Abstract
Cardiovascular disease remains the leading cause of morbidity and mortality worldwide. Emerging evidences suggest that the abnormal mitochondrial fission participates in pathogenesis of cardiac diseases, including myocardial infarction (MI) and heart failure. However, the molecular components regulating mitochondrial network in the heart remain largely unidentified. Here we report that miR-361 and prohibitin 1 (PHB1) constitute an axis that regulates mitochondrial fission and apoptosis. The results show that PHB1 attenuates mitochondrial fission and apoptosis in response to hydrogen peroxide treatment in cardiomyocytes. Cardiac-specific PHB1 transgenic mice show reduced mitochondrial fission and myocardial infarction sizes after myocardial infarction surgery. MiR-361 is responsible for the dysfunction of PHB1 and suppresses the translation of PHB1. Knockdown of miR-361 reduces mitochondrial fission and apoptosis in vivo and in vitro. MiR-361 cardiac-specific transgenic mice represent elevated mitochondrial fission and myocardial infarction sizes upon myocardial ischemia injury. This study identifies a novel signaling pathway composed of miR-361 and PHB1 that regulates mitochondrial fission program and apoptosis. This discovery will shed new light on the therapy of myocardial infarction and heart failure.
Similar content being viewed by others
Main
The heart drives the blood flow in the body and it has a large requirement of energy. Mitochondria meet the high energy demand of the heart by consistently providing large amounts of ATP through oxidative phosphorylation. Thus, mitochondrial malfunction is tightly related to cardiac diseases and contributes to cardiomyocyte injury, cardiomyopathy and heart failure. Mitochondria morphology is also associated with the function. Mitochondria constantly undergo fission and fusion. Fission leads to the formation of small round mitochondria and promotes cell apoptosis,1, 2, 3, 4, 5, 6, 7 whereas fusion results in mitochondria elongation and have a protective role in cardiomyocytes maintenance.8 The above findings strongly suggest that mitochondrial fission and fusion machinery is important for cardiac function. In addition, unveiling the mechanism of mitochondrial network regulation will provide a novel therapeutic strategy for heart failure.
The mitochondrial prohibitin complex is a macromolecular structure at the inner mitochondrial membrane that is composed of prohibitin 1 (PHB1) and prohibitin 2 subunits.9 These two proteins comprise an evolutionary conserved and ubiquitously expressed family of membrane proteins and are implicated in several important cellular processes such as mitochondrial biogenesis and function, cell proliferation, replicative senescence, and cell death.10, 11 The first mammalian PHB1 was identified as a potential tumor suppressor with anti-proliferative activity.12 Recent findings suggest that PHB1 has an important role in regulating mitochondrial morphology. Loss of PHB1 results in accumulation of fragmented mitochondria in MEFs and HeLa cells.13, 14 However, it is not yet clear whether PHB1 participates in the regulation of mitochondrial dynamics in cardiomyocytes.
MicroRNAs (miRNAs) are a class of short single-stranded non-coding endogenous RNAs and act as negative regulators of gene expression by inhibiting mRNA translation or promoting mRNA degradation.15, 16 Although the function of miRNAs has been widely studied in apoptosis, development, differentiation and proliferation, few works have been focused on miRNAs in the mitochondrial network regulation. It has been reported that miR-30b targets to p53 and inhibits mitochondrial fission.17 In addition, other miRNAs also affect the function of mitochondria by targeting to mitochondrial calcium uniporter.18 The study of miRNA function in mitochondria may shed new light on the machinery that underlies mitochondrial regulation.
This study unveils that PHB1 is involved in the regulation of mitochondrial network in cardiomyocytes. PHB1 inhibits mitochondrial fission and apoptosis in cardiomyocytes. In addition, PHB1 transgenic mice exhibit a reduced myocardial infarction sizes upon myocardial ischemia injury in vivo. In searching for the mechanism by which PHB1 is downregulated under pathologic condition, we identify miR-361 participates in the suppression of PHB1 translation. MiR-361 initiates mitochondrial fission, apoptosis and myocardial infarction through downregulating PHB1. Our results reveal a novel mitochondrial regulating model, which is composed of miR-361 and PHB1. Modulation of their levels may represent a novel approach for interventional treatment of myocardial infarction and heart failure.
Results
PHB1 is able to inhibit mitochondrial fission and apoptosis in cardiomyocytes
PHB1 is mainly localized in the mitochondrial inner membrane and has been implicated in a wide variety of functions in many cell types. However, the molecular mechanism of PHB1 function in the heart remains unclear. To test whether PHB1 participates in the regulation of mitochondrial fission in cardiomyocytes, we treated cardiomyocytes with 0.2 mM hydrogen peroxide (H2O2) to induce apoptosis (Supplementary Figures 1A and B). The expression levels of PHB1 in mitochondria were downregulated in response to H2O2 treatment (Figure 1a). Cardiomyocytes underwent mitochondrial fission upon 0.2 mM H2O2 treatment (Figure 1b). We then tested whether PHB1 is involved in the occurrence of mitochondrial fission. Enforced expression of PHB1 (Supplementary Figure 2A) efficiently attenuated the reduction of PHB1 upon H2O2 insult (Figure 1c). Enforced expression of PHB1 attenuated mitochondrial fission induced by H2O2 as revealed by the analysis of mitochondrial morphology (Figure 1d) and the counting of the cells with fission (Figure 1e). Further, PHB1 also attenuated H2O2-induced apoptosis analyzed by terminal deoxynucleotidyl transferase-mediated dUTP nick-end-labeling (TUNEL) assay (Figure 1f) and caspase-3 activity (Figure 1g). Cytochrome c (cyto c) release from mitochondria into the cytosol is a critical event in apoptosis. Therefore, we also detected the effect of PHB1 on cyto c release. Our results showed that H2O2 treatment induced cyto c release from mitochondria and PHB1 attenuated H2O2-induced cyto c release (Supplementary Figure 2B). Taken together, these results suggest that PHB1 participates in the regulation of mitochondrial fission and apoptosis in cardiomyoctyes.
PHB1 is able to inhibit mitochondrial fission and apoptosis in cardiomyocytes. (a) H2O2 induces a reduction of PHB1 levels in mitochondria. Cardiomyocytes were treated with 200 μM H2O2 at indicated time. PHB1 levels were analyzed by immunoblot. (b) H2O2 induces mitochondrial fission. Cardiomyocytes were treated with 200 μM H2O2 at indicated times. Cells were stained with mitotracker-red. In addition, the cells with fragmented mitochondria were counted. *P<0.05 versus control. (c) Enforced expression of PHB1 restores the reduction of PHB1 levels induced by H2O2. Cardiomyocytes were infected with adenoviral PHB1 and β-gal for 24 h, then treated with 200 μM H2O2 for another 24 h. PHB1 levels were analyzed by immunoblot. (d and e) Enforced expression of PHB1 prevents mitochondrial fission induced by H2O2. Cardiomyocytes were treated as described in (c), cells were stained with mitotracker-red (d), bar=20 μm. In addition, the cells with fragmented mitochondria were counted (e). (f and g) Enforced expression of PHB1 reduces apoptosis induced by H2O2. Cardiomyocytes were treated as described in (c), TUNEL was used to analyze apoptotic cells (f, left panel). TUNEL-positive cells were counted and calculated (f, right panel). The caspase-3 activity was analyzed by using an Apo-ONE Homogeneous Caspase-3/7 assay kit (g). *P<0.05 versus H2O2 alone
PHB1 regulates mitochondrial fission and apoptosis in vivo
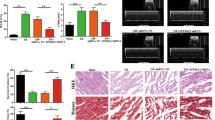
To understand the pathophysiological role of PHB1, we detected whether PHB1 was involved in the pathogenesis of myocardial infarction in the animal model. We used the myocardial ischemic model. Ischemia induced a reduction in PHB1 levels (Figure 2a). To better understand the function of PHB1 in the heart, we generated transgenic mice with cardiac-specific overexpressed PHB1. Four lines of PHB1 transgenic mice demonstrated a high level of PHB1 in the heart (Figure 2b). These mice developed normally to adulthood without significant alterations in terms of phenotype and cardiac function under physiological condition. PHB1 transgenic mice exhibited elevated PHB1 levels, compared with WT group after MI surgery (Figure 2c). Furthermore, PHB1 transgenic mice showed a reduction in mitochondrial fission (Figure 2d) and apoptosis (Figure 2e) upon MI surgery. Myocardial infarction size in PHB1 transgenic mice also showed a remarkable decrease, compared with WT group upon myocardial ischemia injury (Figure 2f). The cardiac function was ameliorated in PHB1 transgenic mice group (Figure 2g). Thus, it appears that PHB1 is able to inhibit mitochondrial fission, apoptosis and myocardial infarction in the heart.
PHB1 regulates mitochondrial fission and apoptosis in vivo. (a) PHB1 is downregulated in response to myocardial ischemia injury. Mice were induced to undergo cardiac ischemia as described in Materials and methods section. PHB1 levels were detected by immunoblot. (b) The expression of PHB1 was analyzed by qRT-PCR from wild-type and PHB1 transgenic mice of different lines. The results were normalized to COX IV. (c) PHB1 transgenic mice restore the levels of PHB1 upon MI surgery. WT and PHB1 transgenic mice were subjected to MI surgery as described in Materials and methods section. PHB1 levels were detected by immunoblot. (d and e) PHB1 transgenic mice attenuate mitochondrial fission and apoptosis upon ischemia injury. WT and PHB1 transgenic mice were subjected to myocardial ischemia injury as described in Materials and methods section. (d) Mitochondrial fission was analyzed. (e) TUNEL assay was performed to detect apoptotic cells. *P<0.05 versus MI+WT. (f) PHB1 transgenic mice attenuates myocardial infarct sizes in response to ischemia injury. WT and PHB1 transgenic mice were subjected to MI surgery as described in Materials and methods section, and infarct sizes were calculated. Left ventricle (LV), infarct area (INF). *P<0.05 versus MI+WT. (g) Echocardiographic analysis. Mice were treated as described in (f), and echocardiography was employed to test cardiac function. Fractional shortening (FS) was calculated. *P<0.05 versus MI+WT
miR-361 participates in the regulation of PHB1 expression
MiRNAs are a class of small non-coding RNAs and act as negative regulators of gene expression. To explore the underlying mechanism by which PHB1 is downregulated upon H2O2 and ischemia injury, we tested whether PHB1 can be regulated by miRNA. We first screened some cardiac-associated miRNAs, which had been reported in past several years by quantitative reverse transcription-polymerase chain reaction (qRT-PCR). Among several miRNAs, miR-361 levels were significantly upregulated upon H2O2 (Figure 3a) and other miRNAs remain unchanged (data not shown). We also analyzed the 3′UTR of PHB1 using the RNAhybrid program and observed that PHB1 is a potential target of miR-361 (Figure 3b), which promoted us to focus on the function of miR-361. We use several approaches to explore whether PHB1 expression can be regulated by miR-361. We first tested whether overexpression or knockdown of miR-361 can alter the expression of endogenous PHB1. Our results showed that enforced expression of miR-361 resulted in a reduction of endogenous PHB1 (Figure 3c). In contrast, knockdown of endogenous miR-361 by antagomir induced an increase in PHB1 expression upon H2O2 treatment (Figure 3d). To understand whether the effect of miR-361 on PHB1 is specific, we then used the target protector (TP) technology.19 The inhibitory effect of miR-361 on PHB1 expression was reduced in the presence of the TP (Figure 3e). PHB1 downregulation was attenuated by the TP in response to H2O2 (Figure 3f). These results indicate that miR-361 can specifically regulate PHB1.
miR-361 participates in the regulation of PHB1 expression. (a) Cardiomyocytes were exposed to H2O2. MiR-361 levels were analyzed by qRT-PCR. *P<0.05 versus control. (b) Putative miR-361 binding site in the 3′UTR region of PHB1. (c) MiR-361 suppresses the expression of PHB1. Cardiomyocytes were infected with adenoviral miR-361 or β-gal. The levels of PHB1 were analyzed by immunoblot. (d) Knockdown of miR-361 attenuates the decrease in PHB1 levels upon H2O2. Cardiomyocytes were transfected with miR-361 antagomir (anta-361) or the antagomir negative control (anta-NC), then exposed to H2O2. The levels of PHB1 were analyzed by immunoblot. (e) PHB1 TP attenuates the reduction of PHB1 induced by miR-361. Cardiomyocytes were infected with adenoviral miR-361, then transfected with the TP (PHB1-TPmiR-361) or the control (PHB1-TPcontrol). PHB1 levels were detected by immunoblot. (f) PHB1 TP inhibits H2O2-induced PHB1 downregulation. Cardiomyocytes were transfected with the PHB1-TPmiR-361 or PHB1-TPcontrol, and then exposed to H2O2. The levels of PHB1 in mitochondria were analyzed by immunoblot. (g) PHB1 wild-type (WT) 3′UTR and the mutated 3′UTR in the miR-361 binding site are shown. (h) MiR-361 suppresses PHB1 translation. HEK293 cells were infected with adenoviral miR-361 or β-gal, then transfected with the luciferase constructs of the wild-type PHB1-3′UTR (PHB1-3′UTR-wt) or a mutated PHB1-3′UTR (PHB1-3′UTR-mut). The luciferase activity was analyzed. *P<0.05
We used the luciferase assay system to test whether miR-361 can influence the translation of PHB1. As shown in Figure 3h, the luciferase reporter assay revealed that the wild-type 3’-UTR of PHB1 exhibited a low translation level in the presence of miR-361, whereas the mutated 3′-UTR (Figure 3g) did not show a significant response to miR-361. Taken together, these data suggest that PHB1 is a specific target of miR-361.
miR-361 can initiate mitochondrial fission and apoptotic program
We explored the functional role of miR-361 in mitochondrial fission and apoptosis. Knockdown of miR-361 by antagomir efficiently attenuated miR-361 levels in response to H2O2 treatment (Figure 4a). Further, knockdown of miR-361 also attenuated mitochondrial fission induced by H2O2 (Figures 4b and c). Concomitantly, apoptosis was reduced in the presence of miR-361 antagomir as revealed by TUNEL assay (Figure 4d), caspase-3 activity (Figure 4e) and cyto c release from mitochondria (Figure 4f). These data indicate that miR-361 can initiate mitochondrial fission and apoptosis in cardiomyocytes.
miR-361 provokes mitochondrial fission and apoptosis in cardiomyocytes. (a) MiR-361 antagomir inhibits the elevation of miR-361 levels upon H2O2. Cardiomyocytes were transfected with anta-361 or anta-NC for 24 h, then exposed to H2O2 for another 24 h. Cells were harvested for qRT-PCR analysis of miR-361 levels. *P<0.05 versus H2O2 alone. (b and c) Knockdown of miR-361 prevents mitochondrial fission induced by H2O2. Cardiomyocytes were treated as described in a, and the cells were stained with MitoTracker Red (b), bar=20 μm. Cells with fragmented mitochondria were counted (c). *P<0.05 versus H2O2 alone. (d–f) Knockdown of miR-361 prevents apoptosis induced by H2O2. Cardiomyocytes were treated as described in a. Apoptosis was analyzed by TUNEL assay (d) and the caspase-3 activity was analyzed by using an Apo-ONE Homogeneous Caspase-3/7 assay kit (e). *P<0.05 versus H2O2 alone. Cyto c distribution in mitochondria-enriched heavy membranes (HMs) or cytosol detected by immunoblot (f)
miR-361 provokes mitochondrial fission and myocardial infarction in mice
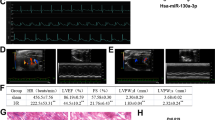
Subsequently, we detected whether miR-361 is involved in the pathogenesis of myocardial infarction in the animal model. MiR-361 was elevated in response to ischemia injury (Figure 5a). Knockdown of miR-361 by antagomir delivery in vivo (Supplementary Figures 3A and B) resulted in an increase in PHB1 expression (Figure 5b) and a reduction in mitochondrial fission (Figure 5c) and apoptosis (Figure 5d). Furthermore, we generated transgenic mice with cardiac-specific overexpression of miR-361 (Supplementary Figures 4A and B). Analysis of adult mice generated from five stable independent transgenic mice founder lines showed that their miR-361 expression was about three to six times as high as that of wild-type hearts (Supplementary Figure 4C). The miR-361 transgenic mice generated from Line#1 were used for this study (Figure 5e). All five transgenic mice founder lines developed normally to adulthood without significant alterations in terms of phenotype (Supplementary Figure 4D) and cardiac function (Supplementary Figure 4E) under physiological condition.
miR-361 regulates mitochondrial fission and apoptosis in vivo. (a) MiR-361 is upregulated in response to ischemia injury. Mice were induced to undergo cardiac ischemia as described in Materials and methods section. MiR-361 levels were detected by qRT-PCR. *P<0.05 versus Sham. (b) Knockdown of miR-361 increases PHB1 expression upon myocardial ischemia injury. Adult male C57BL/6 mice (8 weeks old) were delivered in 3 consecutive days intravenous injections of miR-361 antagomir (anta-361) or antagomir control (anta-NC) at doses of 35 mg/kg body weight. Three days after injection, the mice were exposed to ischemia for 14 days. PHB1 expression was analyzed by immunoblot. (c and d) Knockdown of miR-361 attenuates mitochondrial fission upon myocardial ischemia injury. Mice were treated as described in b. Counting of the fragmented mitochondria was shown in c. Counting of the apoptotic cell was shown in d. TUNEL-positive myocyte nuclei (apoptotic cells) are shown in green. Nuclei stained by DAPI shown in blue. Cardiomyocytes were labeled with α-actinin. n=6, *P<0.05 versus MI alone. (e) qRT-PCR analysis of miR-361 expression levels in different organs or tissues isolated from individual miR-361 transgenic mouse (miR-361 Tg) and wild-type mouse (WT). (f) MiR-361 transgenic mice show a decrease in PHB1 expression. Western blots showing PHB1 expression in mice heart samples from wild-type mice and miR-361 transgenic mice. (g) MiR-361 transgenic mice show a profound reduction in PHB1 protein level compared with WT group after MI surgery. Wild-type mice and miR-361 transgenic mice (8 weeks old) were exposed to 2 weeks of ischemia. PHB1 protein level was analyzed by immunoblot. (h and i) MiR-361 transgenic mice exhibit increased mitochondrial fission, apoptosis and myocardial infarction sizes in response to ischemia injury. Wild-type mice and miR-361 transgenic mice were exposed to 2 weeks of ischemia. Mitochondrial fission (h, upper panel), apoptosis (h, low panel) and myocardial infarction sizes (i) were analyzed. n=8, *P<0.05 versus WT+MI
Next, we demonstrated that miR-361 transgenic mice exhibited an increase in miR-361 expression (Supplementary Figure 5A) and a decrease in PHB1 protein level (Figure 5f). MiR-361 transgenic mice showed an enhanced increase in miR-361 expression (Supplementary Figure 5A) and a profound reduction in PHB1 protein level compared with WT group after MI surgery (Figure 5g). In addition, we also detected PHB1 mRNA level and found that there is no difference of expression between wild-type mice and miR-361 transgenic mice after MI injury (Supplementary Figure 5B), suggesting that miR-361 exerts its inhibitory effect on PHB1 translation level, instead of mRNA level. MiR-361 transgenic mice also potentiated mitochondrial fission and apoptosis upon MI surgery (Figure 5h) and showed an enlarged myocardial infarct sizes, compared with WT group after MI surgery (Figure 5i). These data suggest that miR-361 participates in mediating the signal for mitochondrial fission and apoptosis in the heart.
miR-361 regulates mitochondrial fission and apoptosis through targeting PHB1
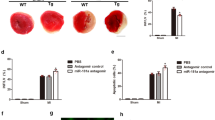
We explored how miR-361 exerts its effect on mitochondrial fission and apoptotic program. As miR-361 is able to suppress PHB1 expression, we thus tested whether PHB1 is a downstream mediator of miR-361. SiRNA-induced gene silencing significantly reduced PHB1 expression levels (Figure 6a). Knockdown of miR-361 inhibited the elevation of miR-361 levels, mitochondrial fission and apoptosis induced by H2O2. However, the inhibitory effect of miR-361 knockdown was attenuated by knockdown of PHB1 (Figures 6b and c). Our results also further showed that the TP of PHB1 inhibited mitochondrial fission (Figure 6d) and apoptosis (Figure 6e) induced by H2O2. These data suggest that miR-361 and PHB1 are functionally linked and miR-361 targets PHB1 in the cascades of mitochondrial fission and apoptosis.
miR-361 regulates mitochondrial fission and apoptosis through targeting PHB1. (a) Expression of PHB1 is knocked down by siRNA-mediated gene silencing. Cardiomyocytes were infected with adenoviral PHB1-siRNA or its scramble form (PHB1-sc) at the indicated MOI. PHB1 levels in mitochondria were detected by immunoblot. (b and c) Knockdown of PHB1 attenuates the inhibitory effect of miR-361 antagomir on mitochondrial fission and apoptosis upon H2O2. Cardiomyocytes were infected with adenoviral PHB1-siRNA or its scramble form, transfected with anta-361 or anta-NC, and then exposed to H2O2. Cells were harvested for the analysis of PHB1 expression levels (b), mitochondrial fission (c, upper panel) and apoptosis (c, low panel). *P<0.05. (d and e) PHB1 TP inhibits mitochondrial fission and apoptosis induced by H2O2. Cardiomyocytes were transfected with the TP (PHB1-TPmiR-361) or the control (PHB1-TPcontrol), and then exposed to H2O2. Mitochondrial fission (d) and apoptosis (e) were analyzed. *P<0.05 versus H2O2 alone
Discussion
Mitochondria are enriched in cardiomyocyte and constantly undergo fusion and fission. Mitochondria form a tubular network and are essential for cardiomyocytes function. Dysfunction in mitochondrial fusion and fission results in cardiac disorders.20, 21 Studying and unveiling the underlying mechanism of miRNA and PHB1 in mitochondrial function may have groundbreaking discoveries in the realm of cell biology and in therapeutics of cardiac diseases as well. Here we present evidence for a critical role of PHB1 in regulating mitochondrial dynamics. PHB1 can inhibit mitochondrial fission and apoptosis in cardiomyocytes and PHB1 transgenic mice showed a reduced myocardial infarction sizes upon ischemia injury in vivo. In addition, we demonstrated that miR-361 is responsible for the downregulation of PHB1, and modulation of miR-361 levels also affects mitochondrial fission, apoptosis and myocardial infarction. MiR-361 exerts its effect on mitochondrial fission and apoptosis through targeting PHB1. Our results provide novel evidence demonstrating that miR-361 and PHB1 constitute an axis in the regulated machinery of mitochondrial network.
Mitochondria undergo extensive fragmentation during apoptosis. The relationship between mitochondrial fission and apoptosis is largely dependent on the type of stimuli and the cell. It is suggested that different lethal stimuli trigger different cellular responses. Perfettini et al.22 also pointed out that mitochondrial fission can either enhance or reduce the probability of mitochondrial outer membrane permeabilization and consequent cell death, depending on the apoptosis stimulus. For example, Drp1-dependent mitochondrial fragmentation lies upstream in several apoptotic pathways in HeLa cells.23 In addition, inhibition of mitochondrial fission reduces COS-7 and SW480 apoptosis.23 Translocation of Bax and Bak to the fission sites enables the fission machinery to promote apoptosis in HeLa cells.24 In oxidative stress-mediated mitochondrial injured neurons, inhibition of the mitochondrial fission machinery inhibits cell death, indicating that it is important in apoptosis.25 Drp1 dysfunction reduces mitochondrial fission and cyto c release, which finally decreases cell death in response to proapoptotic agents in cardiac cell lines.26 At the same time, it is also suggested that mitochondrial fission may be a consequence rather than a cause of apoptosis in several cell lines. For example, in the liver cell, the trigger of mitochondrial fission by Hepatitis C virus attenuates apoptosis.27 In addition, mitochondrial fission uncouples from cyto c release in HeLa cells transfected with expression plasmids encoding Bax.28 Our present work showed that H2O2 treatment led to mitochondrial fission and cyto c release from mitochondria, and consequent cardiomyocyte death. However, PHB1 or miR-361 knockdown attenuated H2O2-induced mitochondrial fission, cyto c release and cell death. Our results indicate that mitochondrial fission promotes apoptosis in the H2O2-induced cardiomyocyte apoptotic pathway.
It has been reported that PHB1 increased apoptosis in gastric cancer cell BGC823 through activating the mitochondrial pathway.29 However, other report shows that prohibitin inhibits apoptosis in rat granulosa cells through the extracellular signal-regulated kinases 1/2 and the Bcl family of proteins.30 In addition, PHB1 inhibits staurosporine-induced apoptosis and promotes survival in ovarian cancer cells.31 Consistent with previous findings, our study shows that PHB1 inhibits H2O2-induced apoptosis in cardiomyocytes. Thus, whether PHB1 is proapoptotic or anti-apoptotic mainly depends on the type of cell. It may be that same factor has different roles in different cell types.
PHB1 has also been suggested to be localized in the nucleus in some mammalian cell lines.32, 33 PHB1 has also been shown to interact with the retinoblastoma protein, resulting in the inhibition of the transcriptional activity of E2F.34 PHB1 protects the cells from camptothecin-induced apoptosis via downregulation of E2F activity in a B-cell lymphoma line.35 These findings suggest the involvement of PHB1 in the regulation of transcription in the nucleus. In this study, we analyzed the mitochondrial PHB1 protein in cardiomyocytes and identified miR-361 as a novel regulator for PHB1. Further investigations into the function and molecular regulation of PHB1 in mitochondria and nucleus should constitute an important area for future research. Our results may have important therapeutic implications for the usage of PHB1 in the treatment of apoptosis-related cardiac diseases.
Some miRNAs have been reported to regulate apoptosis in cardiomyocytes.20, 36, 37 However, few works have been focused on miRNAs in the mitochondrial network regulation. It is critical to identify those miRNAs that can regulate mitochondrial dynamics and apoptosis in the heart and to characterize their signal transduction pathways in the apoptotic cascades. Our present work for the first time demonstrates that miR-361 is able to regulate mitochondrial fission and apoptosis both in vitro and in vivo through targeting PHB1. Accordingly, it can be speculated that the identification of miRNAs that regulate apoptotic proteins may fill in the gap between unknown aspects of cell biology and the pathogenesis of diseases.
Previous studies have showed that miR-361 has important role in various cancerous diseases, such as prostate cancer,38 cervical cancer39 and cutaneous squamous cell carcinoma.40 However, no report shows that miR-361 is involved in the regulation of cardiac diseases. Our results identify that miR-361 initiates mitochondrial fission and apoptotic program by supressing PHB1 expression in cardiomyocytes. In addition, miR-361 cardiac-specific transgenic mice represent elevated myocardial infarction sizes after MI surgery. Our study will shed new light on the miRNA-based therapy for myocardial infarction and heart failure.
Materials and Methods
Generation of cardiac-specific miR-361 transgenic mice and PHB1 transgenic mice
For creating miR-361 transgenic mice, a DNA fragment containing murine miR-361 was cloned to the vector, pαMHC-clone26 (kindly provided by Dr. Zhong Zhou Yang), under the control of the α-myosin heavy chain (α-MHC) promoter. The primers used to generate miR-361 transgenic mice include, forward primer: 5′-GATTCTTTCTTGGGACTCGGAAGCT-3′; reverse primer: 5′-GGGATAAGATGCTAATGAATGTGCT-3′. For generation of PHB1 transgenic mice, the mouse PHB1-coding sequence was synthesized by PCR using mouse cDNA as the template and was cloned to the vector under the control of the α-MHC promoter. The primers used to generate PHB1 transgenic mice include, forward primer: 5′-ATGGCTGCCAAAGTGTTTGAG-3′; reverse primer: 5′-TCACTGGGGAAGCTGGAGAAG-3′. Microinjection was performed following standard protocols.
Cardiomyocyte culture and treatment
Cardiomyocytes were isolated from 1- to 2-day old mice as we described.41 Briefly, after dissection hearts were washed, minced in HEPES-buffered saline solution. Tissues were then dispersed in a series of incubations at 37 °C in HEPES-buffered saline solution containing 1.2 mg/ml pancreatin and 0.14 mg/ml collagenase (Worthington, Lakewood, NJ, USA). After centrifugation, cells were re-suspended in Dulbecco’s modified Eagle medium/F-12 (GIBCO, Grand Island, NY, USA) containing 5% heat-inactivated horse serum, 0.1 mM ascorbate, insulin-transferring-sodium selenite media supplement (Sigma, St. Louis, MO, USA), 100 U/ml penicillin, 100 μg/ml streptomycin and 0.1 mM bromodeoxyuridine. The dissociated cells were pre-plated at 37 °C for 1 h. The cells were then diluted to 1 × 106 cells/ml and plated in 10 μg/ml laminin-coated different culture dishes according to the specific experimental requirements. Cells were treated with 200 μM H2O2 for the indicated periods of time.
TUNEL assays
Apoptosis was determined by the terminal deoxyribonucleotidyl transferase-mediated TUNEL using a kit from Roche (Hamburg, Germany). The detection procedures were in accordance with the kit instructions. Caspase-3 was determined by analyzing its activity using an Apo-ONE Homogeneous Caspase-3/7 assay kit from Promega (Madison, WI, USA) according to the manufacturer’s protocol.
Preparations of subcellular fractions
Subcellular fractions were prepared as we described.42 In brief, the cells were washed twice with PBS and the pellets were suspended in 0.2 ml of buffer A (20 mmol/l HEPES pH 7.5, 10 mmol/l KCl, 1.5 mmol/l MgCl2, 1 mmol/l EGTA, 1 mmol/l EDTA, 1 mmol/l DTT, 0.1 mmol/l PMSF, 250 mmol/l sucrose) containing a protease inhibitor cocktail. The cells were homogenized by 12 strokes in a Dounce homogenizer. The homogenates were centrifuged twice at 750 g for 5 min at 4 °C to collect nuclei and debris. The supernatants were centrifuged at 10 000 g for 15 min at 4 °C to collect mitochondria-enriched heavy membrane pellet. The resulting supernatants were centrifuged to yield cytosolic fractions.
Preparations of miR-361 expression constructs
miR-361 was synthesized by PCR using mouse genomic DNA as the template. The upstream primer was 5′- CTGAGGGAAAACAAATCTTACC-3′; the downstream primer was 5′-CAGGTGTTACAGCATTAGAAAG-3′. The PCR fragment was finally cloned into the Adeno-X Expression System (Clontech, Otsu, Japan) according to the manufacturer's instructions.
Adenoviral constructions and infection
The mouse PHB1 was synthesized by PCR using mouse cDNA as the template. The adenovirus harboring the PHB1 was constructed using the Adeno-X expression system (Clontech). The construction of adenovirus containing β-galactosidase (β-gal) is as we described elsewhere.43 The mouse PHB1 RNA interference (siRNA) target sequence is 5′-TCCTCTTCCGGCCGGTGGC-3′. A scramble form was used as a control, 5′-CCGCTCTCGTCGCTCGTGG-3′. The adenoviruses harboring PHB1-siRNA or its scramble form were constructed using the pSilencer adeno 1.0-CMV System (Ambion, Grand Island, NY, USA) according to the kit’s instructions. All constructs were amplified in HEK293 cells. Adenoviral infection of cardiomyocytes was performed as we described previously.41
Transfection of the antagomir
The chemically modified antagomir complementary to miR-361 was designed to inhibit endogenous miR-361 expression, the antagomir negative control (antagomir-NC) were obtained from GenePharma Co. Ltd (Shanghai, China). The miR-361 antagomir sequence was 5′-GUACCCCUGGAGAUUCUGAUAA-3′. The antagomir-NC sequence was 5′-CAGUACUUUUGUGUAGUACAA-3′. Cells were transfected with the antagomirs or the antagomir-NC using Lipofectamine 2000 (Invitrogen, Grand Island, NY, USA) according to the manufacturer's instruction.
TP preparation and transfection
TP was designed and named as others and we described.43 In brief, PHB1-TPmiR-361 sequence is 5′- AGGCCAGTCTGCAGAGGCACTTGG-3′. PHB1-TPcontrol sequence is 5′-TGACAAATGAGACTCTCTCCTCTCC-3′. They were synthesized by Gene Tools, and were transfected into the cells using the Endo-Porter kit (Gene Tools, Philomath, OR, USA) according to the kit’s instructions.
Immunoblot
Immunoblot was carried out as we previously described.42 Briefly, the cells were lysed for 1 h at 4 °C in a lysis buffer (20 mmol/l Tris pH 7.5, 2 mmol/l EDTA, 3 mmol/l EGTA, 2 mmol/l dithiothreitol (DTT), 250 mmol/l sucrose, 0.1 mmol/l phenylmethylsulfonyl fluoride, 1% Triton X-100) containing a protease inhibitor cocktail. The samples were subjected to 12% SDS-PAGE and transferred to nitrocellulose membranes. Equal protein loading was controlled by Ponceau Red staining of membranes. Blots were probed using the primary antibodies. The anti-PHB1 antibody was from Santa Cruz Biotechnology (Santa Cruz, CA, USA). The anti-COX IV antibody was from Abcam (Grand Island, NY, USA). The antibody to cyto c was from BD Biosciences (San Jose, CA, USA). After four times washing with PBS, the horseradish peroxidase-conjugated secondary antibodies were added. Antigen–antibody complexes were visualized by enhanced chemiluminescence.
Quantitative reverse transcription-PCR
Stem-loop qRT-PCR for mature miR-361 was performed as described on a CFX96 Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA). Total RNA was extracted using Trizol reagent. After DNAse I (Takara, Shiga, Japan) treatment, RNA was reverse transcribed with reverse transcriptase (ReverTra Ace, Toyobo, Kita-ku, Osaka, Japan). The levels of miR-361 analyzed by qRT-PCR were normalized to that of U6. U6 primers were forward: 5′-GCTTCGGCAGCACATATACT-3′; reverse: 5′-AACGCTTCACGAATTTGCGT-3′.
Preparations of the luciferase construct of PHB1-3′UTR and luciferase activity assay
PHB1-3′UTR was amplified by PCR. The forward primer was 5′-CACCCCAGAAAATCACTGTGAA-3′; the reverse primer was 5′- TCTTCCATGGGACTCACATCTCA-3′. To produce mutated 3′UTR, the mutations were generated using QuikChange II XL Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA, USA). The constructs were sequence verified. Wild-type and mutated 3′UTRs were subcloned into the pGL3 vector (Promega) immediately downstream of the stop codon of the luciferase gene.
Luciferase activity assay was performed using the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer’s instructions. Cells were co-transfected with the plasmid constructs of 150 ng per well of pGL3-PHB1-3′UTR or pGL3-PHB1-3′UTR-mut using Lipofectamine 2000 (Invitrogen), then were infected with adenovirus miR-361 or β-gal at a MOI of 80. At 48 h after infection, luciferase activity was measured.
Mitochondrial staining
Mitochondrial staining was carried as we and others described with modifications.4, 44 Briefly, cells were plated onto the cover slips coated with 0.01% poly-L-lysine. After treatment, they were stained for 20 min with 0.02 μM MitoTracker Red CMXRos (Molecular Probes, Eugene, OR, USA). Mitochondria were imaged using a laser scanning confocal microscope (Zeiss LSM510 META, Berkochen, Germany). The percentage of cells with fragmented mitochondria relative to the total number of cells is presented as the mean±S.E.M. of at least three independent experiments, counted by an observer blinded to the experimental conditions. Six distinct fields for each 50 cells were counted. At least 300 cells per group were counted.
Electron microscopy
Heart ultrastructural analyses were performed to quantify mitochondrial fission. Sample preparations and conventional electron microscopy (EM) were carried out as described before.20 Samples were examined at a magnification of 15 000 with a JEOL JEM-1230 transmission electron microscope (JEOL Co. Ltd, Akishima, Tokyo, Japan). For comparison of mitochondrial fission, EM micrographs of thin sections were evaluated. The size of individual mitochondrion was measured by using Image-Pro Plus software (Silver Spring, MD, USA). Approximately 1200–1500 mitochondria were measured to determine the percentages of mitochondria with various sizes. In MI-treated heart tissues, mitochondria disintegrated into numerous small round fragments of varying size, and the number of small mitochondrion was increased. Thus, we determined the mitochondria with size <0.6 mm2 as fission mitochondria. Data represent mean±S.E.M. of at least three independent experiments.
Animal experiments
Male adult C57BL/6 mice (8 weeks old) were obtained from Institute of Laboratory Animal Science of Chinese Academy of Medical Sciences (Beijing, China). All experiments were performed according to the protocols approved by the Institute Animal Care Committee. The mice received on 3 consecutive days intravenous injections of miR-361 antagomir, or its control at a dose of 35 mg/kg body weight in a small volume (0.2 ml) per injection. PHB1 transgenic mice, miR-361 transgenic mice and WT mice were subjected to myocardial ischemic model for 2 weeks. Myocardial ischemic model was established by ligating left anterior descending of coronary artery to cause myocardial ischemia as we previously described.20 Sham-operated group experienced the same procedure except the snare was left untied. Cardiac function of these groups of animals was evaluated by echocardiographic analysis 14 days after the surgery. Evans blue dye was treated as described,20 The areas of infarction (INF) and nonischemic left ventricle (LV) were assessed with computer-assisted planimetry (NIH Image 1.57, Bethesda, MD, USA) by an observer blinded to the sample identity. The ratio of INF/LV was calculated as described.20
Statistical analysis
Data are expressed as the mean±S.E.M. of at least three independent experiments. We used a one-way analysis of variance for multiple comparisons. A value of P<0.05 was considered significant.
Abbreviations
- miRNAs:
-
microRNAs
- H2O2:
-
hydrogen peroxide
- PHB1:
-
prohibitin 1
- TUNEL:
-
terminal deoxynucleotidyl transferase-mediated dUTP nick-end-labeling
- EM:
-
electron microscopy
- MI:
-
myocardial ischemia
- qRT-PCR:
-
quantitative reverse transcription-polymerase chain reaction
- TP:
-
target protector
References
Suen DF, Norris KL, Youle RJ . Mitochondrial dynamics and apoptosis. Genes Dev 2008; 22: 1577–1590.
Tanaka A, Youle RJ . A chemical inhibitor of DRP1 uncouples mitochondrial fission and apoptosis. Mol Cell 2008; 29: 409–410.
McBride HM, Neuspiel M, Wasiak S . Mitochondria: more than just a powerhouse. Curr Biol 2006; 16: R551–R560.
Frank S, Gaume B, Bergmann-Leitner ES, Leitner WW, Robert EG, Catez F et al. The role of dynamin-related protein 1, a mediator of mitochondrial fission, in apoptosis. Dev Cell 2001; 1: 515–525.
Breckenridge DG, Stojanovic M, Marcellus RC, Shore GC . Caspase cleavage product of BAP31 induces mitochondrial fission through endoplasmic reticulum calcium signals, enhancing cytochrome c release to the cytosol. J Cell Biol 2003; 160: 1115–1127.
Wasiak S, Zunino R, McBride HM . Bax/Bak promote sumoylation of DRP1 and its stable association with mitochondria during apoptotic cell death. J Cell Biol 2007; 177: 439–450.
Griffin EE, Graumann J, Chan DC . The WD40 protein Caf4p is a component of the mitochondrial fission machinery and recruits Dnm1p to mitochondria. J Cell Biol 2005; 170: 237–248.
Chen Y, Liu Y, Dorn GW . Mitochondrial fusion is essential for organelle function and cardiac homeostasis. Circulation research 2011; 109: 1327–1331.
Tatsuta T, Model K, Langer T . Formation of membrane-bound ring complexes by prohibitins in mitochondria. Mol Biol Cell 2005; 16: 248–259.
Artal-Sanz M, Tavernarakis N . Prohibitin and mitochondrial biology. Trends Endocrinol Metab 2009; 20: 394–401.
Theiss AL, Sitaraman SV . The role and therapeutic potential of prohibitin in disease. Biochimica et Biophysica Acta 2011; 1813: 1137–1143.
McClung JK, Danner DB, Stewart DA, Smith JR, Schneider EL, Lumpkin CK et al. Isolation of a cDNA that hybrid selects antiproliferative mRNA from rat liver. Biochem Biophys Res Commun 1989; 164: 1316–1322.
Kasashima K, Ohta E, Kagawa Y, Endo H . Mitochondrial functions and estrogen receptor-dependent nuclear translocation of pleiotropic human prohibitin 2. J Biol Chem 2006; 281: 36401–36410.
Merkwirth C, Dargazanli S, Tatsuta T, Geimer S, Lower B, Wunderlich FT et al. Prohibitins control cell proliferation and apoptosis by regulating OPA1-dependent cristae morphogenesis in mitochondria. Genes Dev 2008; 22: 476–488.
Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003; 425: 415–419.
Ohtani K, Dimmeler S . Control of cardiovascular differentiation by microRNAs. Basic Res Cardiol 2011; 106: 5–11.
Li J, Donath S, Li Y, Qin D, Prabhakar BS, Li P . miR-30 regulates mitochondrial fission through targeting p53 and the dynamin-related protein-1 pathway. PLoS Genet 2010; 6: e1000795.
Marchi S, Lupini L, Patergnani S, Rimessi A, Missiroli S, Bonora M et al. Downregulation of the mitochondrial calcium uniporter by cancer-related miR-25. Curr Biol 2013; 23: 58–63.
Choi WY, Giraldez AJ, Schier AF . Target protectors reveal dampening and balancing of Nodal agonist and antagonist by miR-430. Science 2007; 318: 271–274.
Wang JX, Jiao JQ, Li Q, Long B, Wang K, Liu JP et al. miR-499 regulates mitochondrial dynamics by targeting calcineurin and dynamin-related protein-1. Nat Med 2011; 17: 71–78.
Li J, Donath S, Li Y, Qin D, Prabhakar BS, Li P . miR-30 regulates mitochondrial fission through targeting p53 and the dynamin-related protein-1 pathway. PLoS Genet 2010; 6: e1000795.
Perfettini JL, Roumier T, Kroemer G . Mitochondrial fusion and fission in the control of apoptosis. Trends Cell Biol 2005; 15: 179–183.
Frank S, Gaume B, Bergmann-Leitner ES, Leitner WW, Robert EG, Catez F et al. The role of dynamin-related protein 1, a mediator of mitochondrial fission, in apoptosis. Dev Cell 2001; 1: 515–525.
Karbowski M, Lee YJ, Gaume B, Jeong SY, Frank S, Nechushtan A et al. Spatial and temporal association of Bax with mitochondrial fission sites, Drp1, and Mfn2 during apoptosis. J Cell Biol 2002; 159: 931–938.
Sompol P, Ittarat W, Tangpong J, Chen Y, Doubinskaia I, Batinic-Haberle I et al. A neuronal model of Alzheimer's disease: an insight into the mechanisms of oxidative stress-mediated mitochondrial injury. Neuroscience 2008; 153: 120–130.
Ong SB, Subrayan S, Lim SY, Yellon DM, Davidson SM, Hausenloy DJ . Inhibiting mitochondrial fission protects the heart against ischemia/reperfusion injury. Circulation 2010; 121: 2012–2022.
Kim SJ, Syed GH, Khan M, Chiu WW, Sohail MA, Gish RG et al. Hepatitis C virus triggers mitochondrial fission and attenuates apoptosis to promote viral persistence. Proc Natl Acad Sci USA 2014; 111: 6413–6418.
Sheridan C, Delivani P, Cullen SP, Martin SJ . Bax- or Bak-induced mitochondrial fission can be uncoupled from cytochrome C release. Mol Cell 2008; 31: 570–585.
Zhang L, Ji Q, Ni ZH, Sun J . Prohibitin induces apoptosis in BGC823 gastric cancer cells through the mitochondrial pathway. Asian Pac J Cancer Prev 2012; 13: 3803–3807.
Chowdhury I, Thompson WE, Welch C, Thomas K, Matthews R . Prohibitin (PHB) inhibits apoptosis in rat granulosa cells (GCs) through the extracellular signal-regulated kinase 1/2 (ERK1/2) and the Bcl family of proteins. Apoptosis 2013; 18: 1513–1525.
Gregory-Bass RC, Olatinwo M, Xu W, Matthews R, Stiles JK, Thomas K et al. Prohibitin silencing reverses stabilization of mitochondrial integrity and chemoresistance in ovarian cancer cells by increasing their sensitivity to apoptosis. Int J cancer 2008; 122: 1923–1930.
Fusaro G, Dasgupta P, Rastogi S, Joshi B, Chellappan S . Prohibitin induces the transcriptional activity of p53 and is exported from the nucleus upon apoptotic signaling. The Journal of biological chemistry 2003; 278: 47853–47861.
Kurtev V, Margueron R, Kroboth K, Ogris E, Cavailles V, Seiser C . Transcriptional regulation by the repressor of estrogen receptor activity via recruitment of histone deacetylases. J Biol Chem 2004; 279: 24834–24843.
Wang S, Nath N, Adlam M, Chellappan S . Prohibitin, a potential tumor suppressor, interacts with RB and regulates E2F function. Oncogene 1999; 18: 3501–3510.
Fusaro G, Wang S, Chellappan S . Differential regulation of Rb family proteins and prohibitin during camptothecin-induced apoptosis. Oncogene 2002; 21: 4539–4548.
Matkovich SJ, Van Booven DJ, Eschenbacher WH, Dorn GW 2nd . RISC RNA sequencing for context-specific identification of in vivo microRNA targets. Circ Res 2011; 108: 18–26.
Kim HW, Haider HK, Jiang S, Ashraf M . Ischemic preconditioning augments survival of stem cells via miR-210 expression by targeting caspase-8-associated protein 2. J Biol Chem 2009; 284: 33161–33168.
Liu D, Tao T, Xu B, Chen S, Liu C, Zhang L et al. MiR-361-5p acts as a tumor suppressor in prostate cancer by targeting signal transducer and activator of transcription-6(STAT6). Biochem Biophys Res Commun 2014; 445: 151–156.
Wu X, Xi X, Yan Q, Zhang Z, Cai B, Lu W et al. MicroRNA-361-5p facilitates cervical cancer progression through mediation of epithelial-to-mesenchymal transition. Med Oncol (Northwood, London, England) 2013; 30: 751.
Kanitz A, Imig J, Dziunycz PJ, Primorac A, Galgano A, Hofbauer GF et al. The expression levels of microRNA-361-5p and its target VEGFA are inversely correlated in human cutaneous squamous cell carcinoma. PloS One 2012; 7: e49568.
Tan WQ, Wang K, Lv DY, Li PF . Foxo3a inhibits cardiomyocyte hypertrophy through transactivating catalase. J Biol Chem 2008; 283: 29730–29739.
Li PF, Li J, Muller EC, Otto A, Dietz R, von Harsdorf R . Phosphorylation by protein kinase CK2: a signaling switch for the caspase-inhibiting protein ARC. Mol Cell 2002; 10: 247–258.
Lin Z, Murtaza I, Wang K, Jiao J, Gao J, Li PF . miR-23a functions downstream of NFATc3 to regulate cardiac hypertrophy. Proc Natl Acad Sci USA 2009; 106: 12103–12108.
Wang JX, Li Q, Li PF . Apoptosis repressor with caspase recruitment domain contributes to the chemotherapy resistance by abolishing mitochondrial fission mediated by dynamin-related protein-1. Cancer Res 2009; 69: 492–500.
Acknowledgements
This work was supported by National Natural Science Foundation of China (81270160), Beijing Municipal Natural Science Foundation (7142103).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by M Piacentini
Supplementary Information accompanies this paper on Cell Death and Differentiation website
Supplementary information
Rights and permissions
About this article
Cite this article
Wang, K., Liu, CY., Zhang, XJ. et al. miR-361-regulated prohibitin inhibits mitochondrial fission and apoptosis and protects heart from ischemia injury. Cell Death Differ 22, 1058–1068 (2015). https://doi.org/10.1038/cdd.2014.200
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2014.200
This article is cited by
-
MiR-181a protects the heart against myocardial infarction by regulating mitochondrial fission via targeting programmed cell death protein 4
Scientific Reports (2024)
-
The function of prohibitins in mitochondria and the clinical potentials
Cancer Cell International (2022)
-
micro-RNAs dependent regulation of DNMT and HIF1α gene expression in thrombotic disorders
Scientific Reports (2019)
-
Role of noncoding RNAs in regulation of cardiac cell death and cardiovascular diseases
Cellular and Molecular Life Sciences (2018)
-
MicroRNA-143 promotes cardiac ischemia-mediated mitochondrial impairment by the inhibition of protein kinase Cepsilon
Basic Research in Cardiology (2017)