Abstract
The tumor suppressor protein promyelocytic leukemia (PML) is a key regulator of inflammatory responses and tumorigenesis and functions through the assembly of subnuclear structures known as PML nuclear bodies (NBs). The inflammation-related cytokine tumor necrosis factor-α (TNFα) is known to induce PML protein accumulation and PML NB formation that mediate TNFα-induced cell death in cancer cells and inhibition of migration and capillary tube formation in endothelial cells (ECs). In this study, we uncover a novel mechanism of PML gene regulation in which the p38 MAPK and its downstream kinase MAP kinase-activated protein kinase 1 (MNK1) mediate TNFα-induced PML protein accumulation and PML NB formation. The mechanism includes the presence of an internal ribosome entry site (IRES) found within the well-conserved 100 nucleotides upstream of the PML initiation codon. The activity of the PML IRES is induced by TNFα in a manner that involves MNK1 activation. It is proposed that the p38–MNK1–PML network regulates TNFα-induced apoptosis in breast cancer cells and TNFα-mediated inhibition of migration and capillary tube formation in ECs.
Similar content being viewed by others
Main
The pro-inflammatory cytokine, tumor necrosis factor-α (TNFα), was first characterized by its cytotoxic ability to induce cancer cell apoptosis.1 TNFα is known to have a role in the maintenance of tissue homeostasis. Dysregulation of TNFα contributes to the pathogenesis of several diseases, such as cancer, cardiovascular, neurological and metabolic disorders.2, 3 TNFα is primarily produced by activated macrophages in response to microbial infection and inflammation. TNFα signaling is critical for repair of injured lesions. However, sustained TNFα stimulation may also lead to endothelial cell (EC) dysfunction and the inhibition of angiogenesis.4, 5, 6, 7 Moreover, long-term TNFα treatment in breast cancer cells inhibits global protein synthesis and selectively activates apoptotic signals to promote cell death.8, 9, 10, 11, 12, 13 However, a detailed mechanism that links TNFα-regulated protein synthesis and apoptosis is still elusive.
The conjugation of TNFα to its cognate receptors, TNFR1/TNFR2, activates TNFR downstream signaling cascades that include activating NF-κB, caspases, c-Jun N-terminal kinase (JNK), p38 MAPK and extracellular signal-regulated kinase (ERK), depending on cell type-specific responses.3 Generally, caspases, JNK and p38 are apoptotic mediators involved in cytokine and stress-mediated apoptosis, whereas ERK kinase and NF-κB are mainly responsible for cell survival in response to mitogenic stimuli.14, 15, 16 The mechanisms by which p38 promotes apoptosis are complex and involve its ability to regulate transcription, protein synthesis and turnover through its downstream kinases.17
The promyelocytic leukemia (PML) protein is a tumor suppressor that functions as a transcription regulator in response to environmental stimuli.18 A significant fraction of PML is enriched in subnuclear structures known as the PML nuclear bodies (PML NBs). These are nuclear depots for a variety of proteins that move in and out in response to stimuli.19 PML NBs control the storage of nuclear proteins that await recruitment in response to a wide variety of cellular signalings and can serve as sites for post-translational modification of proteins including acetylation and sumoylation.20, 21, 22 Thus, PML and PML NBs regulate fundamental cellular processes such as transcription, DNA replication and cell cycle progression.18 Disruption of PML NBs has serious consequences that ultimately alter cell fate.11 In contrast, overexpression of PML results in cell growth arrest, senescence and apoptosis.8, 22, 23 Histochemical studies indicated that PML is maintained at low levels in most examined tissues and cancer cells, but is expressed at a higher level in inflammation-associated tissues including macrophages and vascular endothelium, suggesting an important role for PML in tumorigenesis, inflammatory responses and endothelial function.4, 24, 25 We and others have shown that PML protein is subjected to extensive stability regulation.26, 27, 28, 29 It was previously reported that TNFα induced PML protein accumulation and PML NB formation in human umbilical endothelial cells (HUVECs).4 Our previous report demonstrated the TNFα induced PML protein accumulation partly through transcriptional control,4 suggesting that post-transcriptional regulation also plays a role in this process.
Internal ribosome entry site (IRES)-mediated translation via mRNA 5′-UTRs is a mechanism to selectively increase expression of apoptosis-associated proteins when cells are under stresses such as oxidative stress and genotoxic stress,30, 31 conditions known to downregulate global translation. For example, c-myc, DAP5, APAF-1, XIAP and p53 have been reported to switch from cap-dependent translation to IRES-mediated translation in response to apoptotic stimulation.31, 32, 33, 34, 35 TNFα downstream MAPK kinases such as p38, ERK and MAP kinase-activated protein kinase 1 (MNK1) selectively increase IRES-mediated translation of a subset of mRNAs.36, 37 As PML is known to promote cell death in response to apoptotic stimulation, it is likely that TNFα induces PML protein accumulation through a translation-dependent mechanism.
In this study, we dissect the mechanisms underlying transcription-independent regulation of PML in response to TNFα stimulation. We demonstrate that MNK1, a p38 downstream kinase, plays a pivotal role in TNFα-mediated accumulation of PML by activating an IRES in the 5′-UTR of PML mRNA to increase PML protein accumulation. Through this mechanism, the TNFα–MNK1–PML axis regulates the expression of histone deacetylase 7 (HDAC7) target genes including MCP-1 and MMP10 and controls migration and capillary tube formation in ECs. Furthermore, activation of the PML IRES by MNK1 promotes TNFα-induced cell death and contributes to MCF7 apoptosis.
Results
TNFα enhances PML protein accumulation via both transcription-dependent and -independent mechanisms
We have previously shown that TNFα induced a twofold increase in PML mRNA, whereas PML protein expression is elevated to an even greater extent. Indeed, in HUVECs, PML protein continued to accumulate throughout the time course of exposure (Figure 1a), but the PML mRNA levels plateaued at 4 h of TNFα treatment (Figure 1b). Similar observations were noted in HeLa cells (Figures 1c and d). These data suggest that TNFα induces PML protein accumulation in both transcription-dependent and -independent manners. To test whether protein stability control is involved in TNFα-mediated PML protein accumulation, we examined the PML protein half-life with or without TNFα stimulation. Using cychloheximide treatment, which blocks translation, we observed little difference in the PML protein half-life with or without TNFα treatment in HeLa cells and HUVECs (Figures 1e and f and Supplementary Figure 1). Taken together, these data suggested that TNFα-induced PML protein accumulation is at least partially due to translational control.
TNFα induces PML protein accumulation without changing protein stability. Accumulation of PML protein (a) and mRNA (b) during TNFα stimulation in HUVECs. HUVECs were treated with TNFα (20 ng/ml) and harvested at the indicated times. The whole-cell lysates and total RNA were prepared for western blotting with the indicated antibodies and qRT-PCR, respectively. β-Actin protein and 18S rRNA were used as an internal control for western blotting and qRT-PCR, respectively. (c and d) The effect of TNFα on PML protein expression in HeLa cells. The experiments were similar to those in (a) and (b) except HeLa cells were used. (d) Quantitative measurement of the TNFα effect on PML protein and mRNA accumulation (n=3). (e) The effects of TNFα on PML protein half-life in HeLa cells were measured as described in the Materials and Methods. (f) The half-life of PML protein with or without TNFα treatment (20 h) was derived from three experiments. All the results shown in graphs are mean±S.D.
TNFα-induced PML accumulation is p38 kinase dependent
Several TNFα downstream MAPK kinases, including JNK, ERK and p38, have been reported to mediate protein or mRNA regulation in mammalian cells.38, 39 To determine which kinase is involved in TNFα-mediated induction of PML protein, we first carried out kinase inhibitor screenings with or without TNFα treatment. We found that the blockade of p38 (SB202190) and ERK2 (U0126) kinases in HUVECs effectively reduced TNFα-induced PML accumulation 7- and 2.3-fold, respectively (Figure 2a). We focused on p38 kinase in the following studies. The efficacy of the kinase inhibitors was confirmed by the disappearance of kinase activation markers in western blots (Figure 2b and Supplementary Figure 3). Using an siRNA approach, we further validated that the induction of PML by TNFα is p38 dependent (Figure 2c). Immunofluorescence microscopy studies also demonstrated that the PML NB formation tightly correlates with the abundance of PML in response to TNFα treatment and p38 knockdown (Supplementary Figure 4a). A previous report indicated that a fraction of PML was found in the endoplasmic reticulum (ER) of mouse embryonic fibroblasts (MEFs).40 Using confocal microscopy and subcellular fractionation experiments, we observed that endogenous PML predominantly localizes in the nucleus with or without TNFα treatment. We did not observe PML in the cytoplasm or ER in HUVECs or MCF7 cells, although we cannot exclude the possibility that a residual fraction of PML is present in the ER (Figure 3 and Supplementary Figure 2). Based on these data, we conclude that p38 is indispensable for TNFα-mediated induction of nuclear PML protein accumulation.
TNFα increases PML protein accumulation in a p38 kinase-dependent manner. (a) The effects of MAPK inhibitors on TNFα-mediated PML protein accumulation. HUVECs were pretreated with kinase inhibitors SP600125 (JNK) (10 μM), SB202195 (p38) (10 μM) or U0126 (ERKs) (10 μM) before 16 h of TNFα (20 ng/ml) treatment. Cells were harvested and whole-cell lysates prepared for western blotting with the indicated antibodies. The relative ratio of PML to β-actin is shown in the bottom. (b) Inhibition of p38 kinase activity by SB202190. HUVECs were treated with DMSO or SB202190 (10 μM) before TNFα treatment and harvested at indicated times for western blotting with the indicated antibodies. α-Tubulin was included as a loading control. (c) The effect of p38 knockdown on TNFα-mediated PML protein accumulation. HUVECs were transiently transfected with a control or two independent p38 siRNAs, treated with or without TNFα, harvested and western blotted with the indicated antibodies
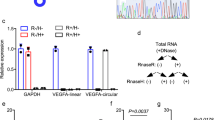
MNKs are p38 downstream kinases that mediate TNFα-induced PML protein accumulation. HUVECs were pretreated with kinase inhibitors or transfected with kinase-specific siRNAs before TNFα treatment, followed by western blotting with the indicated antibodies. (a) HUVECs were treated with p38 kinase inhibitor and TNFα as indicated. Nuclear and cytosolic fractions were separated and subjected to western blotting. C: cytosolic fraction; N: nuclear fraction. (b) The effect of MSK inhibitor on TNFα-mediated PML protein accumulation in HUVECs. (c) The effect of CK2 inhibitor on TNFα-mediated PML protein accumulation in HUVECs. (d) The effect of knockdown of MK2 on TNFα-mediated PML protein accumulation in HUVECs. (e) The effect of a MNK inhibitor on TNFα-mediated PML protein accumulation in HUVECs. (f) The effect of knockdown of MNK1 on TNFα-mediated PML protein abundance and mRNA accumulation in HUVECs. The mRNA levels of PML in each sample were quantified by qRT-PCR (n=3). (g) Knockdown of MNK1 had little or no effect on PML protein half-life in TNFα-treated HUVECs. Quantitation of PML protein abundance from western blots is shown on the right (n=4). (h–j) The effect of knockdown of MNK2 on TNFα-mediated induction of PML protein in HUVECs. The mRNA levels of PML and MNK2 in each sample were quantified by qRT-PCR (n=3). (k) Polysome profiling analysis demonstrates that TNFα–MNK1 axis controls heavy polysome occupancy on PML mRNA
MNK1 is a p38 downstream kinase responsible for TNFα-mediated PML upregulation
In Figure 2c and Supplementary Figure 4a, we observed that PML protein in HUVECs accumulated in PML NBs and decreased in p38 knockdown cells. Furthermore, subcellular fractionation also indicated that most PML protein was detected in the nuclear fraction and was upregulated by TNFα and inhibition of p38 compromised TNFα-mediated accumulation of PML protein (Figure 3a). Four kinases, including casein kinase 2 (CK2), MNK1, mitogen and stress-activated protein kinase (MSK1) and MAPKAP kinase 2 (MK2), are p38 downstream kinases that mediate various aspects of p38 activity.15, 41 To further determine which kinase plays a key role in TNFα regulation of PML, we blocked the p38 downstream kinase activity individually, either by kinase inhibitors or siRNAs. From these inhibition assays, we concluded that inhibition of CK2, MSK1 and MK2 had little or no effect on TNFα-mediated PML protein accumulation (Figures 3b–d). In contrast, inhibition of MNK1 kinase (MNKi) activity by CGP57380 (Supplementary Figure 3c) attenuated TNFα effects on the accumulation of nuclear PML protein (Figure 3e). This is similar to the effect of the p38 inhibitor, SB202190 (Figures 3a and e). To further confirm a role of MNK1 kinase in TNFα-mediated regulation of PML, we carried out knockdown studies and found that the depletion of MNK1 reduced TNFα effects on PML protein accumulation but not on its mRNA abundance (Figure 3f). The TNFα-induced and MNK1-dependent PML protein accumulation is not due to an increase in PML protein stability because knockdown of MNK1 had little or no effect on PML protein half-life after TNFα stimulation (Figure 3g). In addition to MNK1, we also found that knockdown of the MNK1 paralog, MNK2, in HUVECs reduced TNFα-induced PML protein accumulation without affecting its mRNA levels (Figures 3h–j). Furthermore, polysome analyses demonstrated that TNFα stimulation increased the presence of PML mRNA in heavy polysome but this increase is abolished by MNK1 inhibitor CGP57380 (Figure 3k).
MNK1 kinase regulates the TNFα-induced activity of an IRES located within the PML 5′-UTR
MNK1 kinase has been reported to promote protein synthesis through the activation of a subset of IRESs, although the detailed mechanism remains unknown.36, 42 We first explored whether human PML 5′-UTR has an IRES using a bicistronic luciferase reporter system (Figure 4a). This reporter contains an upstream Renilla luciferase (RLuc) gene, which is translated in a cap-dependent manner, fused with the Firefly luciferase (FLuc) gene, which can only be translated when the intra-cistronic sequence contains a functional IRES. One needs to be cautious as the putative intra-cistronic IRES may harbor a putative cryptic promoter.43 To test this possibility, we generated expression constructs in which several truncated human PML 5′-UTR fragments were inserted immediately upstream of the FLuc gene in a pGL3-basic reporter plasmid that does not contain any promoter sequence. We were able to determine that the -100->-1 region of the PML 5′-UTR (140 nucleotides) contains no detectable promoter activity when compared with an empty vector in HeLa cells (Supplementary Figure 5a) and HUVECs (Supplementary Figure 5b). In contrast, -140->-1 of PML 5′-UTR construct expresses significant reporter activity. Together, these data suggest that -140->-100 of PML 5′-UTR contains a cryptic promoter. When introducing pRF bicistronic reporter into the RLuc knockdown HeLa cells, we observed that the FLuc activity driven by PML 5′-UTR (-100->-1) and EMCV IRES are largely decreased in parallel to Renilla luciferase reduction. This result suggests that neither PML 5′-UTR (-100->-1) nor EMCV IRES contains a cryptic promoter (Figure 4d).44 Importantly, the PML 5′-UTR (-100->-1)-pRF exhibited substantially higher levels of IRES activity than both positive controls, EMCV IRES or the p53 IRES in HeLa cells (Supplementary Figure 5c) and HUVECs (Figure 4b). Furthermore, we demonstrated that the detected FLuc reporter activity in our bicistronic system is not derived from spurious splicing or a leaky ribosomal read-through (Figures 4c and e and Supplementary Figure 6).45 To further strengthen our conclusion, we transfected in vitro transcribed bicistronic mRNAs into HeLa cells and demonstrated that the Firefly luciferase activity driven by EMCV IRES and PML 5′-UTR (-100->-1) were substantially higher than pRF, regardless of whether the mRNA was capped (Figure 4f). In summary, we conclude that the PML 5′-UTR (-100->-1) contains an IRES.
Identification of PML 5′-UTR (-100->-1) as an IRES activated by the TNFα–MNK1 axis. (a) A schematic representation of bi-cistronic pRF plasmids used in the transient transfection reporter assays. (b) The effects of PML 5′-UTR (-100->-1) on reporter activity. The relative ratio of Firefly luciferase/Renilla luciferase intensity is plotted. The results are mean±S.D. in triplicates (n=3). (c) Determination of possible alternative splicing of the transcripts of PML 5′-UTR (-100->-1)-pRF. The cDNAs prepared from pRF or PML 5′-UTR (-100->-1)-pRF transfected HUVECs subjected to PCR using primer pairs as indicated in (a). The final PCR products were separated on a 0.8% agarose gel, stained by EtBr and images recorded. (d) Knockdown of RLuc in bicistronic system reduces FLuc driven by EMCV IRES and PML 5′-UTR (-100->-1) but not by PML 5′-UTR (-140->-1). (e) The reporter activity of PML 5′-UTR (-100->-1) is not derived from ribosomal read-through. HeLa cells were transiently transfected with engineered pRF and β-gal plasmids as indicated. At 48 h post transfection, the Firefly luciferase and Renilla luciferase activity were measured and normalized to the transfection control, β-gal. The luciferase activity of PML 5′-UTR (-100->-1)-pRF was set as 1. The results are mean ±S.D. in triplicates. (f) EMCV IRES and PML 5′-UTR (-100->-1) harbors IRES activity as evidenced by transfection of in vitro transcribed mRNAs into HeLa cells. The relative activity of RLuc and FLuc was normalized to the corresponding mRNA levels 9 h post transfection. (g) TNFα enhances PML 5′-UTR (-100->-1) activity in a MNK1-dependent manner. shCtrl and shMNK1 knockdown HeLa cells were transiently transfected with PML 5′-UTR (-100->-1)-pRF plasmid for 72 h, treated with TNFα at the indicated times and cells harvested. The relative activity of Firefly luciferase over Renilla luciferase was plotted. The results are mean ±S.D. in triplicates (n=3). (h and i) Ectopic expression of eIF4E inhibitor, 4E-BP or a mutant defective in phosphorylation by mTORC1, 4E-BP (5A), does not compromise TNFα-mediated PML protein accumulation or PML IRES activation. HeLa cells were transiently transfected with PML 5′-UTR (-100->-1)-pRF plasmid with the indicated plasmids for 48 h, treated with TNFα for 20 h and cells harvested. (h) Endogenous PML protein in pEBG, pEBG-4E-BP and pEBG-4E-BP (5A) transfected cells were determined by western blottings. The relative PML protein expression was normalized by β-actin levels and (PML)/(β-actin) in the absence of TNFα was set as 1. The relative fold induction of PML protein abundance by TNFα is shown in each sample. Both long (L.E.) and short (S.E.) exposures of PML western blot are shown. (i) The relative effect of TNFα on (FLuc)/(RLuc) of PML 5′-UTR (-100->-1) reporter in transfected samples is plotted. The results are shown as mean±S.D. (n=4)
To determine whether the PML IRES responds to TNFα, the PML 5′-UTR (-100->-1)-pRF reporter construct was transfected into HeLa cells with or without TNFα treatment. Upon TNFα stimulation, PML 5′-UTR (-100->-1)-pRF showed 1.8- and 2.3-fold increases in reporter activity at 24 and 48 h of TNFα treatment, respectively, but the increase in FLuc/RLuc by TNFα was attenuated when MNK1 was knocked down. This result indicated that the PML 5′-UTR (-100->-1) is activated through a MNK-dependent mechanism in response to TNFα treatment (Figure 4g). The translation initiation factor eIF4E is a major substrate of MNK1. However, it is unclear whether eIF4E is involved in this process. To examine this possibility, we co-transfected PML 5′-UTR (-100->-1)-pRF with the eIF4E inhibitor, 4E-BP, and a mutant defective in mTORC1 phosphorylation, 4E-BP(5A). This 4E-BP mutant sequesters and reduces eIF4E-mediated cap-dependent mRNA translation.46, 47 As shown in Figure 4h, 4E-BP or 4E-BP (5A) had little or no effect on TNFα-mediated induction of endogenous PML protein accumulation (Figure 4h). Reporter assays further suggested that TNFα-mediated PML IRES activation is eIF4E independent (Figure 4i). These data suggest that eIF4E has a minimal role in TNFα-induced, IRES-mediated PML mRNA translation.
MNK1 kinase contributes to TNFα-induced PML accumulation for its function in the inhibition of HUVEC migration and in vitro network formation
We have previously shown that PML is essential for TNFα-mediated inhibition of EC migration and capillary network formation,4, 25 both of which are key processes in angiogenesis. In parallel with the ability of TNFα to induce PML protein accumulation (Figure 1a), TNFα inhibits EC migration and capillary tube formation in a time-dependent manner (Figures 5a–c). To dissect whether MNK1 plays a role in these two EC activities, we knocked down MNK1 and PML in HUVECs and compared their effects on TNFα-mediated inhibition of angiogenesis. As expected, knockdown of PML in HUVECs largely attenuated TNFα-mediated inhibition of HUVEC migration and in vitro capillary network formation (Figures 5d–f). Consistently, knockdown of MNK1 in HUVECs also compromised TNFα inhibitory effects on cell migration and capillary network formation (Figures 5e and f). Similar to previous reports, TNFα treatment in our assays did not induce HUVEC apoptosis or senescence (Supplementary Figure S8).7, 48 Taken together, these data suggested that the MNK1–PML axis contributes to the TNFα-mediated inhibition of angiogenesis.
MNK1 mediates TNFα-induced inhibition of cell migration and capillary tube formation in ECs. The effect of TNFα on HUVEC network formation (a and b) and cell migration (c) in vitro. HUVECs were subjected to capillary tube formation and wound healing assays during TNFα treatment as described in the Materials and Methods. Representative images in capillary tube formation assays are shown in (a). (b and c) All the results are mean±S.D. (n=6). (d) Knockdown efficiency of MNK1 or PML in aliquots of (e) and (f). (e) The effect of MNK1 or PML knockdown on TNFα-mediated inhibition of EC migration. The left panel shows representative images of wound healing assays and the right panel shows cell migration rates. (f) The effect of MNK1 or PML knockdown on TNFα-mediated inhibition of in vitro capillary network formation. Representative images are shown in the left panel and the numbers of branches in each field were counted and quantified as shown in the right panel. (e and f) The statistical results are mean±S.D. (n=6)
MMP10 and MCP-1 are TNFα-induced genes targeted by p38–MNK1–PML axis
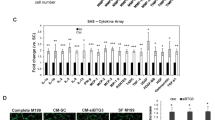
Our finding that MNK1 potentiates TNFα-mediated induction of PML protein accumulation raised the possibility that MNK1 plays a role in TNFα–PML axis to regulate expression of PML-associated genes, thus contributing to TNFα-mediated regulation of EC biology. To test this, we first analyzed the gene expression profiles in HUVECs and identified a list of chemokine and angiostatic genes that are positively regulated by both TNFα and PML.25 In addition to MMP10, expression of PDGFB, CXCL11, CXCLR4 and MCP-1 was downregulated in PML knockdown HUVECs (Supplementary Figure 9a). We focused on two genes, MMP10 and MCP-1, for further analyses because the functions of these two genes in ECs are well characterized.49, 50, 51 Using two independent PML shRNAs, we further confirmed that MMP10 and MCP-1 were PML downstream target genes (Figure 6a). Moreover, TNFα-mediated upregulation of MMP10 and MCP-1 mRNAs was significantly reduced in cells pretreated with inhibitors of p38 or MNK1 (Figure 6b) and cells in which MNK1 was knocked down (Figure 6c). Based on these data, we conclude that MMP10 and MCP-1 mRNAs are induced by TNFα in a p38-, MNK1- and PML-dependent manner.
Inhibition of PML, p38 or MNK1 decreases TNFα-inducible MCP-1 and MMP10 mRNA expression. (a) The effect of PML knockdown on MCP-1 and MMP10 mRNAs. The PML knockdown efficiency was shown in the left panel. (b) The effect of inhibition of p38 and MNK kinases on TNFα-mediated induction of MCP-1 and MMP10 mRNAs. The levels of MCP-1 and MMP10 mRNAs in each sample were measured and normalized to 18S rRNA. (c) The effect of MNK1 knockdown on TNFα-mediated accumulation of MCP-1 and MMP10 mRNAs. The statistical results are mean±S.D. (n=3)
The expression of MMP10 and MCP1 is controlled by TNFα-MNK1-PML axis via HDAC7
The induction of PML and PML NBs by stress stimuli can increase the sequestration of several PML-associated transcription regulators, preventing access to chromatin and thus contributing to the alterations in the expression of a subset of PML-responsive genes.18 Our previous studies suggested that HDAC7 is a PML-associated protein and that upregulation of PML causes the derepression of HDAC7 target genes (Supplementary Figure 9b).5, 26 We therefore investigated whether HDAC7 participates in TNFα- and PML-mediated induction of MMP10 and MCP-1 mRNA expression. By analyzing the promoters of MCP-1 from human, mouse and chimpanzee, we identified putative binding sites for transcription factors MEF-2, GATAs and BCL6 that have been shown to bind class II HDACs (Figure 7a).52 Using chromatin immunoprecipitation (ChIP) assays, we observed HDAC7 binding to the MCP-1 promoter at two regions, approximately −1.2 and −1.6 kb upstream of MCP-1 transcription start site. However, these associations were disrupted by TNFα treatment (Figure 7a). Furthermore, knockdown of PML increased HDAC7 occupancy on both MMP10 and MCP-1 promoters and neutralized TNFα effects on the recruitment of HDAC7 to the MCP-1 and MMP10 promoters. This indicates that PML negatively regulates the recruitment of HDAC7 to the MMP10 and MCP-1 promoters (Figures 7b and c). Finally, we carried out double knockdown experiments followed by TNFα treatment and found that further knockdown of HDAC7 under a PML or MNK1 knockdown background rescued MCP-1 and MMP10 expression in HUVECs (Figure 7d). In summary, our data demonstrated that TNFα-induced MCP-1 and MMP10 expression is regulated by the MNK1–PML–HDAC7 regulatory network.
TNFα-mediated MCP-1 and MMP10 mRNA expression is controlled by MNK1–PML–HDAC7 axis. (a, left panel) A schematic representation of the MCP-1 promoter showing putative transcription factor binding sites that are conserved in human, chimpanzee and mouse. (a, right panel) The effect of TNFα on the association of HDAC7 with the MCP-1 promoter analyzed by ChIP assays. (b) The effect of PML knockdown on the association of HDAC7 with the MCP-1 promoter upon TNFα stimuli. The knockdown efficiency is shown in the left. (c) The effect of PML knockdown on the association of HDAC7 with the MMP10 promoter upon TNFα stimuli. (d) The effect of HDAC7 knockdown on TNFα-induced accumulation of MCP-1 and MMP10 mRNAs in MNK1 and PML knockdown cells. HUVECs were infected with lentiviruses carrying the indicated shRNAs and treated with TNFα. The normalized expression of MCP-1 and MMP10 in HUVECs is shown. The shCtrl knockdown cells were set at 1. The knockdown efficiency of gene-specific shRNA is shown
MNK1 contributes to TNFα-induced PML protein accumulation and inhibition in MCF7 breast cancer
Because PML is a well-known tumor suppressor, we next examined whether this TNFα-mediated regulation of PML also occurs in cancer cells. We first examined the TNFα-induced PML accumulation in MCF7 breast cancer cells because of the ability of this cancer cell line to respond to TNFα treatment.9, 53 Similar to what we observed in HUVECs, the PML protein level was significantly upregulated in TNFα-treated MCF7 cells, and was attenuated by inhibitors of p38 and MNK1 (Figure 8a and Supplementary Figure 11). In addition, knockdown of MNK1 by shMNK1 blocked TNFα-mediated induction of PML, without decreasing PML mRNA (Figure 8b). Consistently, PML 5′-UTR (-100->-1)-pRF showed a 1.8-fold increase in relative reporter activity after 24 h of TNFα treatment in MCF7 cells. However, the increase in FLuc/RLuc by TNFα was attenuated when MNK1 was inactivated. This result indicates that the PML 5′-UTR (-100->-1) IRES activity is induced through a MNK1-dependent mechanism in response to TNFα treatment (Figure 8c). To determine biological significance of TNFα–MNK1–PML axis in MCF7 cells, we performed viability assays by Trypan blue exclusion. We found that TNFα treatment of MCF7 cells results in 50% of the cells undergoing cell death, and knockdown of MNK1 or PML partially alleviated this effect (Figure 8d). Furthermore, knockdown by shMNK1 or shPML mitigated TNFα-induced cleavage of PARP1, a PML-associated apoptotic effector (Figure 8e).20 In addition, inhibition of MNK1 activity by CGP57380 also attenuated TNFα cytotoxicity on MCF7 cells (Figure 8f).
MNK1 mediates TNFα-induced PML protein accumulation and apoptosis in MCF7 breast cancer cells. (a) The effect of inhibition of p38 and MNK1 activity on TNFα-induced PML protein expression in MCF7 cells. MCF7 cells were pretreated with kinase inhibitors followed by TNFα treatment, harvested, whole-cell lysates prepared and western blotted with the indicated antibodies. (b) The effect of knockdown of MNK1 on TNFα-induced PML protein and mRNA accumulation. (c) Inhibition of MNK1 activity reduces TNFα-mediated PML IRES activation in MCF7 cells. MCF7 cells were transiently transfected with PML 5′-UTR (-100->-1)-pRF plasmid for 72 h, treated with or without MNK1 inhibitor and TNFα and the cells were harvested. The relative activity of Firefly luciferase to Renilla luciferase was plotted as shown. The results are mean±S.D. in triplicates (n=3). (d and e) The effect of knockdown of MNK1 or PML on TNFα-mediated cell cytotoxicity in MFC7 cells (d) and the cleavage of PARP1 (e). MCF7 cells were infected with lentiviruses expressing a control, MNK1 or PML shRNA followed by TNFα treatment. Aliquots of cells were used to determine cell death rate by Trypan blue exclusion assays (d) or cleavage of PARP1 by western blotting (e). The results are mean±S.D. (n=6). (f) The effect of TNFα and the MNK1 inhibitor CGP57380 on TNFα-mediated inhibition of MCF7 cell proliferation determined by colony formation assays. (g and h) The effect of TNFα and the MNK1 inhibitor CGP57380 on TNFα-mediated cell cytotoxicity in HL60 and NB4 cells by WST-1 cell proliferation assays. (i) A model by which TNFα induces anti-angiogenesis and apoptosis activity through enhancing PML mRNA translation that involves PML IRES and the p38–MNK1 axis
The effect of TNFα and MNK1 inhibitor on proliferation of HL60 and NB4 cells
To extend our findings, we determined the effect of TNFα, CGP57380 or both on the survival of hematopoietic cancer cells, HL60 and NB4 cells. HL60 cells expressing wild-type PML were sensitive to TNFα treatment as previously reported and CGP57380 attenuated TNFα effects on the inhibition of HL60 cell proliferation (Figure 8g).54 In contrast, NB4 cells, which express a PML–RARα fusion, demonstrated resistance to TNFα and CGP57380 treatment. Interestingly, cotreatment of both reagents led to a significant inhibition of cell growth in NB4 cells, suggesting that the TNFα–MNK1–PML axis regulates proliferation of these cells (Figure 8h). In summary, these data support a model in which the p38–MNK1–PML axis mediates TNFα-induced MCF7 cell death in good correlation with regulation of the activity of an IRES that was described for the first time in this report to be present within 100 nucleotides upstream of the PML translational initiation codon.
Discussion
In the present study, we have identified an IRES in human PML 5′-UTR spanning -100->-1 nucleotides upstream of the translational initiation codon. Notably, this IRES is activated by the TNFα–p38–MNK1 axis, in agreement with induced translation of the PML mRNA and PML protein, thus contributing to TNFα-regulated cancer cell apoptosis and inhibition of EC migration and capillary tube formation (Figure 8i).
Several reports have suggested that MNK1-mediated phosphorylation of eIF4E contributes to protein synthesis and cell growth.55, 56 Interestingly, Mnk1−/− Mnk2−/− mice are viable, fertile and developmentally normal despite the absence of phosphorylated eIF4E, and neither de novo protein synthesis nor cap-dependent translation was affected in Mnk1−/− Mnk2−/− MEFs.56 These results suggest that MNK1 may not directly participate in cap-dependent translation. In contrast, several lines of evidence have demonstrated that MNK1 may have a role in cap-independent translational control.36, 57 It has been shown that overexpression of MNK1 increased cap-independent translation compared with cap-dependent translation.58 In this study, we propose that MNK1 is a critical effector that potentiates TNFα-mediated PML mRNA translation through the PML IRES (Figures 4 and 8 and Supplementary Figure 5). It has been previously shown that PML has the ability to sequester eIF4E to PML NBs, thereby blocking eIF4E-mediated nuclear export of a subset of mRNAs.59, 60 We speculate that MNK1 promotes IRES-dependent PML mRNA translation and PML-mediated eIF4E nuclear retention contributes to the MNK1-mediated inhibition of a subset of mRNA translation.
It has been shown that TNFα stimulation in T cells forms a positive feed-forward loop that activates TNFα mRNA translation via MNK1 and its 3′-UTR.61 This finding raises the possibility that IRES translation is part of a positive feed-forward loop that contributes to TNFα-mediated PML protein accumulation in HUVECs. Unlike T cells, our data suggested that TNFα-mediated PML protein accumulation is eIF4E independent. First, TNFα induces PML protein accumulation in a time-dependent manner that continues beyond 4 h of TNFα treatment, when the active form of eIF4E (p-eIF4E) is significantly decreased (Figures 1b and d and 3 and Supplementary Figure 7c). Second, a previous report has shown that TNFα stimulation in MCF7 cells results in a decrease in 4E-BP phosphorylation that promotes the ability of 4E-BP to sequester and suppress eIF4E.9, 62 However, we demonstrated that with an ectopically expressed 4E-BP mutant, 4E-BP (5A) that is devoid of phosphorylation by mTORC1, TNFα-mediated PML protein accumulation and PML IRES activation behaves similarly to the 4E-BP-transfected cells (Figures 4h–i).
Mouse PML has been found localized in the ER of MEFs and contributes to cell apoptosis.40 In our study, we were unable to detect endogenous PML in the ER of HUVECs or MCF7 cells (Figure 3 and Supplementary Figure 2). This observation suggests that the ability of PML-mediated TNFα cytotoxic activity in MCF7 cells does not involve the previously described cytoplasmic PML. The mouse Pml 5′-UTR has been shown to mediate oncogenic RAS-induced PML mRNA translation through an mTOR kinase cascade,63 but the detailed mechanism is not clear. The human PML 5′-UTR is 140 nucleotides long, 40 nucleotides longer than the mouse Pml. Using bicistronic reporter assays, we have mapped a bona fide IRES to human PML 5′-UTR (-100->-1) region. We used stringent criteria to determine the human functional PML IRES. First, we excluded the possibility that human PML 5′-UTR (-100->-1) region contains any cryptic promoter activity by employing a promoterless reporter assay (Supplementary Figure 5). Furthermore, direct transfection of in vitro synthesized bicistronic mRNAs demonstrated that PML 5′-UTR (-100->-1) harbors an IRES (Figure 4f). Sequence alignment of PML 5′-UTR from several mammals revealed that the PML 5′-UTR (-100->-1) region is highly conserved, with the exception of the mouse PML 5′-UTR that is loosely conserved (Supplementary Figure 10a). A free energy and secondary structure prediction revealed that the PML 5′-UTR (-100->-1) region contains two structural elements, namely two stem loops connected by an open loop, that may be the possible structural requirement for ribosome entry (Supplementary Figure 10b).64, 65 All known PML isoforms are the products of alternative splicing of C-terminal exons from a primary PML transcript and there have been no reports of PML isoforms with a different 5′-UTR.66 Thus, it is likely that IRES-mediated translational regulation applies to all isoforms. The IRES-mediated mRNA translation mainly occurs when cells confront different stresses, such as hypoxia and a cytotoxic reagent treatment.67 The IRES-mediated PML mRNA translation observed here is proposed to have a dominant role in PML protein production for cell growth control under these conditions. In summary, we conclude that the PML 5′UTR (-100->-1) region contains a functional IRES and is likely part of the underlying mechanism that controls the accumulation of PML protein.
Several studies suggested that targeting MNK1 represents a novel strategy to inhibit tumor growth.68, 69, 70 Breast cancer cell lines such as SKBr3 or BT474 showed sensitivity to MNK1 inhibitor CGP57380-mediated anti-proliferation.68 However, MCF7 cells are resistant to CGP57380 treatment. In our study, CGP57380 treatment in MCF7 cells attenuated TNFα-induced cell death and PML accumulation and caused a decrease in the PML IRES response to TNFα (Figures 4 and 8 and Supplementary Figure 11). In addition, knockdown of MNK1 by shRNAs reduced TNFα-mediated PARP1 activation and subsequently protected MCF7 cells from TNFα-induced cell death (Figures 8d–f). Taken together, these results suggested that the antibreast cancer activity by MNK1 inhibitors is cell subtype dependent. This precaution is also likely to be true when treating leukemia patients with IFNα or IFNγ because inhibition of MNK1 activity also compromised IFN-mediated suppression of leukemic progenitor cells.71 Consistent with a previous report, we demonstrate that TNFα significantly inhibits HL60 cell growth, whereas it has little or no effect on NB4 cells that express PML–RARα fusion protein.72 Interestingly, TNFα and CGP57380 synergistically inhibit NB4 cell proliferation. We suspect that the CGP57380-mediated increase in cytotoxicity to TNFα is, in part, due to the inhibition of PML–RARα mRNA translation through the 5′-UTR IRES (Figure 8h).73 Accordingly, inhibitors of MNK1 could be a promising adjuvant agent against some subtypes of leukemia or cancers in which the PML gene harbors dominant-negative, gain-of-function mutations.
Our data also indicate that MNK1 plays a key role in TNFα-mediated suppression of angiogenesis and EC migration (Figure 5). However, unlike its cytotoxic effects on cancer cells, TNFα does not induce apoptosis or senescence in primary cells, including endothelial cells under our assay conditions (Supplementary Figure 8).7, 9, 54, 74 We propose that a potential mechanism to regulate angiogenesis by TNFα is through the regulation of angiogenesis-associated gene expression.4, 25 Indeed, using ChIP assays and qRT-PCR, we demonstrated that MMP10 and MCP-1 are TNFα-inducible genes that are regulated by the MNK1–PML–HDAC7 axis in HUVECs (Figures 6 and 7 and Supplementary Figure 9). Notably, MCP-1 and MMP10 are multifunctional effectors involved in various physiological and pathological processes, including sustained EC activation, endothelium dysfunction and vascular permeability.49–51, 75, 76 Our study reveal a novel mechanism by which the MNK1–PML–HDAC7 cascade mediates TNFα-induced gene expression and provides several possible routes targeting MCP-1 to attenuate the vascular complications in different inflammation-associated diseases.
Materials and Methods
Plasmids and DNA constructs
The expression plasmids are listed in Supplementary Table 2. The PCR fragments of PML 5′-UTR were amplified using HEK293T genomic DNA as a template. For bicistronic plasmids, PCR products were digested by restriction enzymes, EcoRI and NcoI, and subcloned into the pRF plasmid to replace the EMCV IRES, a gift from Dr. Mazumder (Cleveland, OH, USA). The p53 IRES and hairpin were generated as previously reported and subcloned into the pRF plasmid.77, 78 The shRNA plasmids were purchased from Sigma (St. Louis, MO, USA) and their related information is listed in Supplementary Table 1. All clones were sequenced to confirm their identity.
Cell lines, reagents and antibodies
HEK293T, MCF7 and HeLa cells were purchased from ATCC (Manassas, VA, USA) and cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum (FBS), 50 U of penicillin G/ml and 50 μg of streptomycin sulfate at 37 °C in 5% CO2. HUVECs were purchased from Lonza and cultured in endothelial cell basal medium (EBM-2; Lonza, Boston, MA, USA) with EGM-2 SingleQuot growth supplements (Lonza, Madison, WI, USA). Cells that were <6 passages were used in this study. TNFα was purchased from Promega (G5421, Madison, WI, USA). The kinase inhibitors used in this study are: p38 kinase (SB202190; Sigma), CKII (TBCA; Calbiochem, San Diego, CA, USA), MSK (SB747651; Tocris Bioscience, Bristol, UK), JNKs (SP600125; Sigma); ERKs (U0126; Sigma) and MNK (CGP57380; Cayman Chemical, Ann Arbor, MI, USA; or cercosporamide; Tocris Bioscience). The siRNAs were purchased from Thermo Scientific (Waltham, MA, USA) and the siRNA target sequences are listed in Supplementary Table 1. Antibodies used in this study are listed in the Supplementary Table 3. The transfection reagents DharmaFECT1 (T-2001) and Lipofectamine 2000 (LF2000) were purchased from Thermo Scientific and Invitrogen (Grand Island, NY, USA), respectively.
Cycloheximide treatment
Cells were treated with or without TNFα followed by 40 μM cycloheximide (Sigma) treatment for the indicated times and harvested 16 h after TNFα treatment. Whole-cell lysates were subjected to western blotting with anti-PML and anti-β-actin antibodies. The intensity was quantified by ImageJ (National Institute of Health, Bethesda, MD, USA).
Kinase inhibition and TNFα treatment
Cells were transiently transfected with kinase-specific siRNAs for 48 h or pretreated with kinase inhibitors for 1 h before TNFα treatment. The kinase inhibitor concentration used in this study is 10 μM unless specifically noted. For HUVECs, the cells were treated with TNFα (20 ng/ml) for 16 h. MCF7 and HeLa cells were treated with TNFα 60 ng/ml for 48 h or an indicated time.
siRNA knockdown and cellular protein extraction
For siRNA knockdown, a nontargeting siRNA or two independent siRNAs against target genes (Thermo Scientific) were transfected into HUVECs using DF1 transfection reagent (Thermo Scientific) according to the manufacture’s protocol. Cells were treated with TNFα for 16 h and harvested 72 h after siRNA transfection before preparation of total RNA or whole-cell extracts. Renilla luciferase knockdown by pBS/U6-RLi was performed as previously reported.44 For whole-cell extracts, cells were resuspended in RIPA buffer (1 × PBS, 1% NP-40, 0.5% sodium deoxycholate and 0.1% SDS) plus protease inhibitors (Roche, Nutley, NJ, USA) on ice for 35 min and vortexed every 5 min until completely homogenized. The cell lysates were mixed with an equal volume of 2 × SDS buffer (100 mM Tris-Cl, pH 6.8, 4% SDS, 0.2% bromophenol blue, 20% glycerol and 2% β-mercaptoethanol) and boiled in a hot pot for 10 min. For western blots, β-actin or α-tubulin were included as internal loading controls.
Lentivirus preparation for shRNA knockdown
The shRNAs expressing plasmids were purchased from Sigma and the targeted sequences are listed in Supplementary Table 1. To generate infectious viral particles expressing shRNAs, we individually cotransfected the shRNA plasmid into HEK293T cells with packaging vectors pMD2G and ps-PAX-2 by Lipofectamine 2000 according to the manufacture’s protocol (Invitrogen). The conditioned medium containing the lentivirus was collected 40 h after transfection. After removing cell debris by passage through a 0.45 μm filter, the medium was diluted with culture medium in a 1 : 4 ratio for viral infection. At 6 h after viral infection, the medium was replaced with fresh medium and cells were allowed to recover overnight followed by puromycin selection for an additional 48 h. After puromycin selection, the medium was removed and replaced with fresh medium. Cells were cultured overnight in regular medium before further experiments.
Total RNA extraction, RT-PCR and quantitative real-time PCR (qRT-PCR)
At 72 h after transient transfection or infection with siRNAs or shRNAs respectively, HUVECs, HeLa and MCF7 cells were harvested, total RNA prepared using PrepEase RNA Spin kits (USB/Affymetrix, Cleveland, OH, USA) and quantified by A260/A280 spectrometry. The cDNA pool was generated using Superscript 3 Reverse Transcriptase (Life Technologies, Grand Island, NY, USA) according to the manufacturer’s instructions. The cDNAs of interest and internal controls were quantified by qPCR using an iCycler (Bio-Rad, Hercules, CA, USA) platform with 2 × iQ SYBR Green Supermix (Bio-Rad) and appropriate primer sets. The PCR program was set for 40 cycles with three steps of 95 °C for 30 s, 55 °C for 30 s and 72 °C for 30 s. Melting curves were obtained after PCR to ensure the homogeneity of the PCR products. The relative abundance of genes of interest was normalized to an internal control (18S rRNA) and depicted as mean±S.D. from three independent experiments.
Immunofluorescence microscopy
HUVECs were cultured in 12-well plates, transfected with a control or two independent p38 siRNA and followed by TNFα treatment. Immunofluorescence microscopy was performed using anti-PML antibodies and secondary antibodies conjugated with Alexa Fluor 594 (Invitrogen). Cells were mounted with DAPI (Vectashield; Vector Laboratories, Burlingame, CA, USA) and images were visualized using a fluorescent microscope (Leica Biosystems Inc., Buffalo Grove, IL, USA). For confocal microscopy, anti-PML and anti-protein disulfide isomerase (PDI) antibodies were used to visual whether PML localizes in the ER. The images were taken using a confocal microscope (Leica Biosystems Inc.).
Subcellular fractionation
HUVECs were cultured in 60 mm plates and treated with kinase inhibitors, siRNAs and TNFα as indicated. Before harvest, HUVECs were washed with 1 × PBS one time and spun down using a desktop centrifuge at 4 °C, 5000 r.p.m. for 5 min. The cell pellets were resuspended in five times their volume in cytosolic fractionation buffer (10 mM Tris pH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.5% NP-40 and 5% glycerol) and kept on ice for 10 min. The nuclei were collected by centrifugation at 4000 r.p.m. for 6 min and the supernatants (first cytosolic fraction) were transferred to a new tube. The precipitated pellets were resuspended a second time in cytosolic fractionation buffer and spun down again. The supernatant was collected and combined with the first extract as the cytosolic fraction. The final nuclear pellets were resuspended in nuclear fraction buffer (10 mM HEPES pH 7.5, 400 mM NaCl, 5 mM EDTA, 0.5% NP-40, 1 mM DTT and 5% glycerol) on ice for 15 min and vortexed every 5 min until completely homogenized.
Chromatin immunoprecipitation
ChIP assays were modified from our published protocol.79 Briefly, HUVECs were treated with 1% formaldehyde for 10 min at room temperature and the crosslinking reaction was stopped by adding glycine to a final concentration of 0.125 M and incubated for 5 min. Cells were harvested and chromatin was sheared by sonication to an average length of 300–700 bp. Anti-HDAC7 antibody was confirmed (Supplementary Figure 9b) and used for immunoprecipitation and the immune complex was subsequently pulled down by protein A beads and washed at least 5 times by ChIP wash buffer. As a control experiment (Mock), no antibody was added. The DNA–protein complex was eluted with a freshly prepared elution buffer (50 mM NaHCO3, 1% SDS) at room temperature for 20 min. The eluted chromatin was incubated with 300 mM NaCl at 68 °C to reverse the crosslinks. The protein was subsequently digested with 10 mg/ml proteinase K at 45 °C for 3 h. The DNA was extracted using a DNA purification kit (Roche) and subjected to PCR analysis after RNA digestion. The results were calculated from three independent real-time PCR experiments and presented as mean±S.D. of the relative fold enrichment as the percentage of input signal. The primers for real-time PCR are listed in the Supplementary Table 1.
Dual luciferase reporter assay for bicistronic reporter and cryptic promoter analysis
The bicistronic reporter plasmids or promoterless constructs were individually transfected into HUVECs, MCF7 or HeLa cells by LF2000 (Invitrogen). At 48 h post transfection, cells were harvested by adding 0.3 ml lysis buffer and mixed gently for 10 min on ice. The activities of Renilla (Renilla reniformis) luciferase (RLuc) and Firefly (Photinus pyralis) luciferase (FLuc) were measured using dual luciferase reporter assay (E1910; Promega) by FB12 luminometer (Berthold Technologies, Bad Wildbad, Germany) according to the manufacturer’s instructions. Briefly, 50 μl of lysate was mixed with 50 μl of Luciferase Assay Reagent II to determine luminescent signal for FLuc. After the luminescence was quantified, the FLuc activity was quenched and RLuc activity was measured by adding 50 μl Stop & Glo Reagent (E1910 Promega). For Figures 4d and e, a CMX-β-galactosidase (β-gal) plasmid was cotransfected with modified pRF constructs as a transfection efficiency control and the intensity of β-gal was measured as previously described.5
In vitro mRNA transcription and transfection
The DNAs containing 5′ T7 polymerase promoter and 3′ 35 nucleotide polyA tail were amplified from pRF bicistronic plasmids by specific primers as listed in the Supplementary Table 1. The amplified DNA fragments were used to in vitro transcribe uncapped mRNA using T7 RNA polymerase (M0251) (New England Biolabs, Ipswich, MA, USA) according to the manufacture’s protocol. To generate capped mRNA, a 10-fold excess of RNA cap structure analog (3′-0-Me-m7G(5')ppp(5')G) was used to substitute GTP during in vitro transcription. After overnight transcription reaction at 37 °C, the mRNAs were purified by high pure RNA purification kit (Roche). Then, 3 μg mRNA was transfected into HeLa cells in 12-well plates by LF2000 for 9 h as previously reported and the activities of RLuc and FLuc were measured as described above.44 Total mRNAs were extracted from an aliquot of the transfected cells and the Renilla and Firefly luciferase mRNAs in each sample were determined by qRT-PCR and normalized to 18S rRNA.
Capillary tube formation assay
HUVECs were transiently infected with the indicated virus expressing shRNAs. Cells were treated with trypsin and 104 cells were plated on Matrigel (Millipore, Temecula, CA, USA) in 96-well plates. After seeding on the gel for 2 h, cells were treated with or without TNFα (20 ng/ml) in the culture medium for 12 h before image capture. Six fields per experimental group were randomly picked and the branch points in each field were counted for statistical analysis. A fraction of knockdown cells was collected to examine the knockdown efficacy by western blotting. All results are shown as mean±S.D.
Wound healing assay
HUVECs were transiently infected with virus expressing shRNAs. The next day, the cells were plated on six-well dishes. Before they become confluent, the cells were treated with TNFα (20 ng/ml) for 4 h and the wounds were generated by scratching with sterile pipet tips. At 0 and 8 h after scratching, the images of six randomly chosen fields were captured by a camera equipped with microscope and the width of the same wounds was measured by ImageJ (National Institute of Health). The cell migration rate was quantified by measuring the ratio of the wound closure between 0 and 8 h. The results are shown as mean±S.D.
Trypan blue exclusion assay
MCF7 cells were transiently infected with lentivirus expressing shRNAs. The next day, cells were plated on 35 mm dishes followed by TNFα treatment. Cells were kept in DMEM containing reduced 2% FBS for 1 h before TNFα treatment. At 48 h after TNFα (60 ng/ml) treatment, cells were treated with trypsin and resuspended with 0.2% Trypan blue in 1 × PBS to examine cell viability. An aliquot of cells was loaded onto a hemacytometer and the stained dead cells and the total cells were counted in triplicate experiments. The results are shown as mean±S.D.
WST-1 cell proliferation assays
5 × 104 cells were seeded in 96-well plates and treated as described. WST-1 (Roche) reagents were added to the medium according to the manufacture’s protocol. After 4 h, the number of live cells was determined by absorbance at 450–650 nm.
Colony formation assay
3 × 103 MCF7 cells were plated in 35 mm plates and treated with CGP57380 (10 μM) and TNFα (60 ng/ml). Before treatment, medium was replaced with DMEM medium with 2% serum. Cell culture was maintained in fresh medium with CGP57380, TNFα or both and medium changed every 2 days. After 2 weeks, the colonies were visualized by crystal violet staining and the numbers of colonies was analyzed by ImageJ (National Institute of Health).
Detection of apoptotic and senescent cells
HUVECs were treated with TNFα (20 ng/ml) at the time points indicated and fixed in 4% paraformaldehyde. The apoptotic cells were determined by immunostaining using antibodies against cleaved caspase 3. Approximately 200 cells were scored in each experiment. For detection of senescent cells, senescence-associated β-galactosidase expression was determined by staining cells with 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-gal) (B4252) (Sigma).
Polysome profiling analysis
HUVECs were treated with TNFα and CGP57380 at the indicated time. The polysome profiling analysis was performed according to a previous publication.80 The cell lysates were layered over 15–54% of cold sucrose gradients in buffer (10 mM HEPES-KOH (pH 7.5), 2.5 mM MgCl2, 100 mM KCl). Gradients were centrifuged at 26 000 r.p.m. in a Beckman (Brea, CA, USA) SW28 rotor for 4 h at 4 °C. The relative quantity of mRNA was measured by qPCR.
Statistical analysis
Data are presented as mean±S.D. of three independent experiments. Two compared groups were analyzed by two-tailed Student’s t test. Statistical significance is presented as: n.s., not significant, *P<0.05, **P<0.01 and ***P<0.001.
Abbreviations
- TNFα:
-
tumor necrosis factor-α
- EC:
-
endothelial cell
- MNK1:
-
MAP kinase-activated protein kinase 1
- IRES:
-
internal ribosome entry site
- JNK:
-
c-Jun N-terminal kinase
- ERK:
-
extracellular signal-regulated kinase
- PML:
-
promyelocytic leukemia
- HDAC7:
-
histone deacetylase 7
- CK2:
-
casein kinase 2
- MSK1:
-
mitogen and stress-activated protein kinase
- MK2:
-
MAPKAP kinase 2
- HUVEC:
-
human umbilical endothelial cell
- PML NB:
-
PML nuclear body
- APL:
-
acute promyelocytic leukemia
References
Carswell EA, Old LJ, Kassel RL, Green S, Fiore N, Williamson B . An endotoxin-induced serum factor that causes necrosis of tumors. Proc Natl Acad Sci USA 1975; 72: 3666–3670.
Pasparakis M . Regulation of tissue homeostasis by NF-kappaB signalling: implications for inflammatory diseases. Nat Rev Immunol 2009; 9: 778–788.
Aggarwal BB, Gupta SC, Kim JH . Historical perspectives on tumor necrosis factor and its superfamily: 25 years later, a golden journey. Blood 2012; 119: 651–665.
Cheng X, Liu Y, Chu H, Kao HY . Promyelocytic leukemia protein (PML) regulates endothelial cell network formation and migration in response to tumor necrosis factor alpha (TNFalpha) and interferon alpha (IFNalpha). J Biol Chem 2012; 287: 23356–23367.
Gao C, Cheng X, Lam M, Liu Y, Liu Q, Chang KS et al. Signal-dependent regulation of transcription by histone deacetylase 7 involves recruitment to promyelocytic leukemia protein nuclear bodies. Mol Biol Cell 2008; 19: 3020–3027.
Zhang Y, Herbert BS, Rajashekhar G, Ingram DA, Yoder MC, Clauss M et al. Premature senescence of highly proliferative endothelial progenitor cells is induced by tumor necrosis factor-alpha via the p38 mitogen-activated protein kinase pathway. FASEB J 2009; 23: 1358–1365.
Janzen C, Sen S, Cuevas J, Reddy ST, Chaudhuri G . Protein phosphatase 2A promotes endothelial survival via stabilization of translational inhibitor 4E-BP1 following exposure to tumor necrosis factor-alpha. Arterioscler Thromb Vasc Biol 2011; 31: 2586–2594.
Wu W-S, Xu Z-X, Hittelman WN, Salomoni P, Pandolfi PP, Chang K-S . Promyelocytic leukemia protein sensitizes tumor necrosis factor α-induced apoptosis by inhibiting the NF-κB survival pathway. J Biol Chem 2003; 278: 12294–12304.
Jeffrey IW, Bushell M, Tilleray VJ, Morley S, Clemens MJ . Inhibition of protein synthesis in apoptosis: differential requirements by the tumor necrosis factor alpha family and a DNA-damaging agent for caspases and the double-stranded RNA-dependent protein kinase. Cancer Res 2002; 62: 2272–2280.
Bernardi R, Pandolfi PP . Role of PML and the PML-nuclear body in the control of programmed cell death. Oncogene 2003; 22: 9048–9057.
Wang ZG, Ruggero D, Ronchetti S, Zhong S, Gaboli M, Rivi R et al. PML is essential for multiple apoptotic pathways. Nat Genet 1998; 20: 266–272.
Clemens MJ, Bushell M, Jeffrey IW, Pain VM, Morley SJ . Translation initiation factor modifications and the regulation of protein synthesis in apoptotic cells. Cell Death Differ 2000; 7: 603–615.
Quignon F, De Bels F, Koken M, Feunteun J, Ameisen JC, de The H . PML induces a novel caspase-independent death process. Nat Genet 1998; 20: 259–265.
Pages G, Lenormand P, L'Allemain G, Chambard JC, Meloche S, Pouyssegur J . Mitogen-activated protein kinases p42mapk and p44mapk are required for fibroblast proliferation. Proc Natl Acad Sci USA 1993; 90: 8319–8323.
Wagner EF, Nebreda AR . Signal integration by JNK and p38 MAPK pathways in cancer development. Nat Rev Cancer 2009; 9: 537–549.
Beg AA, Baltimore D . An essential role for NF-kappaB in preventing TNF-alpha-induced cell death. Science 1996; 274: 782–784.
Zarubin T, Han J . Activation and signaling of the p38 MAP kinase pathway. Cell Res 2005; 15: 11–18.
Zhong S, Salomoni P, Pandolfi PP . The transcriptional role of PML and the nuclear body. Nat Cell Biol 2000; 2: E85–E90.
Lallemand-Breitenbach V, de Thé H . PML nuclear bodies. Cold Spring Harb Perspect Biol 2010; 2: a000661.
Lapi E, Di Agostino S, Donzelli S, Gal H, Domany E, Rechavi G et al. PML, YAP, and p73 are components of a proapoptotic autoregulatory feedback loop. Mol Cell 2008; 32: 803–814.
Van Damme E, Laukens K, Dang TH, Van Ostade X . A manually curated network of the PML nuclear body interactome reveals an important role for PML-NBs in SUMOylation dynamics. Int J Biol Sci 2010; 6: 51–67.
Pearson M, Carbone R, Sebastiani C, Cioce M, Fagioli M, Saito S et al. PML regulates p53 acetylation and premature senescence induced by oncogenic Ras. Nature 2000; 406: 207–210.
Mu ZM, Le XF, Vallian S, Glassman AB, Chang KS . Stable overexpression of PML alters regulation of cell cycle progression in HeLa cells. Carcinogenesis 1997; 18: 2063–2069.
Gurrieri C, Capodieci P, Bernardi R, Scaglioni PP, Nafa K, Rush LJ et al. Loss of the tumor suppressor PML in human cancers of multiple histologic origins. J Natl Cancer Inst 2004; 96: 269–279.
Cheng X, Kao HY . Microarray analysis revealing common and distinct functions of promyelocytic leukemia protein (PML) and tumor necrosis factor alpha (TNFalpha) signaling in endothelial cells. BMC Genomics 2012; 13: 453.
Gao C, Ho CC, Reineke E, Lam M, Cheng X, Stanya KJ et al. Histone deacetylase 7 promotes PML sumoylation and is essential for PML nuclear body formation. Mol Cell Biol 2008; 28: 5658–5667.
Reineke EL, Lam M, Liu Q, Liu Y, Stanya KJ, Chang KS et al. Degradation of the tumor suppressor PML by Pin1 contributes to the cancer phenotype of breast cancer MDA-MB-231 cells. Mol Cell Biol 2008; 28: 997–1006.
Yuan WC, Lee YR, Huang SF, Lin YM, Chen TY, Chung HC et al. A Cullin3-KLHL20 ubiquitin ligase-dependent pathway targets PML to potentiate HIF-1 signaling and prostate cancer progression. Cancer Cell 2011; 20: 214–228.
Jeanne M, Lallemand-Breitenbach V, Ferhi O, Koken M, Le Bras M, Duffort S et al. PML/RARA oxidation and arsenic binding initiate the antileukemia response of As2O3. Cancer Cell 2010; 18: 88–98.
Li W, Thakor N, Xu EY, Huang Y, Chen C, Yu R et al. An internal ribosomal entry site mediates redox-sensitive translation of Nrf2. Nucleic Acids Res 2010; 38: 778–788.
Yang DQ, Halaby MJ, Zhang Y . The identification of an internal ribosomal entry site in the 5'-untranslated region of p53 mRNA provides a novel mechanism for the regulation of its translation following DNA damage. Oncogene 2006; 25: 4613–4619.
Coldwell MJ, Mitchell SA, Stoneley M, MacFarlane M, Willis AE . Initiation of Apaf-1 translation by internal ribosome entry. Oncogene 2000; 19: 899–905.
Holcik M, Lefebvre C, Yeh C, Chow T, Korneluk RG . A new internal-ribosome-entry-site motif potentiates XIAP-mediated cytoprotection. Nat Cell Biol 1999; 1: 190–192.
Henis-Korenblit S, Strumpf NL, Goldstaub D, Kimchi A . A novel form of DAP5 protein accumulates in apoptotic cells as a result of caspase cleavage and internal ribosome entry site-mediated translation. Mol Cell Biol 2000; 20: 496–506.
Stoneley M, Chappell SA, Jopling CL, Dickens M, MacFarlane M, Willis AE . c-Myc protein synthesis is initiated from the internal ribosome entry segment during apoptosis. Mol Cell Biol 2000; 20: 1162–1169.
Shi Y, Frost P, Hoang B, Yang Y, Fukunaga R, Gera J et al. MNK kinases facilitate c-myc IRES activity in rapamycin-treated multiple myeloma cells. Oncogene 2013; 32: 190–197.
Shi Y, Sharma A, Wu H, Lichtenstein A, Gera J . Cyclin D1 and c-myc internal ribosome entry site (IRES)-dependent translation is regulated by AKT activity and enhanced by rapamycin through a p38 MAPK- and ERK-dependent pathway. J Biol Chem 2005; 280: 10964–10973.
de Nadal E, Ammerer G, Posas F . Controlling gene expression in response to stress. Nat Rev Genet 2011; 12: 833–845.
Roux PP, Blenis J . ERK and p38 MAPK-activated protein kinases: a family of protein kinases with diverse biological functions. Microbiol Mol Biol Rev 2004; 68: 320–344.
Giorgi C, Ito K, Lin HK, Santangelo C, Wieckowski MR, Lebiedzinska M et al. PML regulates apoptosis at endoplasmic reticulum by modulating calcium release. Science 2010; 330: 1247–1251.
Scaglioni PP, Yung TM, Cai LF, Erdjument-Bromage H, Kaufman AJ, Singh B et al. A CK2-dependent mechanism for degradation of the PML tumor suppressor. Cell 2006; 126: 269–283.
Brown MC, Bryant JD, Dobrikova EY, Shveygert M, Bradrick SS, Chandramohan V et al. Induction of viral, m7G-cap-independent translation and oncolysis by MAPK interacting kinase (MNK)-mediated effects on the Ser-Arg rich protein kinase, SRPK. J Virol 2014; 88: 13135–13148.
Thompson SR . So you want to know if your message has an IRES? Wiley Interdiscip Rev RNA 2012; 3: 697–705.
Van Eden ME, Byrd MP, Sherrill KW, Lloyd RE . Demonstrating internal ribosome entry sites in eukaryotic mRNAs using stringent RNA test procedures. RNA 2004; 10: 720–730.
Holcik M, Graber T, Lewis SM, Lefebvre CA, Lacasse E, Baird S . Spurious splicing within the XIAP 5' UTR occurs in the Rluc/Fluc but not the betagal/CAT bicistronic reporter system. RNA 2005; 11: 1605–1609.
Rapley J, Oshiro N, Ortiz-Vega S, Avruch J . The mechanism of insulin-stimulated 4E-BP protein binding to mammalian target of rapamycin (mTOR) complex 1 and its contribution to mTOR complex 1 signaling. J Biol Chem 2011; 286: 38043–38053.
Mothe-Satney I, Yang D, Fadden P, Haystead TA, Lawrence JC Jr . Multiple mechanisms control phosphorylation of PHAS-I in five (S/T)P sites that govern translational repression. Mol Cell Biol 2000; 20: 3558–3567.
Pohlman TH, Harlan JM . Human endothelial cell response to lipopolysaccharide, interleukin-1, and tumor necrosis factor is regulated by protein synthesis. Cell Immunol 1989; 119: 41–52.
Chang S, Young BD, Li S, Qi X, Richardson JA, Olson EN . Histone deacetylase 7 maintains vascular integrity by repressing matrix metalloproteinase 10. Cell 2006; 126: 321–334.
Saunders WB, Bayless KJ, Davis GE . MMP-1 activation by serine proteases and MMP-10 induces human capillary tubular network collapse and regression in 3D collagen matrices. J Cell Sci 2005; 118: 2325–2340.
Stamatovic SM, Keep RF, Kunkel SL, Andjelkovic AV . Potential role of MCP-1 in endothelial cell tight junction 'opening': signaling via Rho and Rho kinase. J Cell Sci 2003; 116: 4615–4628.
Martin M, Kettmann R, Dequiedt F . Class IIa histone deacetylases: regulating the regulators. Oncogene 2007; 26: 5450–5467.
Burow ME, Weldon CB, Tang Y, Navar GL, Krajewski S, Reed JC et al. Differences in susceptibility to tumor necrosis factor alpha-induced apoptosis among MCF-7 breast cancer cell variants. Cancer Res 1998; 58: 4940–4946.
Munker R, Koeffler P . In vitro action of tumor necrosis factor on myeloid leukemia cells. Blood 1987; 69: 1102–1108.
Waskiewicz AJ, Johnson JC, Penn B, Mahalingam M, Kimball SR, Cooper JA . Phosphorylation of the cap-binding protein eukaryotic translation initiation factor 4E by protein kinase Mnk1 in vivo. Mol Cell Biol 1999; 19: 1871–1880.
Ueda T, Watanabe-Fukunaga R, Fukuyama H, Nagata S, Fukunaga R . Mnk2 and Mnk1 are essential for constitutive and inducible phosphorylation of eukaryotic initiation factor 4E but not for cell growth or development. Mol Cell Biol 2004; 24: 6539–6549.
Brown MC, Dobrikov MI, Gromeier M . MAPK-interacting kinase regulates mTOR/AKT signaling and controls SRPK-responsive type 1 IRES-mediated translation and viral oncolysis. J Virol 2014; 88: 13149–13160.
Knauf U, Tschopp C, Gram H . Negative regulation of protein translation by mitogen-activated protein kinase-interacting kinases 1 and 2. Mol Cell Biol 2001; 21: 5500–5511.
Culjkovic B, Topisirovic I, Skrabanek L, Ruiz-Gutierrez M, Borden KL . eIF4E is a central node of an RNA regulon that governs cellular proliferation. J Cell Biol 2006; 175: 415–426.
Cohen N, Sharma M, Kentsis A, Perez JM, Strudwick S, Borden KL . PML RING suppresses oncogenic transformation by reducing the affinity of eIF4E for mRNA. EMBO J 2001; 20: 4547–4559.
Buxade M, Parra JL, Rousseau S, Shpiro N, Marquez R, Morrice N et al. The Mnks are novel components in the control of TNF alpha biosynthesis and phosphorylate and regulate hnRNP A1. Immunity 2005; 23: 177–189.
Gingras AC, Raught B, Sonenberg N . eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem 1999; 68: 913–963.
Scaglioni PP, Rabellino A, Yung TM, Bernardi R, Choi S, Konstantinidou G et al. Translation-dependent mechanisms lead to PML upregulation and mediate oncogenic K-RAS-induced cellular senescence. EMBO Mol Med 2012; 4: 594–602.
Komar AA, Hatzoglou M . Cellular IRES-mediated translation: the war of ITAFs in pathophysiological states. Cell Cycle 2011; 10: 229–240.
Baird SD, Turcotte M, Korneluk RG, Holcik M . Searching for IRES. RNA 2006; 12: 1755–1785.
Cheng X, Kao HY . Post-translational modifications of PML: consequences and implications. Front Oncol 2013; 2: 210.
Young RM, Wang SJ, Gordan JD, Ji X, Liebhaber SA, Simon MC . Hypoxia-mediated selective mRNA translation by an internal ribosome entry site-independent mechanism. J Biol Chem 2008; 283: 16309–16319.
Wheater MJ, Johnson PW, Blaydes JP . The role of MNK proteins and eIF4E phosphorylation in breast cancer cell proliferation and survival. Cancer Biol Ther 2010; 10: 728–735.
Lim S, Saw TY, Zhang M, Janes MR, Nacro K, Hill J et al. Targeting of the MNK-eIF4E axis in blast crisis chronic myeloid leukemia inhibits leukemia stem cell function. Proc Natl Acad Sci USA 2013; 110: E2298–E2307.
Ramalingam S, Gediya L, Kwegyir-Afful AK, Ramamurthy VP, Purushottamachar P, Mbatia H et al. First MNKs degrading agents block phosphorylation of eIF4E, induce apoptosis, inhibit cell growth, migration and invasion in triple negative and Her2-overexpressing breast cancer cell lines. Oncotarget 2014; 5: 530–543.
Joshi S, Kaur S, Redig AJ, Goldsborough K, David K, Ueda T et al. Type I interferon (IFN)-dependent activation of Mnk1 and its role in the generation of growth inhibitory responses. Proc Natl Acad Sci USA 2009; 106: 12097–12102.
Testa U, Grignani F, Samoggia P, Zanetti C, Riccioni R, Lo Coco F et al. The PML/RARalpha fusion protein inhibits tumor necrosis factor-alpha-induced apoptosis in U937 cells and acute promyelocytic leukemia blasts. J Clin Invest 1998; 101: 2278–2289.
Dolniak B, Katsoulidis E, Carayol N, Altman JK, Redig AJ, Tallman MS et al. Regulation of arsenic trioxide-induced cellular responses by Mnk1 and Mnk2. J Biol Chem 2008; 283: 12034–12042.
Sedger LM, McDermott MF . TNF and TNF-receptors: from mediators of cell death and inflammation to therapeutic giants - past, present and future. Cytokine Growth Factor Rev 2014; 25: 453–472.
Gerszten RE, Garcia-Zepeda EA, Lim YC, Yoshida M, Ding HA, Gimbrone Jr MA et al. MCP-1 and IL-8 trigger firm adhesion of monocytes to vascular endothelium under flow conditions. Nature 1999; 398: 718–723.
Yang J, Park Y, Zhang H, Gao X, Wilson E, Zimmer W et al. Role of MCP-1 in tumor necrosis factor-alpha-induced endothelial dysfunction in type 2 diabetic mice. Am J Physiol Heart Circ Physiol 2009; 297: H1208–H1216.
Basu A, Das P, Chaudhuri S, Bevilacqua E, Andrews J, Barik S et al. Requirement of rRNA methylation for 80S ribosome assembly on a cohort of cellular internal ribosome entry sites. Mol Cell Biol 2011; 31: 4482–4499.
Ray PS, Grover R, Das S . Two internal ribosome entry sites mediate the translation of p53 isoforms. EMBO Rep 2006; 7: 404–410.
Hsu KS, Kao HY . beta-Transducin repeat-containing protein 1 (beta-TrCP1)-mediated silencing mediator of retinoic acid and thyroid hormone receptor (SMRT) protein degradation promotes tumor necrosis factor alpha (TNFalpha)-induced inflammatory gene expression. J Biol Chem 2013; 288: 25375–25386.
Guan BJ, Krokowski D, Majumder M, Schmotzer CL, Kimball SR, Merrick WC et al. Translational control during endoplasmic reticulum stress beyond phosphorylation of the translation initiation factor eIF2alpha. J Biol Chem 2014; 289: 12593–12611.
Acknowledgements
This work was supported, in whole or in part, by R01 DK078965 and HL093269 (to H-YK), R37 DK060596 and R01 DK053307 (to MH), P30 AR-039750 to the Case Western Reserve University Skin Diseases Research Center and the Ohio Department of Development Center for Innovative Immunosuppressive Therapeutics (Prime Award, TECH09-023). We also thank Drs. David Samols and William Merrick for their comments on the manuscript. We also thank Drs. Avruch, Lloyd and Mazumder for their kind gifts, the pEBG-4EBP, pBS/U6-RLi and pRF plasmids.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by V Stambolic
Supplementary Information accompanies this paper on Cell Death and Differentiation website
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Hsu, KS., Guan, BJ., Cheng, X. et al. Translational control of PML contributes to TNFα-induced apoptosis of MCF7 breast cancer cells and decreased angiogenesis in HUVECs. Cell Death Differ 23, 469–483 (2016). https://doi.org/10.1038/cdd.2015.114
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2015.114
This article is cited by
-
Kindlin-2 controls angiogenesis through modulating Notch1 signaling
Cellular and Molecular Life Sciences (2023)
-
TRIMs: selective recruitment at different steps of the NF-κB pathway—determinant of activation or resolution of inflammation
Cellular and Molecular Life Sciences (2021)
-
PML: Regulation and multifaceted function beyond tumor suppression
Cell & Bioscience (2018)
-
Prolyl isomerase Pin1: a promoter of cancer and a target for therapy
Cell Death & Disease (2018)
-
More than just scanning: the importance of cap-independent mRNA translation initiation for cellular stress response and cancer
Cellular and Molecular Life Sciences (2017)