next examined whether the abnormal osteogenesis in mTORosx mice was a result of inadequate osteoblast differentiation. We cultured calvarial cells from mTOROsx and WT mice and found that mTOROsx calvarial cells showed reduced osteoblast differentiation, revealed by decreased alkaline phosphatase (ALP) activity and fewer calcified nodules, measured by alizarin red staining (Figure 2f). Consistent with the reduced ossification and osteoblast differentiation, expression of the characteristic osteoblast marker genes, collagen 1α1 (Col1α1, Figures 2i and j) and osteocalcin (Ocn, Figures 2i and k) were reduced in osteoblasts from mTORosx mice in vivo accompanied with the decrease of Runx2 expression (Figures 2g and h). Taken together, the CCD phenotype and reduced bone mass, possibly secondary to impaired osteoblastic differentiation in mTORosx mice, supported the hypothesis that mTOR is critical for bone development.
Raptor deficiency in preosteoblasts also causes a CCD phenotype and reduced bone mass with impaired osteoblast differentiation
To further investigate the specific contribution of mTORC1 in osteoblasts, we generated conditional Raptor knockout mice (Rapfl/fl;Osx-cre, hereafter RapOsx) by mating Rapfl/fl mice with Osx-cre mice (Figure 3a). Western blot assay revealed the loss of Raptor protein in parietal bones from RapOsx mice (Figure 3b). Similar to mTORosx mice, Raposx mice showed a slower growth pattern after birth (Figures 3c and d). The newborn RapOsx mice also exhibited the CCD phenotype with clavicular hypoplasia and hypomineralization of the calvarium (Figure 3f) although RapOsx mice had similar skeletal size to their WT littermates (Figure 3e). Hypocalcification of calvarial and clavical bones were confirmed in 4-week-old RapOsx mice by micro-CT and X-ray analysis (Figure 3g). Moreover, 4-week-old RapOsx mice had an osteopenic phenotype with decreased BV/TV, Tb.N., Tb.Th. and Ct.Th. in the femur (Figures 4a–e). Further, 6-month-old RapOsx mice showed CCD phenotype with clavicular hypoplasia and hypomineralization of the calvarium (Supplementary Figure 1A). Micro-CT scanning confirmed calvarial hypoplasia (Supplementary Figure 1B) and showed deceased BV/TV, Tb.N., Tb.Th. and Ct.Th. in RapOsx mice in comparison with WT mice (Supplementary Figure 1C–G). The significant difference between RapOsx mice and RapOsx/+ mice accompanied with the skeletal defects in 6-month-old RapOsx mice confirmed that it is the loss of Raptor in osteoblasts, not the presence of the Cre transgene, that is responsible for the bone defects in RapOsx mice.
Ablation of Raptor in preosteoblasts resulted in CCD-like phenotype. (a) Illustration of Raptor deletion in Osx-expressing preosteoblasts. (b) Expression of Raptor in parietal bone from WT and RapOsx newborns. (c) Representative view of 4-week-old WT, RapOsx/+ and RapOsx mice. (d) Body weight of male WT, RapOsx/+ and RapOsx littermates measured at different age points (n=5 for each group, t-test). Data represent means±s.d. *P<0.05 in comparison with WT mice, #P<0.05 when compared with RapOsx/+ mice. (e) Representative images of skeleton from WT, RapOsx/+and RapOsx newborn littermates by alcian blue and alizarin red staining. (f) Clavicular hypoplasia and calvarium hypomineralization of RapOsx newborns. Arrow indicates calvarial lesions in mTOROsx mice. (g) Micro-CT of skulls and X-ray of clavicles from 4-week-old WT, RapOsx/+ and RapOsx mice. Arrow indicates calvarial lesions and hypoplastic clavicule in mTOROsx mice
Raptor deficiency in osteoblasts displayed reduced bone mass with impaired osteoblast differentiation. (a) Micro-CT images of femur from 4-week-old WT, RapOsx/+ and RapOsx littermates. (b–e) Quantitative parameters of Micro-CT. Data represent means±s.d. *P<0.05, n=5. (f) ALP staining and alizarin red staining of WT and RapOsx calvarial cells induced by osteogenic medium. Data represent means±s.d. *P<0.05, n=3. (g) Immunohistochemistry for Runx2 of distal femur from 7-day-old WT and RapOsx mice. (h) Number of Runx2-positive osteoblasts counted in the trabecular bone of distal femur from WT and RapOsx mice. Data represent means±s.d. *P<0.05, n=3. (i) In situ hybridizationfor Col1α1 and Ocn in femur from 7-day-old WT and RapOsx littermaters. (j) Number of Col1α1-positive osteoblasts counted in the trabecular bone of distal femur of 7-day-old WT and RapOsx littermaters. Data represent means±s.d. *P<0.05, n=3. (k) Number of Ocn-positive osteoblasts counted along the cortical bone of distal femur from 7-day-old WT and RapOsx littermaters. Data represent means±s.d. *P<0.05, n=3. (l) Representative images of alizarin red and calcein double labeling of femur from WT and RapOsx littermates. (m,n) Quantitative parameters mineral apposition rate (MAR) and bone formation rate (BFR) in WT and RapOsx mice. Data represent means±s.d. *P<0.05, n=3
Primary osteoblast cultures from RapOsx mice exhibited decreased osteoblast differentiation, determined by decreased ALP activity and mineralized matrix production (Figure 4f). Consistent with this, RapOsx mice displayed decreased Col1α1 and Ocn mRNA expression in vivo (Figures 4i–k) as well as reduced Runx2 protein expression (Figures 4g and h). The similar bone phenotype between RapOsx mice and mTOROsx mice supported the hypothesis that mTOR drives skeletal development mainly via the mTORC1 complex. The decreased bone mass in both mTOROsx and RapOsx mice might arise through increased bone resorption. To address this, we analyzed femurs from 4-week-old mice for the presence of tartrate-resistant acid phosphatase (TRAP)-positive osteoclasts. In comparison with WT littermates, both mTOROsx and RapOsx mice displayed a modest decrease in TRAP-positive osteoclasts (Supplementary Figures 2A and B), indicating that the bone lesions in mTOROsx or RapOsx mice are attributable to impaired bone formation as opposed to decreased bone resorption. Further confirming the decrease in osteoblast activity, 4-week-old RapOsx mice demonstrated a decreased bone formation rate (BFR) and mineral apposition rate (MAR) as determined by alizarin red and calcein labeling (Figures 4l and m). Collectively, these data support the hypothesis that mTOR/Raptor (mTORC1) is critical for osteoblast activity and anabolic bone formation.
S6K1 is a downstream factor of mTOR/Raptor in osteoblasts
S6K1 is the most important downstream regulator of mTORC1 and plays crucial roles in development and aging. To determine whether S6K1 could function downstream of mTOR/Raptor in osteoblasts, we analyzed the level of phospho-S6K1in 7-day-old mice by immunohistochemistry. An antibody specific to phospho-S6K1 (T389) demonstrated robust signal in osteoblast-lining cells in subchondral trabecular bone and in adjacent osteocytes (Figures 5a and d). At the same time, both mTOROsx and RapOsx mice showed decreased S6K1 phosphorylation in comparison with their WT littermates (Figures 5a). We also analyzed lysates from parietal bone of mTOROsx and RapOsx newborn mice. As shown in Figures 5c and f, although the expression of S6K1 is comparable between WT mice and mTOROsx or RapOsx mice, the level of phosphorylation of S6K1 in mTOROsx or RapOsx mice was dramatically decreased. To examined whether the reduction of S6K1 phosphorylation could be responsible for impaired osteoblast differentiation, the expression of constitutively-active S6K1 (CAS6K1, T390E) was enforced in Raptor-deficient calvarial osteoblasts. CAS6K1 overexpression was determined by immunoblotting (Figure 5h) and the efficiency of CAS6K1 was confirmed by increased phosphorylation of ribosomal protein S6 (P-S6, S 235/236). Furthermore, the impaired osteoblast differentiation of RapOsx calvarial cells was significantly improved by CAS6K1 overexpression as demonstrated by increased ALP activity and mineralized nodule formation (Figure 5g). Similarly, enforced expression of CAS6K1 rescued the decreased expression of Runx2 (Figure 5i), Col1α1 (Figure 5j) and Ocn (Figure 5k) in Raptor-deficient calvarial cells. These data demonstrate that the impaired S6K1 activity is responsible for the impaired osteoblast differentiation of Raptor-deficient calvarial osteoblasts.
S6K1 is the downstream factor of mTOR/Raptor in osteoblasts. Immunohistochemistry for P-S6K1(T389) in femur from 7-day-old WT and mTOROsx littermaters. (a) Number of P-S6K1-positive osteoblast counted in the distal femur of WT and mTOROsx mice. Data represent means±s.d. *P<0.05, n=3. (b) Western blot assay for cell lysates from parietal bone of WT and TOROsx newborn mice. IHC for phospho-S6K1 in femur from P7 WT and RapOsx littermaters. (c) Immunohistochemistry for P-S6K1(T389) in femur of 7-day-old WT and RapOsx littermaters. (d) Number of P-S6K1-positive osteoblast counted in the distal femur of WT and RapOsx mice. Data represent means±s.d. *P<0.05, n=3. (e) Western blot assay for cell lysates from parietal bone of WT and RapOsx newborns. (f) ALP staining and alizarin red staining of WT and RapOsx calvarial cells that were infected with lentivirus expressing CAS6K1 or GFP and induced by osteogenic medium. (g) Western blot assay for cell lysates from WT and RapOsx calvarial cells that were infected with lentivirus expressing CAS6K1 or GFP followed by osteogenic medium. (i–k) The relative mRNA levels of Runx2, Col1α1 and Ocn were quantified by qPCR. Data represent means±s.d. *P<0.05, n=3
The mTOR-Raptor-S6K1 axis signaling regulates Runx2 expression in osteoblasts
Both mTOROsx and RapOsx mice displayed a striking similarity to Runx2+/− mice with features including CCD phenotype and reduced bone mass, suggesting that mTORC1 may regulate Runx2 expression. This hypothesis was supported by the facts that both mTOROsx and RapOsx mice displayed decreased Runx2 protein expression in vivo (Figures 2g and h and Figures 4g and h) and that enforced expression of CAS6K1 increased Runx2 mRNA level (Figure 5i). Likewise, CAS6K1 was able to rescue the decreased Runx2 expression in Raptor-deficient osteoblasts at the protein level (Figure 5h). To exclude the possibility that the decreased Runx2 expression is due to the different cell populations, we cultured BMSCs from Raptorfl/fl mice and then infected the cells with adenovirus expressing GFP and CRE recombinase. As shown in Figure 6a, CRE adenovirus led to reduced expression of Raptor accompanied by decreased levels of Runx2 protein. Thus, the effect of Raptor on regulating Runx2 expression cannot be attributed to differences in cell population. Consistent with the lower Runx2 protein level, CRE adenovirus caused impaired osteoblast differentiation (Figure 6b). The change of Runx2 protein level was reflected in a dramatic decrease of Runx2 mRNA level by CRE adenovirus (Figure 6c), which was accompanied by a decrease of Runx2 downstream osteogenic genes such as Col1α1(Figure 5d) and Ocn (Figure 5e). These data suggest that mTORC1 could regulate Runx2 expression at the transcript level. To further determine whether the impaired osteoblast differentiation induced by inactivation of mTORC1 was due to the reduction of Runx2 expression, we analyzed the effects of enforced Runx2 expression on Raptor-deficient osteoblasts. Calvarial osteoblastic cells from RapOsx mice were infected with lentivirus expressing Runx2 (Lenti-Runx2) or GFP (Lenti-GFP). Western blot assay confirmed Runx2 overexpression in RapOsx osteoblasts (Figure 6f). The decreased osteoblast differentiation in RapOsx calvarial cells was rescued by Runx2 overexpression as determined by ALP activity and mineralized nodule formation (Figure 6g). Consistent with this, the decreased expression of Runx2 downstream genes including Col1α1and Ocn was rescued by Runx2 overexpression (Figures 6h and i). These results indicated that mTOR/Raptor-S6K1 signaling promoted skeletal development and osteoblast differentiation through regulation of Runx2 expression.
mTOR/Raptor-S6K1 signaling regulates Runx2 expression in osteoblast. (a) Western blot assay for cell lysates from Raptorfl/fl BMSCs that were infected with adenovirus expressing GFP and CRE recombinase. (b) ALP staining and alizarin red staining of BMSCs induced by osteogenic medium. (c–e) The relative mRNA levels of Runx2, Col1α1 and Ocn were quantified by qPCR. Data represent means±s.d. *P<0.05, n=3. (f) Western blot assay for cell lysates from WT and RapOsx calvarial cells infected with lentivirus expressing Runx2 or GFP. (g) ALP staining and alizarin red staining of WT and RapOsx calvarial cells that were infected with lentivirus expressing Runx2 or GFP and induced by osteogenic medium. (h–i) The relative mRNA levels of Col1α1 and Ocn were quantified by qPCR. Data represent means±s.d. *P<0.05, n=3
S6K1 regulates Runx2 expression via its enhancer
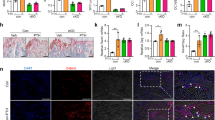
Next, we intended to investigate the mechanisms by which mTORC1 regulates Runx2 expression. First, we examined the effects of CAS6K1 on activity of the Runx2 promoter by an in vitro luciferase assay. As shown in Supplementary Figure 3A, CAS6K1 had no effects on activation of the Runx2 promoter. Kawane et al. demonstrated that the enhancer of Runx2 plays an important role in directing Runx2 expression in osteoblasts.23 Consequently, we next examined the effects of CAS6K1 on the Runx2 enhancer. As shown in Figure 7a, the activity of the luciferase reporter driven by the 3x 89 bp Runx2 core enhancer (Runx2 enhancer) was promoted by CAS6K1 expression. Furthermore, we found Runx2 enhancer activity decreased in RapOsx calvarial cells compared with WT group (Figure 7b), which confirmed mTORC1-S6K1may regulate Runx2 expression through its enhancer. The Runx2 core enhancer is bound by Distal-less homeobox 5 (DLX5) and MEF2C,23 so we examined the effects of CAS6K1 on DLX5- and MEF2C-induced Runx2 enhancer activity. As shown in Figure 7c, CAS6K1 could increase DLX5-induced Runx2 enhancer activity, but not MEF2C (Supplementary Figure 3B). However, we were unable to detect any interaction between S6K1 and DLX5 by co-immunoprecipitation (coIP) experiments (Supplementary Figure 1C). This is consistent with the fact that DLX5 does not contain the S6K1 phosphorylation motif RxRxxS/T (where x could be any amino acid). Thus, S6K1 may regulate Runx2 enhancer activity by phosphorylating proteins other than DLX5. It has been reported that S6K1 promotes estrogen receptor α (ERα) activity by phosphorylating it on S167 in cancer cells9, 24 and ERα can regulate Runx2 expression in osteoblast progenitors.25, 26, 27 We hypothesized that S6K1 may regulate Runx2 enhancer activity by phosphorylating ERα. First, we confirmed the protein interaction between S6K1 and ERα (Figure 7d). Next, we found that ERα could increase the activity of Runx2 enhancer, while co-transfection of CAS6K1 was able to promote this effect (Figure 7e).We then examined whether mutation of the S6K1 phosphorylation motif renders ERα refractory to activation by S6K1. The S6K1 phosphorylation site of ERα was mutated by converting Ser171 into alanine.28 We found that when compared with WT ERα, ERα-S171A was resistant to CAS6K1 effects on the activity of Runx2 enhancer (Figure 7f). These data indicate that S6K1 regulates Runx2 enhancer activity by phosphorylation of ERα. To further test this hypothesis, we analyzed the phosphorylation of ERα (P-ERα, S167) in 7-day-old mTOROsx and RapOsx mice by immunohistochemistry. As shown in Supplementary Figures 3D,E and Figures 7g and h, P-ERα-positive cells decreased in both mTOROsx and RapOsx mice compared to WT mice, which confirmed that S6K1 regulates Runx2 enhancer activity by phosphorylation of ERα.
S6K1 regulates Runx2 expression via its enhancer. (a) Effects of CAS6K1 on Runx2 enhancer activity. 3X 89 bp Runx2 enhancer corn sequence-driven luciferase reporter and pRL-TK were co-transfected with or without CAS6K1 in C3H10T1/2 cells. Data represent means±s.d. *P<0.05, n=3. (b) Effects of Raptor deficiency on Runx2 enhancer activity. 3X 89 bp Runx2 enhancer corn sequence-driven luciferase reporter and pRL-TK were co-transfected in WT and RapOsx calvarial cells. Data represent means±s.d. *P<0.05, n=3. (c) CAS6K1 promoted DLX5-induced Runx2 enhancer activity. 3X 89 bp Runx2 enhancer corn sequence-driven luciferase reporter and pRL-TK were co-transfected with DLX5 and different concentration of CAS6K1 in C3H10T1/2 cells. Data represent means±s.d. *P<0.05, n=3. (d) Co-immunoprecipitation analysis of S6K1 and ERα. Flag-CAS6K1 and HA-ERα expression constructs were co-transfected in 293Tcells for 48 h and co-immunoprecipitation analysis was performed. (e) CAS6K1 promoted ERα-induced Runx2 enhancer activity. 3X 89 bp Runx2 enhancer corn sequence-driven luciferase reporter and pRL-TK were co-transfected with ERα and CAS6K1 in C3H10T1/2 cells. Data represent means±s.d. *P<0.05, n=3. (f) Effects of mutated ERα of S6K1 phosphorylation motif on Runx2 enhancer activity. Luciferase assay was performed by co-translating CAS6K1 plasmid with either ERα or ERα S171A, the later one could not be phosphorylated by S6K1. Data represent means±s.d. *P<0.05, NS, P>0.05 n=3. (g) Immunohistochemistry for P-ERα (S167) in femur from 7-day-old WT and RapOsx littermaters. (h) Number of P-ERα-positive osteoblasts counted in the distal femur of WT and RapOsx mice. Data represent means±s.d. *P<0.05, n=3. (i) Effects of CAS6K1, DLX5 and ERα on Runx2 enhancer activity. DLX5 and ERα could synergistically increase the activity of Runx2 enhancer and CAS6K1 could further promote the effects. Data represent means±s.d. *P<0.05, n=3. (j) Co-immunoprecipitation analysis of ERα and DLX5. Flag-ERα and HA-DLX5 expressing plasmid were co-transfected into 293T cells. After 48 h, immunoprecipitation was performed with whole cell lysate and followed by western blot assay with indicated antibodies. (k) Co-immunoprecipitation analysis of ERα and DLX5 in calvarial osteoblasts. Immunoprecipitation of ERα and IgG was performed with whole cell lysate of calvarial osteoblasts and followed by western blot assay with DLX5 and ERα. (l) Co-immunoprecipitation analysis of DLX5 with ERα and phosphorylation site mutant ERα. Flag-DLX5 was co-transfected with HA-ERα, HA-ERα-S171E or HA-ERα-S171A respectively in 293T cells for 48 h. Immunoprecipitation of Flag and HA was conformed respectively and western blot assay with indicated antibodies. (m) CHIP-qPCR analysis of ERα in Runx2 enhancer region using WT and RapOsxcalvarial osteoblasts. Data represent means±s.d. *P<0.05, n=3. (n) CHIP-qPCR analysis of DLX5 in Runx2 enhancer region using WT and RapOsxcalvarial osteoblasts. Data represent means±s.d. *P<0.05, n=3
Interestingly, we found that DLX5 and ERα could synergistically increase the activity of Runx2 enhancer and that CAS6K1 could further promote these effects (Figure 7i). As the 89 bp Runx2 core enhancer lacks an ERα binding motif, we hypothesized that ERα might augment Runx2 enhancer activity through interaction with DLX5. Indeed ERα and DLX5 could interact when both were ectopically expressed in 293T cells (Figure 7j). Next, we confirmed this protein interaction between ERα and DLX5 in calvarial osteoblasts (Figure 7k). To further analyze whether the interaction of ERα and DLX5 was dependent on phosphorylation of ERα, WT and phosphorylation site mutant ERα (ERα-WT, ERα-S171E, ERα-S171A) were co-transfected with DLX5 in 293T cells. As shown in Figure 7l, DLX5 could interact with all three ERα protein, which suggested that the interaction of ERα and DLX5 was independent of ERα phosphorylation and excluded the possibility that mTORC1 regulate Runx2 enhancer activity by affecting the interaction of ERα and DLX5. Next we analyze the binding of ERα and Dlx5 to Runx2 enhancer by CHIP-qPCR in RapOsx calvarial osteoblasts. As shown in Figures 7m and n, the binding of ERα and Dlx5 to Runx2 enhancer decreased in RapOsx calvarial osteoblasts in comparison to the WT groups, which indicated that mTORC1-S6K1 signaling can promote the binding of the transcriptional factors ERα and Dlx5 to Runx2 enhancer. These data support the hypothesis that the mTOR-Raptor-S6K1 axis can regulate Runx2 expression via its enhancer, probably by phosphorylating ERα which interacts with DLX5 and augments Runx2 enhancer activity.
mTORC1 interacts genetically with Runx2 in vivo
Based on the in vitro results above, we next questioned whether mTORC1 is an upstream regulator of Runx2 in vivo. We postulated that if mTORC1 indeed regulates expression of Runx2, then reduced activity of mTORC1 in parallel with Runx2 haploinsufficiency in vivo should aggravate the bone defects in Runx2+/− mice. To test this hypothesis, we generated RapOsx/+Runx2+/− mutant mice by crossing RapOsx/+and Runx2+/−mice and analyzed skeletal preparations from newborn WT, RapOsx/+, Runx2+/− and RapOsx/+Runx2+/− mice. As shown in Figure 8a, the body weights of Runx2+/− and RapOsx/+Runx2+/− mice were slightly lower than those of WT and RapOsx/+mice and the skeletons of Runx2+/− and RapOsx/+Runx2+/− mice were consistently smaller (Figure 8b). Runx2+/− mice displayed the previously reported CCD-like phenotype. Heterozygous deletion of Raptor resulted in minor skeletal defects in both the clavicle and calvarium (Figures 8c–e). However, RapOsx/+Runx2+/− mice showed far more severe bone defects than Runx2+/− mice, including a larger hypoplastic area of the cranial bones (Figures 8c and d) and barely visible clavicles (Figures 8c and e). Taken together, these results suggest a genetic relationship between mTORC1 and Runx2, and provide in vivo evidence that mTORC1 can regulate osteoblast function through Runx2.
mTORC1 genetically interact with Runx2. (a) Body weight analysis of neonatal WT, RapOsx/+, Runx2+/−, and RapOsx/+Runx2+/−mice. (b) Alcian blue and alizarin red stained skeletal preps of neonatal WT, RapOsx/+, Runx2+/−, and RapOsx/+Runx2+/−mice. (c) Aggravated clavicular hypoplasia and calvarium hypomineralization in RapOsx/+Runx2+/−mice. (d) Relative non-calcified area of calvarial bones of neonatal WT, RapOsx/+, Runx2+/−, and RapOsx/+Runx2+/−mice. (e) Length of clavicles of neonatal WT, RapOsx/+, Runx2+/−, and RapOsx/+Runx2+/−mice. Data represent means±s.d. *P<0.05, n=3
Discussion
In the present study, we revealed the critical role of mTOR/Raptor signaling in osteoblasts and bone development through conditional knockout of mTOR and Raptor, respectively. Deletion of mTOR in preosteoblasts induced marked skeletal defects, including dwarfism with short limbs, impaired ossification of the cranial bones, hypoplasia of the clavicles and reduced bone mass, which support the conclusion that mTOR is essential for both endochondral and intramembranous ossification. Further, preosteoblast-specific loss of Raptor reproduced almost all skeletal phenotypes of mTOROsx mice, suggesting that mTOR functions mainly though mTORC1 in osteoblasts and osteogenesis. On the other hand, other researchers have found that continuous mTORC1 activation led to bone defects.18, 19 These results suggested that mTOR signaling is regulated precisely during bone development, and either upregulation or downregulation of mTORC1 signaling may result in bone diseases. Although the precise balance point of mTORC1 signaling in osteoblasts will need to be investigated in further studies, the possible side effects on bone of changes in mTORC1 signaling caused by any agent which has been or is intended to be used as a treatment target in a range of diseases11 should be taken into account.
MSCs can differentiate into multiple lineages including the osteoblast, chondrocyte and adipocyte lineages. While it has been demonstrated that mTOR signaling is essential for both chondrocyte20, 29 and adipocyte30 differentiation, there is limited and controversial information available regarding the independent role of mTOR in osteoblast and osteogenesis.12 In this study, we revealed several lines of evidence showing that inactivation of mTORC1 inhibited osteoblast differentiation and bone formation. First, osteoblast-specific deletion of either mTOR or Raptor led to decreased expression of osteoblastic markers in vivo. Second. BFR was reduced in RapOsx mice. Third, osteoblast differentiation of RapOsx parietal cells was impaired. Taken together, we believe that physiological mTORC1 signaling is essential for osteoblast differentiation and bone formation.
There is limited information available about the mechanisms by which mTORC1 promotes osteoblast differentiation. It has been demonstrated that S6K1can positively regulate the differentiation of both chondrocytes29 and adipocytes.10 In our current study, we provided evidence indicating that S6K1 is the major downstream regulator of mTORC1 in osteoblasts and that the mTOR/Raptor-S6K1 axis could promote osteoblast differentiation and osteogenesis.
Mutations of Runx2 result in CCD in humans and Runx2+/− mice exhibited CCD-like phenotypes except for supernumerary teeth.4, 5, 6 However, some CCD patients do not carry Runx2 mutations, and a decrease to 70% of wild-type Runx2 levels can result in CCD syndrome in mice.31 These results indicate that CCD may result from other mechanisms that regulate Runx2 expression. Here, we provided evidence that mTOR/Raptor-S6K1 signaling promotes Runx2 expression. Preosteoblast-specific deletion of either mTOR or Raptor results in bone defects, resembling the CCD phenotype and decreased Runx2 expression in vivo. Moreover, Raptor-deficient BMSCs and parietal cells displayed reduced Runx2 expression, and the impaired osteoblast differentiation in RapOsx parietal cells can be rescued by Runx2 overexpression. The reduced Runx2 expression in RapOsx parietal cells can be rescued by enforced CAS6K1 expression. Molecular experiments demonstrated that CAS6K1 can augment the activity of Runx2 enhancer, but not the Runx2 promoter. Finally, heterozygous Raptor in osteoblasts aggravates skeletal phenotypes of Runx2+/− mice, supporting a genetic link between mTORC1 signaling and Runx2. Taken together, these data suggest that physiological mTORC1 signaling is essential for Runx2 expression. Meanwhile, hyperactivation of mTORC1 in osteoblasts-induced bone defects were also related to decreased Runx2 expression.18, 19
Our study clarifies the critical role of mTOR/Raptor signaling in osteoblasts and bone development and demonstrates that mTOR-Raptor-S6K1 can regulate Runx2 expression, providing new sights into skeletal dysplasias such as CCD.
Materials and methods
Mice
mTORfl/fl mice bearing loxP sites flanking exons 1–5 of the mTOR gene (Stock No: 011009) and Rapfl/fl mice bearing loxP sites flanking exons 6 of the Raptor gene (Stock No: 013188) were purchased from the Jackson Laboratory (Bar Harbor, Maine, USA). mTORfl/fl mice were crossed with Osx-cre mice (Osx1-GFP::Cre mice were purchased from the Jackson Laboratory (Stock No: 006361)) to generate mTORfl/fl;Osx-cre (mTOROsx) and mTORfl/+;Osx-cre (mTOROsx/+) mice. Rapfl/fl were crossed with Osx-cre mice to generate Raptorfl/fl;Osx-cre (RapOsx) and Raptorfl/+;Osx-cre (RapOsx+) mice. Runx2+/− mice were kindly provided by Professor Gerard Karsenty’s laboratory. All mice were C57BL/6 background. All mice were bred and maintained under specific pathogen-free conditions in the institutional animal facility of the Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences. All experiments were performed with the protocol approved by the Animal Care and Use Committee of the Shanghai Institute of Biochemistry and Cell Biology.
Skeletal whole mount staining
Skeletal whole mount staining with Alcian blue and Alizarin red was performed as described previously.32 Mice were killed with CO2, and all skin was carefully removed. Specimens were dehydrated in 95% alcohol for 24 h, followed by cartilage staining in Alcian blue solution for 42 h at 37 °C. After staining, specimens were washed twice in 95% alcohol for 2 h, cleared in 1% KOH for 5 h and stained in Alizarin red solution for 1 h. They were then cleared through 20, 50, and 80% glycerine in 1% KOH, then stored in glycerine.
X-ray and micro-CT analysis
Four-week-old mice were anesthetized with chloral hydrate and subjected to X-ray scanning at 30 KV (Faxitron X-ray, Tucson, AZ, USA). Skulls of 4-week-old and 6-month-old WT and RapOsx mice were used for micro-CT analysis (μCT80, SCANCO Medical AG, Bassersdorf, Switzerland) with a 10-μm voxel size. The femur of 4-week-old mTOROsx, mTOROsx/+, RapOs, RapOsx/+x mice, 6-month-old RapOsx mice and corresponding WT littermates were collected for micro-CT scanning with a 10-μm voxel size. One hundred slices (total 1 voxel size. One hundred slices (total 1lected for micro-CT scanning witrabecular microarchitecture parameters including bone volume fraction (BV/TV), trabecular thickness (Tb.Th.) and trabecular number (Tb.N.) following the introductions of the manufacturer.33, 34 Fifty slices from the middle of the femur were used to analyze cortical thickness (Ct.Th.).35
Histological analysis
Femurs from 7-day-old and 4-week-old mice were fixed in 4% paraformaldehyde for 48 h followed by decalcification in 10% EDTA for 2–4 weeks. Specimens were embedded in paraffin then stained with hematoxylin–eosin and TRAP (Sigma, St. Louis, MO, USA) according to previously described methods.36, 37
Immunohistochemical staining was performed following a previously described protocol.37 Sections were de-waxed and rehydrated. A solution of 3% H2O2 was used to block the activity of endogenous peroxidase. Antigen retrieval was performed with protease K at 37 °C for 15 min. Antibodies to Runx2 (1:200, Santa Cruz Biotechnology, Santa Cruz, CA, USA) and P-S6K1 (T389, 1:200, Merck Millipore, Darmstadt, Germany), P-ERα (S167, 1:200, ABclonal, Boston, MA, USA) were added and incubated overnight at 4 °C. Corresponding biotinylated secondary antibodies were then added and incubated for 1 h at room temperature, followed by color development with an ABC kit (Vector Labs, Peterborough, UK). We counted the number of Runx2-, P-S6K1- and P-ERα-positive cells along the trabecular bone of the distal femur excluding the area within 0.25 mm from the growth plate.
In situ hybridization was performed as previously described.37 Briefly, DIG-labeled RNA probes were used to detect mRNA expression in femurs of 7-day-old mTOROsx, RapOsx and corresponding WT littermates. Probes used in this study: probes for mouse Col1α1 (nucleotides 4466-4783, NM_007742, subcloned in pBlueScript), probes for mouse Ocn (nucleotides 39-342, NM_007541, subcloned in pBlueScript). After hybridization, probes were visualized by anti-DIG biotin-conjugated antibody and an ABC kit (Vector Labs). Then, samples were counterstained with hematoxylin. We counted Col1α1-positive cells along trabecular bone of the distal femur excluding the area within 0.25 mm from the growth plate and Ocn-positive cells along cortical bone of the distal femur.
Cell culture
Four-week-old Raptorfl/fl mice were killed and the hindlimbs were collected. Bone marrow cells were washed out of the long bones and centrifuged at 500 × g for 10 min. The collected BMSCs were cultured in α-MEM with 10% fetal calf serum and 1% penicillin/streptomycin. After 14 days, BMSCs were reseeded at 2.5 × 105/cm2. Twenty-four hours later, BMSCs were infected with adenovirus expressing either CRE recombinase or GFP at a MOI of 10. Then, BMSCs were cultured in osteogenic medium (α-MEM with 10% FBS and 1% penicillin/streptomycin, 100 nM dexamethasone, 50 μM L-ascorbic acid, and 10 mm β-glycerophosphate) until required.
Parietal bones of P5 mice (mTOROsx, RapOsx and WT) were digested in 1 mg/ml collagenase (Sigma) and 2 mg/ml Dispase II (Sigma) in α-MEM for 5 min, three times. The released calvarial cells were cultured in α-MEM with 10% FBS and 1% penicillin/streptomycin. After 7 days, calvarial cells were reseeded at 5 × 104/cm2 and cultured in osteogenic medium. For overexpression of CAS6K1 and Runx2, RapOsx calvarial cells were infected with lentivirus expressing CAS6K1 or Runx2. At the same time WT and RapOsx calvarial cells infected with lentivirus expressing GFP were used as control groups. Then these cells were cultured in osteogenic medium.
Alkaline phosphatase staining and alizarin red staining
ALP assay was performed after 7 days of osteoblast differentiation according to the specification of the manufacturer (Beyotime Institute of Biotechnology, Shanghai, China). Mineralized nodule formation was detected by Alizarin red staining 14 days after osteoblast differentiation (Cyagen Biosciences, Santa Clara, CA, USA).
Quantitative PCR (qPCR)
Total RNA was extracted from osteoblasts differentiated from BMSCs after 7 days and from calvarial cells using TRIzol, following a standard protocol (Invitrogen, Carlsbad, CA, USA). An aliquot of 500 ng RNA was reverse-transcribed to cDNA using TaKaRa PrimeScript Reverse Transcriptase (TaKaRa Bio Inc., Shiga, Japan). qPCR was performed using a SYBR green mixture (TaKaRa) to detect expression of osteogenic genes. Primers used in this study: Hprt-F:5′-GTTAAGCAGTACAGCCCCAAA-3′, Hprt-R: 5′-AGGGCATATCCAACAACAAACTT-3′, β-actin-F: 5′-GTGACGTTGACATCCGTAAAGA-3′, β-actin-R: 5′-GCCGGACTCATCGTACTCC-3′, Runx2-F: 5′-CCAACCGAGTCATTTAAGGCT-3′, Runx2-R: 5′-GCTCACGTCGCTCATCTTG-3′, Col1α1-F: 5′-GCTCCTCTTAGGGGCCACT-3′, Col1α1-R: 5′-CCACGTCTCACCATTGGGG-3′, Ocn-F: CTTGGTGCACACCTAGCAGA-3′, Ocn-R: 5′- CTCCCTCATGTGTTGTCCCT-3′,
Plasmids
cDNA of S6K1, ERα and DLX5 were cloned into a phage-based plasmid. Constitutively active S6K1 was constructed following previously described methods by converting Thr390 to glutamic acid.38 Non-phosphorylatable ERα was constructed by converting Ser171 into alanine and constitutively phosphorylated ERα was constructed by converting Ser171 into glutamic acid. A 3X 89 bp Runx2 enhancer corn sequence was synthesized (by Genescript, Piscataway, NJ, USA) and cloned into a phage-based luciferase reporter.23 The Runx2 plasmid was a gift from Dr. Gerard Karsenty’s lab. The Runx2 promoter-driven pGL3-based luciferase reporter was kindly provided by Dr. Zhang Feng.39
Transient transfection and luciferase reporter assay
Mouse MSC C3H10T1/2 cells were seeded at 4 × 104 cells/well into a 12-well plate, allowed to settle overnight, then transfected using PEI with a luciferase reporter plasmid and Renilla luciferase (Promega, Madison, WI, USA) along with various combinations of expression plasmids as indicated. Empty expression vector plasmids were used as required to make up the total amount of transfected DNA. At 48 h post-transfection, the cells were lysed with lysis buffer (1% NP-40, 10% glycerol, 135 mM NaCl, 20 mM Tris, pH 8.0) with protease inhibitor and lysates were used for the dual-luciferase reporter assay (Promega). In our study, promoter activity was expressed as relative light units normalized to the activity of cotransfected Renilla luciferase.
Co-immunoprecipitation and western blot
For western blot analysis of parietal bone, total proteins were obtained from P0 mice with SDS buffer (TaKara). For western blot analysis of BMSCs and calvarial cells, total proteins were obtained from BMSCs and calvarial cells after 7 days of differentiation in osteogenic medium. Proteins (60 μg of parietal bone protein or 30 μg of cells) were separated by 10% SDS-PAGE followed by western blotting according to a standard protocol. Antibodies used were: mTOR (Cell Signaling Technology, Danvers, MA, USA), Raptor (Cell Signaling Technology), P-S6K1 (T389, Merck Millipore), S6K1 (Cell Signaling Technology), P-S6 (S235/236, Cell Signaling Technology), S6 (Cell Signaling Technology), Runx2 (Santa Cruz Biotechnology), β-actin (Santa Cruz Biotechnology).
CoIP was performed following a method previously described.37 Briefly, 293T cells were seeded into a 10 cm dish at a concentration of 3 × 106 cells/dish and allowed to settle overnight. At 48 h post-transfection with PEI, cells were lysed and whole cell lysates were used for immunoprecipitation by Flag or HA antibody (Sigma) at 4 °C overnight. Western blot assay was performed with the indicated antibodies (anti-HA and anti-Flag, Sigma). For CoIP analysis of ERα and DLX5 in calvarial osteoblast, whole cell lysates were incubated with IgG and ERα (Sangon Bioteck, Shanghai, China) at 4 °C overnight. Western blot assay was performed with DLX5 (Abcam, Cambridge, UK) and anti-ERα(Abcam).
Chromatin immunoprecipitation (ChIP) and qPCR
CHIP analysis in WT and RapOsx calvarial osteoblast was performed following by an Enzymatic Chromatin Immunoprecipitation kit (Cell Signaling Technology) following the instructions of the manufacturer. Briefly, calvarial osteoblasts were cross-linked with 1% formaldehyde for 10 min at room temperature followed by quenched with glycine. Chromatin digestion was performed to obtain DNA fragments from 150 bp to 900 bp by Micrococcal Nuclease. Immunoprecipitation was performed with DLX5 (Abcam) and ERα(Abcam), and IgG was used as a negative control. Precipitated DNA was detected by qPCR with specific primers. Primers for Runx2 enhancer: F:5′-CTGCTTTAGGTAGAGGGCTT-3′, R: 5′-AATCAGAGTGGAGTCTCAGC-3′.
Statistical analysis
All quantitative data are presented as mean±s.d. from at least three independent samples. Student’s t-test was used for statistical evaluations of two group comparisons. Statistical analysis with more than two groups was performed with one-way analysis of variance (ANOVA). P<0.05 was considered statistically significant.
References
Harada S, Rodan GA . Control of osteoblast function and regulation of bone mass. Nature 2003; 423: 349–355.
Guntur AR, Rosen CJ . The skeleton: a multi-functional complex organ. New insights into osteoblasts and their role in bone formation: the central role of PI3Kinase. J Endocrinol 2011; 211: 123–130.
Ducy P, Zhang R, Geoffroy V, Ridall AL, Karsenty G . Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell 1997; 89: 747–754.
Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, Rosewell IR et al. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell 1997; 89: 765–771.
Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K et al. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell 1997; 89: 755–764.
Mundlos S, Otto F, Mundlos C, Mulliken JB, Aylsworth AS, Albright S et al. Mutations involving the transcription factor CBFA1 cause cleidocranial dysplasia. Cell 1997; 89: 773–779.
Bhaskar PT, Hay N . The two TORCs and Akt. Dev cell 2007; 12: 487–502.
Nojima H, Tokunaga C, Eguchi S, Oshiro N, Hidayat S, Yoshino K et al. The mammalian target of rapamycin (mTOR) partner, raptor, binds the mTOR substrates p70 S6 kinase and 4E-BP1 through their TOR signaling (TOS) motif. J biol chem 2003; 278: 15461–15464.
Yamnik RL, Digilova A, Davis DC, Brodt ZN, Murphy CJ, Holz MK . S6 kinase 1 regulates estrogen receptor alpha in control of breast cancer cell proliferation. J biol chem 2009; 284: 6361–6369.
Carnevalli LS, Masuda K, Frigerio F, Le Bacquer O, Um SH, Gandin V et al. S6K1 plays a critical role in early adipocyte differentiation. Dev cell 2010; 18: 763–774.
Zoncu R, Efeyan A, Sabatini DM . mTOR: from growth signal integration to cancer, diabetes and ageing. Nat rev Mol cell biol 2011; 12: 21–35.
Xiang X, Zhao J, Xu G, Li Y, Zhang W . mTOR and the differentiation of mesenchymal stem cells. Acta Biochim Biophys Sin (Shanghai) 2011; 43: 501–510.
Singha UK, Jiang Y, Yu S, Luo M, Lu Y, Zhang J et al. Rapamycin inhibits osteoblast proliferation and differentiation in MC3T3-E1 cells and primary mouse bone marrow stromal cells. J Cell Biochem 2008; 103: 434–446.
Yeh LC, Ma X, Ford JJ, Adamo ML, Lee JC . Rapamycin inhibits BMP-7-induced osteogenic and lipogenic marker expressions in fetal rat calvarial cells. J Cell Biochem 2013; 114: 1760–1771.
Lee KW, Yook JY, Son MY, Kim MJ, Koo DB, Han YM et al. Rapamycin promotes the osteoblastic differentiation of human embryonic stem cells by blocking the mTOR pathway and stimulating the BMP/Smad pathway. Stem cells dev 2010; 19: 557–568.
Fang F, Sun S, Wang L, Guan JL, Giovannini M, Zhu Y et al. Neural crest-specific TSC1 deletion in mice leads to sclerotic craniofacial bone lesion. J bone miner res 2015; 30: 1195–1205.
Riddle RC, Frey JL, Tomlinson RE, Ferron M, Li Y, DiGirolamo DJ et al. Tsc2 is a molecular checkpoint controlling osteoblast development and glucose homeostasis. Mol cell biol 2014; 34: 1850–1862.
Huang B, Wang Y, Wang W, Chen J, Lai P, Liu Z et al. mTORC1 prevents preosteoblast differentiation through the notch signaling pathway. PLoS genet 2015; 11: e1005426.
Chen C, Akiyama K, Wang D, Xu X, Li B, Moshaverinia A et al. mTOR inhibition rescues osteopenia in mice with systemic sclerosis. J Exp Med 2014; 212: 73–91.
Chen J, Long F . mTORC1 signaling controls mammalian skeletal growth through stimulation of protein synthesis. Development 2014; 141: 2848–2854.
Chen J, Long F . mTORC1 signaling promotes osteoblast differentiation from preosteoblasts. PloS one 2015; 10: e0130627.
Martin SK, Fitter S, Dutta AK, Matthews MP, Walkley CR, Hall MN et al. Brief report: the differential roles of mTORC1 and mTORC2 in mesenchymal stem cell differentiation. Stem Cells 2015; 33: 1359–1365.
Kawane T, Komori H, Liu W, Moriishi T, Miyazaki T, Mori M et al. Dlx5 and mef2 regulate a novel runx2 enhancer for osteoblast-specific expression. J bone miner res 2014; 29: 1960–1969.
Yamnik RL, Holz MK . mTOR/S6K1 and MAPK/RSK signaling pathways coordinately regulate estrogen receptor alpha serine 167 phosphorylation. FEBS lett 2010; 584: 124–128.
Kammerer M, Gutzwiller S, Stauffer D, Delhon I, Seltenmeyer Y, Fournier B . Estrogen Receptor a (ERa) and Estrogen Related Receptor a (ERRa) are both transcriptional regulators of the Runx2-I isoform. Mol Cell Endocrinol 2013; 369: 150–160.
Melville KM, Kelly NH, Khan SA, Schimenti JC, Ross FP, Main RP et al. Female mice lacking estrogen receptor-alpha in osteoblasts have compromised bone mass and strength. J bone miner res 2014; 29: 370–379.
Almeida M, Iyer S, Martin-Millan M, Bartell SM, Han L, Ambrogini E et al. Estrogen receptor-alpha signaling in osteoblast progenitors stimulates cortical bone accrual. J clin invest 2013; 123: 394–404.
Guo J-P, Shu S-K, Esposito NN, Coppola D, Koomen JM, Cheng JQ . IKKε phosphorylation of estrogen receptor α Ser-167 and contribution to tamoxifen resistance in breast cancer. J Biol Chem 2010; 285: 3676–3684.
Yan B, Zhang Z, Jin D, Cai C, Jia C, Liu W et al. mTORC1 regulates PTHrP to coordinate chondrocyte growth, proliferation and differentiation. Nat commun 2016; 7: 11151.
Polak P, Cybulski N, Feige JN, Auwerx J, Ruegg MA, Hall MN . Adipose-specific knockout of raptor results in lean mice with enhanced mitochondrial respiration. Cell metab 2008; 8: 399–410.
Lou Y, Javed A, Hussain S, Colby J, Frederick D, Pratap J et al. A Runx2 threshold for the cleidocranial dysplasia phenotype. Hum mol genet 2009; 18: 556–568.
Kelly WL, Bryden MM . A modified differential stain for cartilage and bone in whole mount preparations of mammalian fetuses and small vertebrates. Stain technol 1983; 58: 131–134.
Yuan Q, Jiang Y, Zhao X, Sato T, Densmore M, Schuler C et al. Increased osteopontin contributes to inhibition of bone mineralization in FGF23-deficient mice. J bone miner res 2014; 29: 693–704.
Liu W, Zhou L, Zhou C, Zhang S, Jing J, Xie L et al. GDF11 decreases bone mass by stimulating osteoclastogenesis and inhibiting osteoblast differentiation. Nat commun 2016; 7: 12794.
Guo Y, Sun N, Duan X, Xu X, Zheng L, Seriwatanachai D et al. Estrogen deficiency leads to further bone loss in the mandible of CKD mice. PloS one 2016; 11: e0148804.
Andersson GN, Marks S . Tartrate-resistant acid ATPase as a cytochemical marker for osteoclasts. J Histochem Cytochem 1989; 37: 115.
Zou W, Greenblatt MB, Brady N, Lotinun S, Zhai B, de Rivera H et al. The microtubule-associated protein DCAMKL1 regulates osteoblast function via repression of Runx2. J Exp Med 2013; 210: 1793–1806.
Xia T, Cheng Y, Zhang Q, Xiao F, Liu B, Chen S et al. S6K1 in the central nervous system regulates energy expenditure via MC4R/CRH pathways in response to deprivation of an essential amino acid. Diabetes 2012; 61: 2461–2471.
Zhang F, Xu L, Xu Q, Li D, Yang Y, Karsenty G et al. JMJD3 promotes chondrocyte proliferation and hypertrophy during endochondral bone formation in mice. J mol cell biol 2015; 7: 23–34.
Acknowledgements
The authors thank Drs Gerard Karsenty, Feng Zhang, Minghan Tong for kindly providing reagents. We thank the members of the Zou lab for useful discussions. This work was supported in part by grants from 973 Program from the Chinese Ministry of Science and Technology (MOST) (2014CB964704 and 2015CB964503), the Science and Technology Commission of Shanghai (124119b0101), the National Natural Science Foundation of China (NSFC) (31371463, 31501170, 81371121 and 81570950), the Strategic Priority Research Program of the Chinese Academy of Sciences, Grant No. XDB19000000, Shanghai Summit & Plateau Disciplines, the ‘Chen Xing’ project from Shanghai Jiaotong University and WZ is a scholar of the ‘National 1000 Young Talents Program of China’ and ‘the National Science Fund for Excellent Young Scholars’ (NSFC) (81322027).
Author contributions
Conceived and designed the experiments: JW, WZ. Performed the experiments: QD, ZX, NN, XM, SZ, FX. Analyzed the data: QD, WZ. Contributed reagents/materials/analysis tools: XZ, LJ. Wrote the paper: QD, WZ.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by M Piacentini
Supplementary Information accompanies this paper on Cell Death and Differentiation website
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
About this article
Cite this article
Dai, Q., Xu, Z., Ma, X. et al. mTOR/Raptor signaling is critical for skeletogenesis in mice through the regulation of Runx2 expression. Cell Death Differ 24, 1886–1899 (2017). https://doi.org/10.1038/cdd.2017.110
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2017.110
This article is cited by
-
Bone marrow mesenchymal stem cell-derived exosomes loaded with miR-26a through the novel immunomodulatory peptide DP7-C can promote osteogenesis
Biotechnology Letters (2023)
-
Why SNP rs3755955 is associated with human bone mineral density? A molecular and cellular study in bone cells
Molecular and Cellular Biochemistry (2022)
-
STAT3 is critical for skeletal development and bone homeostasis by regulating osteogenesis
Nature Communications (2021)
-
The mTORC1 complex in pre-osteoblasts regulates whole-body energy metabolism independently of osteocalcin
Bone Research (2021)
-
Activating PIK3CA mutation promotes osteogenesis of bone marrow mesenchymal stem cells in macrodactyly
Cell Death & Disease (2020)