Abstract
The growth of new blood vessels by angiogenesis is essential for normal development, but can also cause or contribute to the pathology of numerous diseases. Recent studies have shown that BIM, a pro-apoptotic BCL2-family protein, is required for endothelial cell apoptosis in vivo, and can contribute to the anti-angiogenic effect of VEGF-A inhibitors in certain tumor models. Despite its importance, the extent to which BIM is autonomously required for physiological endothelial apoptosis remains unknown and its regulation under such conditions is poorly defined. While the transcription factor FOXO3 has been proposed to induce Bim in response to growth factor withdrawal, evidence for this function is circumstantial. We report that apoptosis was reduced in Bim−/− primary endothelial cells, demonstrating a cell-autonomous role for BIM in endothelial death following serum and growth factor withdrawal. In conflict with in vitro studies, BIM-dependent endothelial death in vivo did not require FOXO3. Moreover, endothelial apoptosis proceeded normally in mice lacking FOXO-binding sites in the Bim promoter. Bim mRNA was upregulated in endothelial cells starved of serum and growth factors and this was accompanied by the downregulation of miRNAs of the miR-17∼92 cluster. Bim mRNA levels were also elevated in miR-17∼92+/− endothelial cells cultured under steady-state conditions, suggesting that miR-17∼92 cluster miRNAs may contribute to regulating overall Bim mRNA levels in endothelial cells.
Similar content being viewed by others
Main
The growth of new blood vessels by angiogenesis is fundamentally important for matching vascular supply to the metabolic needs of growing tissues. While normally quiescent in adults, the angiogenic program can be reactivated in pathological form, facilitating the progression of numerous diseases including cancer, chronic inflammatory diseases, diabetic retinopathy and macular degeneration.1 Angiogenesis involves the proliferation, migration and differentiation of endothelial cells, followed by the remodeling of immature new vessels and the recruitment of peri-vascular support cells to form a mature vascular network. Vascular endothelial growth factor A (VEGF-A) is a pro-angiogenic cytokine essential for normal vascular development and angiogenesis2 and is the major target for anti-angiogenic agents currently in clinical use.3 While such agents are effective in the treatment of intraocular vascular diseases,4 they have proven less effective against tumors, which display intrinsic and acquired resistance to VEGF-A inhibitors.5 The exact mechanism(s) by which VEGF-A inhibition prevents pathological vessel growth, particularly in the context of tumors, is the subject of debate. One effect of VEGF-A inhibition may be to impede endothelial cell survival: inhibition of VEGF-A signaling can trigger extensive endothelial cell apoptosis in both normal6, 7 and tumor-associated vessels,8, 9 and exogenous VEGF-A can protect endothelial cells from apoptosis under conditions of stress such as in response to serum deprivation10 and oxygen-induced retinopathy (OIR).11 Understanding the molecular regulation of endothelial cell death may therefore provide useful information for the development of new agents for targeting pathological angiogenesis or the improvement of existing therapeutic strategies.
Apoptosis, or programmed cell death, is a genetically encoded program by which redundant and potentially harmful cells are eliminated from the body. Two pathways sense and transduce apoptotic signals: the intrinsic, BCL2 family-regulated pathway and the extrinsic, death receptor-activated pathway. The BCL2 family of cell death regulators consists of both pro-survival and pro-apoptotic members.12 Apoptotic stimuli including cellular stresses such as growth factor withdrawal, loss of contact with support matrices (‘anoikis’) and DNA damage activate the ‘BH3-only’ subclass of pro-apoptotic proteins (BAD, BID, BIK, BIM, BMF, HRK, noxa and PUMA). BH3-only proteins bind and neutralize the pro-survival members of the family (BCL2, BCLX, BCLW, MCL1 and A1) and either directly or indirectly trigger the activation of the multi-BH domain pro-apoptotic BCL2 family members BAK and BAX. Once unleashed, BAK and BAX cause the release of apoptogenic factors from the mitochondria including cytochrome C, ultimately resulting in the activation of cysteine proteases such as caspase-3, that cleave vital cellular constituents and activate DNases thereby demolishing the cell.12
The BCL2-regulated pathway is essential for apoptosis regulation in endothelial cells. BCL2 expression in endothelial cells is induced by VEGF-A,13, 14 Bcl2−/− mice display increased endothelial apoptosis in vivo,15 whereas mice lacking both BAX and BAK display less.16 The BH3-only protein BIM has a central role in the initiation of endothelial cell death and to date is the only BH3-only protein demonstrated to be necessary for this process.9, 17 VEGF-A has a key role in suppressing the activity of BIM: BIM is required for the apoptotic death of retinal endothelial cells during OIR,17 a process suppressed by ectopic VEGF-A,11 and BIM is required for the apoptotic death of tumor endothelial cells that occurs following VEGF-A inhibition.9 As BIM is required for apoptosis in a broad range of cell types,18 the use of germline Bim knockouts in these studies makes it hard to separate its specific role in endothelial cells from possible indirect effects via other cell types. Furthermore, how any endothelial cell-autonomous activity of BIM might be regulated in vivo is not clear.
The PI3K/Akt pathway is important for endothelial cell survival including in response to the pro-angiogenic growth factors VEGF-A and angiopoietin-1 (Ang1),10, 19 as well as blood flow shear stress.20 In the presence of growth factors, Akt promotes cell survival by phosphorylating the class O forkhead box (FOXO) transcription factors, leading to their expulsion from the nucleus, thus shutting down expression of their pro-apoptotic target genes.21 Bim is a direct transcriptional target of FOXO3 and is induced in response to cytokine deprivation.22, 23, 24, 25 FOXO3 is expressed in endothelial cells and is repressed by PI3K/Akt signaling, including in response to VEGF-A in vitro.26, 27 While endothelial cells express multiple FOXO family members,26, 27, 28 in vitro studies implicate FOXO3 as the predominant regulator of Bim in this cell type: knockdown of Foxo3 but not Foxo1 resulted in reduced Bim expression in endothelial cells,27 whereas overexpression of constitutively active FOXO3 induced Bim expression.29 The requirement for FOXO3 in endothelial cell death in a physiologically relevant context, however, remains to be proven.
Here we present evidence that BIM is intrinsically responsible for the initiation of apoptosis in endothelial cells following serum and growth factor withdrawal. We show that BIM-dependent endothelial apoptosis in vivo proceeds normally in the absence of FOXO3, and does not require direct binding of FOXO factors to the Bim promoter. Upregulation of Bim mRNA in endothelial cells following serum and growth factor withdrawal was accompanied by a corresponding reduction in miR-17-5p and miR-92a, miRNAs known to suppress BIM expression. Accordingly, Bim mRNA levels were elevated in miR-17∼92+/− endothelial cells. Finally, through quantitative assessment of apoptosis using a range of endothelial cell death models, we show that, while BIM is an essential initiator of endothelial cell death in a range of circumstances, it is not the sole inducer of endothelial apoptosis.
Results
BIM is a cell-autonomous inducer of apoptosis in endothelial cells
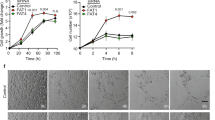
We established a novel method for the culture of primary mouse endothelial cells to assess the cell-intrinsic role of BIM in endothelial cell death in the absence of other cell types. Endothelial cells were isolated from E9.5 embryos by FACS sorting based on expression of the endothelial markers PECAM1 and ICAM2, and lack of the hematopoietic markers CD45 and CD41 (PECAM1Hi ICAM2Hi CD45− CD41−). Sorted cells were plated on fibronectin and cultured for 3 days, after which time >97% of all cells expressed PECAM1 (Figure 1a). To assess the requirement of BIM for apoptosis in these cells, we sought to mimic conditions under which BIM is required for apoptosis in vivo. Given the known role for VEGF-A in repressing BIM-dependent endothelial cell death,9 we assessed viability in response to serum and growth factor deprivation as an approximation of VEGF-A withdrawal. Endothelial cells freshly isolated from Bim−/− and control (Bim+/+ and Bim+/−) littermate embryos were cultured for 3 days without passage, then deprived of serum and growth factors, and viability monitored by time-lapse imaging over 36 h. Although endothelial cells from control embryos had undergone extensive apoptosis after 36 h of serum and growth factor deprivation, significantly less death was observed in Bim−/− endothelial cells from 21 h onward (Figure 1b). These results demonstrated a cell-autonomous requirement for BIM in endothelial cell death following withdrawal of serum and growth factors.
BCL2-interacting mediator of cell death (BIM) is autonomously required for endothelial cell apoptosis in response to serum and growth factor withdrawal. (a) Endothelial cells sorted from E9.5 embryos, cultured for 3 days then stained for platelet/endothelial cell adhesion molecule 1 (PECAM1) (green) and MitoTracker Red (red) (upper panel). Lower panel shows MitoTracker Red signal alone. Shown are representative images of cultures from Bim+/+, Bim+/− and Bim−/− embryos. The total number of cells was counted in each field based on MitoTracker signal and endothelial cells identified as PECAM1+ MitoTracker Red+. Arrows indicate cells that were negative for PECAM1. Scale bar, 200 μm. (b) Time course of endothelial cell death following serum and growth factor withdrawal. Loss of viability was determined by counting the number of cells that became permeable to 4′,6-diamidino-2-phenylindole (DAPI) every hour for 36 h by time-lapse imaging and expressing this as a percentage of the number of viable cells observed at t=0. Viability was determined for Bim+/+ (n=2 embryos), Bim+/− (n=4 embryos) and Bim−/− (n=3 embryos) littermates (*P<0.05, Bim−/− versus Bim+/+, **P<0.05 Bim−/− versus Bim+/−, Student’s t-test). Data are presented as mean±S.E.M.
BIM is expressed in hyaloid endothelial cells and is required for hyaloid vessel regression
Having established that BIM acted autonomously to induce endothelial cell apoptosis, we sought to define how its activity was regulated in a physiological setting. Hyaloid vessels are a transient vessel network that services the developing eye before retina vascularization. In mice, these vessels begin to undergo apoptotic regression beginning at postnatal day (P) 5, resulting in a substantial reduction in vessel density by P8 and complete clearance by around P14. Hyaloid vessel regression requires the intrinsic apoptosis pathway as it is prevented in the combined absence of BAK and BAX.16 Hyaloid endothelial apoptosis is initiated by pericyte-derived Ang2, which blocks Ang1-dependent activation of Akt30 and requires macrophage-derived WNT7b.30, 31 BIM has been implicated in hyaloid vessel regression,17 but its expression, the extent of its requirement, and its regulation in this system have not been determined.
In situ hybridization at P6 demonstrated that Bim mRNA was indeed expressed in hyaloid vessels during their active regression phase (Figure 2a). Using BimlacZ/+ mice,32 we found that in addition to endothelium, Bim was expressed in pericytes and ocular macrophages—cell types with active roles in hyaloid regression (Figure 2b).30 Quantification of hyaloid vessels at P8 showed that Bim−/− pups contained significantly more hyaloid vessels than control littermates, demonstrating that BIM was indeed required for hyaloid vessel regression (Figure 3a). BIM deficiency afforded less protection than that reported in the absence of Ang2, WNT7b or ocular macrophages however,30, 31 and examples of apoptotic vessels were still observed in BIM-deficient mice (n=5) (Figure 3b). Nonetheless, loss of BIM provided long-term protection from regression as those vessels that failed to regress during the neonatal period persisted into adulthood (Figure 3c). These results demonstrate that BIM is required for hyaloid vessel regression, but similar to the extent of its role in endothelial cells deprived of serum and growth factors, it is not solely responsible for apoptosis in this system.
Bim is expressed in hyaloid endothelial cells. (a) In situ hybridization analysis of Bim expression in eyes of P6 wild-type (WT) C57BL/6 mice. Left panel shows bright-field image, right panel shows dark-field image. Arrows indicate representative examples of blood vessels labeling positive for the Bim probe. L, lens; R, retina. Scale bar, 200 μm. (b) Analysis of Bim expression by staining for β-galactosidase activity in P7 eyes from BimlacZ/+ mice. Representative images from BimlacZ/+ (n=2) and littermate Bim+/+ (n=3) mice are shown. Arrows and arrowheads indicate representative examples of macrophages and pericytes, respectively. Scale bar, 50 μm
BCL2-interacting mediator of cell death (BIM) is required for hyaloid blood vessel regression. (a) Hyaloid vessel number was quantified in eyes from Bim−/− (n=4) and Bim+/+ (n=3) littermates at P8. 4′,6-diamidino-2-phenylindole (DAPI) images of representative hyaloid membrane whole-mount spread preparations from Bim−/− and Bim+/+ mice are shown. Bim−/− mice had significantly more vessels per eye than littermates (*P=0.018, Student’s t-test). Data are presented as mean±S.E.M. (b) Representative image of a hyaloid vessel whole mount from a P8 Bim−/− pup (n=5), stained by TUNEL DNA fragmentation assay (red) and for the endothelial marker platelet/endothelial cell adhesion molecule 1 (PECAM1) (gray). Arrows demarcate TUNEL+ vessels undergoing synchronous apoptosis and vessel regression. (c) Representative images of eyes from Bim−/− and wild-type (WT) adult eyes. Arrows indicate persisting hyaloid vessels
Bim mRNA is elevated in hyaloid endothelial cells undergoing regression
Inhibition of VEGF-A in vivo leads to enhanced Bim gene transcription in endothelial cells.9 To determine whether Bim was regulated at the mRNA level during hyaloid regression, we compared Bim mRNA levels in regressing (P5) and non-regressing (P1) vessels. As our expression analysis showed that Bim was expressed in ocular macrophages and pericytes in addition to endothelial cells, we performed our analysis on FACS-sorted hyaloid endothelial cells. Quantitative RT-PCR showed that Bim levels were 2.4 fold higher in hyaloid endothelial cells from P5 vessels compared with those at P1 (Figure 4a). This result demonstrated that Bim expression during hyaloid vessel regression was regulated at the mRNA level.
BCL2-interacting mediator of cell death (BIM)-dependent hyaloid vessel regression occurs independently of FOXO regulation. (a) Bim mRNA levels in P1 and P5 hyaloid endothelial cells determined by quantitative reverse transcriptase (RT)-PCR. Bim levels were normalized to Pgk1 and expressed as fold change relative to P1. Data are presented as mean±S.E.M. from three independent mRNA preparations at both P1 and P5. (b) Hyaloid vessel number was quantified in eyes from Foxo3+/+ (n=4) and Foxo3−/− (n=4) littermates at P8. Data are presented as mean±S.E.M. (c) Semi-quantitative RT-PCR analysis of Foxo1, Foxo3 and Foxo4 mRNA levels in fluorescence-activated cell sorting (FACS)-sorted P5 hyaloid endothelial cells. Hprt is included as a loading control. Negative (water) and positive controls for each gene are included. (d) Hyaloid vessel number was quantified in eyes from BimΔFoxo/ΔFoxo (n=4) and wild-type (WT) (n=4) mice at P8. Data are presented as mean±S.E.M.
BIM-dependent hyaloid vessel regression occurs independently of FOXO3 or direct FOXO binding to the Bim promoter
As FOXO3 is a reported transcriptional regulator of Bim, we sought to determine whether it was required for BIM-dependent hyaloid vessel regression. Quantitative comparison of hyaloid vessel numbers in Foxo3−/− and Foxo3+/+ littermates at P8 showed that hyaloid vessel regression proceeded normally in the absence of FOXO3, demonstrating that it was not required for endothelial apoptosis in this system (Figure 4b). FOXO1 and FOXO4 are two related family members to FOXO3 and bind the same consensus DNA sequence.33 FOXO1 and 4 are expressed in endothelial cells, where they are targets for repression by PI3K/Akt signaling.26, 27, 28 Semi-quantitative RT-PCR performed on FACS-sorted, P5 hyaloid endothelial cells showed that Foxo1, and to a lesser extent Foxo4, were more abundantly expressed in hyaloid endothelium than Foxo3 (Figure 4c). To establish whether BIM-dependent hyaloid regression was due to the redundant activity of FOXO transcription factors, we assessed hyaloid vessel regression in mice in which the four FOXO-binding sites located between positions −625 –+2509 of the Bim transcriptional start site had been mutated (BimΔFoxo).34 Mutation of these four binding sites prevents transcriptional induction of Bim by constitutively active FOXO3.34 Quantitative assessment at P8 showed that hyaloid regression proceeded normally in BimΔFoxo/ΔFoxo mice (Figure 4d). Together these results demonstrate that BIM-dependent hyaloid vessel regression proceeds independently of FOXO3, and does not require direct FOXO-dependent regulation of the Bim promoter.
BIM-dependent endothelial cell apoptosis during hypoxia-induced angiogenesis occurs independently of FOXO3 or direct FOXO binding to the Bim promoter
In light of the previous results, we investigated whether FOXO-dependent regulation of Bim was required for endothelial cell apoptosis in other physiologically relevant contexts. Endothelial apoptosis has been reported in retinal vessels at P5,35 where it is thought to account for vessel pruning around arteries in response to local reductions in VEGF-A levels.36 We investigated whether endothelial apoptosis in the P5 retinal vasculature was affected in Bim−/−, Foxo3−/− or BimΔFoxo/ΔFoxo mice by staining for activated caspase-3, then imaging the entire retinal vasculature by confocal microscopy and quantifying the total number of apoptotic endothelial cells in each eye. BIM-deficient mice showed a significant reduction in the number of apoptotic endothelial cells per retina at P5, indicating that BIM is required for much (but not all) of the apoptosis occurring in the angiogenic retinal vasculature at this time (Figure 5a). In contrast, Foxo3−/− and BimΔFoxo/ΔFoxo mice showed normal rates of endothelial apoptosis in the P5 retinal vasculature (Figures 5b and c). As for hyaloid vessel regression, these results demonstrate that physiological, BIM-dependent endothelial cell death in vivo occurs independently of FOXO3, and does not require direct regulation of the Bim promoter by FOXO transcription factors.
BCL2-interacting mediator of cell death (BIM)-dependent endothelial apoptosis during hypoxia-driven angiogenesis occurs independently of FOXO regulation. (a) Quantitative analysis of apoptotic endothelial cells in retinas of Bim mutants at P5 by immunohistochemical detection of active caspase-3. Representative confocal immunofluorescence microscopy images from Bim+/− and Bim−/− retina whole mounts are shown. Apoptotic cells were detected by staining for active caspase-3 (red) and endothelial cells using isolectin B4 (gray). The total number of apoptotic endothelial cells per retina was determined from control genotypes (combined Bim+/+ and Bim+/−) (n=3) and Bim−/− (n=3) littermates. Data are presented as mean±S.E.M. (*P=0.016, Student’s t-test). Arrows indicate apoptotic endothelial cells. Scale bar, 50 μm. (b) Quantitative analysis of apoptotic endothelial cells in retinas of Foxo3 mutants at P5, as described in a. The total number of apoptotic endothelial cells per retina was determined from Foxo3+/− (n=4) and Foxo3−/− (n=4) littermates. (c) Quantitative analysis of apoptotic endothelial cells in retinas of BimΔFoxo/ΔFoxo mutants at P5, as described in a. The total number of apoptotic endothelial cells per retina was determined from wild type (WT) (n=3) and BimΔFoxo/ΔFoxo (n=4) animals. Data are presented as mean±S.E.M.
Elevated Bim mRNA in endothelial cells following serum and growth factor withdrawal is accompanied by reductions in miRNAs of the miR-17∼92 cluster
The miR-17∼92 cluster is a well-established regulator of Bim expression at the posttranscriptional level.37, 38, 39 To determine whether miR-17∼92 miRNAs might contribute to regulating Bim mRNA levels in endothelial cells, we investigated the expression of two members of this cluster: miR-17 and miR-92a in endothelial cells 24 h after serum and growth factor withdrawal. Quantitative RT-PCR performed using TaqMan probes showed that Bim mRNA was strongly induced (∼15 fold) in response to serum and growth factor deprivation (Figure 6a). In contrast, miR-17-5p was reduced by ∼60% and miR-92a-3p more subtly by ∼25% (Figure 6b), suggesting Bim mRNA levels in serum- and growth factor-starved endothelial cells may be regulated at the posttranscriptional level by members of the miR-17∼92 cluster.
Bim mRNA induction in serum- and growth factor-starved endothelial cells is accompanied by reduction in miR-17∼92 cluster miRNAs. (a) Quantitative reverse transcriptase (RT)-PCR analysis of Bim in endothelial cells grown in the presence (unstarved) or absence (starved) of serum and growth factors for 24 h. Caspase inhibitor quinoline-Val-Asp-CH2-O-Ph (QVD-OPH) was added to all cultures to prevent the loss of cells undergoing apoptosis. Expression of Bim mRNA was normalized to hmbs. Data are represented as fold change relative to unstarved and presented as mean±S.D. of two independent experiments (three replicates per experiment). (b) Quantitative RT-PCR analysis of miR-17-5p and miR-92a-3p in 24 h unstarved and starved endothelial cells as described in a. (c) Quantitative RT-PCR analysis of Bim in mir-17∼92 control genotypes (controls) and mir-17∼92+/− endothelial cells. Cells were grown in the presence (unstarved) or absence (starved) of serum and growth factors for 24 h along with QVD-OPH caspase inhibitor to prevent the loss of cells that have initiated apoptosis. Data are represented as fold change relative to unstarved mir-17∼92 control genotypes and presented as mean±S.E.M. (*P=0.013, n.s., not significant, Student’s t-test). Cells from individual embryos were used to generate data for each genotype and condition: mir-17∼92 control unstarved (n=3 embryos), mir-17∼92+/− unstarved (n=3 embryos), mir-17∼92 control starved (n=6 embryos), mir-17∼92+/− starved (n=3 embryos).
To validate a role for the miR-17∼92 cluster in regulating Bim expression, we investigated Bim mRNA levels in endothelial cells cultured from E9.5 miR-17∼92+/− embryos37 by quantitative RT-PCR using TaqMan probes. miR-17∼92 miRNAs are reduced by around 50% in miR-17∼92+/− animals,37 therefore miR-17∼92+/− endothelial cells are expected to contain comparable levels of mir-17∼92 miRNAs to the cells starved of serum and growth factors described above. Under steady-state conditions (unstarved), we found that Bim mRNA levels were elevated in miR-17∼92+/− endothelial cells compared with littermate controls, consistent with a role for miR-17∼92 miRNAs in repressing Bim mRNA levels at steady state (Figure 6c). In contrast, the effect of miR-17∼92 cluster loss on Bim levels was not observed in endothelial cells that had been serum- and growth factor-starved for 24 h (Figure 6c). It should be noted that the control group in this experiment was composed of the following genotypes: miR-17∼92flox/+without the cre-recombinase and miR-17∼92flox/+with the cre-recombinase present. In the latter case, the cre-recombinase was found to be inefficient, resulting in retention of the functional miR-17∼92flox allele in the majority of the cells. Therefore, these animals were classified as controls. In addition, all control animals had one wild-type miR-17∼92+ allele. Animals displaying complete heterozygous deletion of the miR-17∼92 allele were classified as miR-17∼92+/− embryos. Bim levels in endothelial cells of all the control animals were uniformly low. In contrast, complete heterozygous deletion of the miR-17∼92 allele resulted in a threefold increase in Bim mRNA above controls (Figure 6c).
Discussion
We have extended the range of cell types known to express Bim to include pericytes and tissue-specific macrophages. Both these cell types influence endothelial cell viability, and depending on the context may act to promote30, 31, 35 or suppress apoptosis40, 41 This reinforces the need to address the extent to which BIM acts cell-autonomously to promote endothelial cell apoptosis, independently of effects on other cell types that influence the vasculature. As BIM is essential for apoptosis in many hematopoietic cell types42 and many endothelial cell-specific Cre lines are highly active in hematopoietic cells,43, 44 we established an in vitro viability assay for primary mouse endothelial cells to formally assess the cell-intrinsic role of BIM in endothelial cell death in the absence of other cell types. While previous in vitro analysis of apoptosis in Bim−/− endothelial cells has implicated a cell-autonomous role for BIM, the use of immortalized cells and an extremely high dose of 5-fluorouracil (5 mM) as a death stimulus means these results may not reflect a physiologically relevant role for BIM in endothelial apoptosis.45 In contrast, we developed an assay using non-immortalized, un-passaged, primary endothelial cells and a death stimulus (serum and growth factor withdrawal) to verify that BIM is indeed autonomously required for endothelial cell apoptosis under conditions relevant to the known in vivo role of BIM downstream of VEGF-A.9
Our detailed time-lapse imaging approach revealed that BIM was dispensable for endothelial cell apoptosis in the first 14 h following serum starvation, during which time approximately 40% of endothelial apoptosis occurred. This may reflect a biphasic response of endothelial cells to serum and growth factor withdrawal, for which BIM is required only in the second phase. Alternatively, there may be heterogeneity among the endothelial cells in our culture, with only some requiring BIM to undergo apoptosis. Either way, this result implicates factors in addition to BIM for the initiation of apoptotic death in endothelial cells. Indeed, in none of the contexts we investigated was endothelial death initiated by BIM alone. We have yet to identify other BH3-only proteins that act in concert with BIM to elicit endothelial apoptosis. Potential candidates with known expression in endothelial cells in vivo include BIK in hyaloid endothelial cells,46 and PUMA in retinal endothelial cells.47 PUMA seems particularly attractive given its known role in serum and growth factor withdrawal-induced apoptosis in other cell types.48 Paradoxically, however, it has been reported to have a pro-survival, pro-proliferative role in endothelial cells.47 Given that extensive tissue-specific redundancy exists among BH3-only proteins,49 endothelial cell death will likely depend on multiple BH3-only proteins. Therefore, complete blockade of the intrinsic apoptotic pathway in endothelial cells, for example, through deletion of both BAK and BAX, will be required to determine the full extent to which this pathway is responsible for endothelial cell death.
Despite in vitro evidence that FOXO3 is sufficient to induce Bim-dependent apoptosis in endothelial cells,27, 29 we found no evidence that FOXO3 was required for BIM-dependent endothelial cell apoptosis in vivo. It has been reported that FOXO3 can induce BIM expression in endothelial cells independently of binding to the FOXO consensus sequence.29 Although we can exclude an indirect role for FOXO3 in BIM-dependent endothelial apoptosis, we cannot exclude such a role for FOXO1 or FOXO4. The importance of FOXO1 and FOXO4 in Bim-dependent endothelial death is questionable, however. Unlike Foxo3, knockdown of Foxo1 in endothelial cells did not affect levels of Bim,27 and while expression of constitutively active FOXO4 could induce Bim expression and apoptosis in endothelial progenitor cells, it did not induce death in mature endothelial cells.50 Taken in combination with recent findings in hematopoietic cells,34 it would appear that direct FOXO binding to the Bim promoter is not critical for BIM-dependent apoptosis in general. However, our analysis of endothelial apoptosis was by no means exhaustive, and we cannot exclude the possibility that FOXO3-dependent Bim activity in endothelial cells will be required in specific circumstances for cell killing, for example, in response to reactive oxygen species.51
Given the central importance of BIM in both developmental and stress-induced endothelial cell death in vivo, it is important to resolve how its activity in these cells is regulated independent of FOXO3. A major component of miRNA-mediated posttranscriptional gene repression is through target mRNA degradation.52 Bim is a well-defined target of the miR-17∼92 microRNA cluster37, 38 and VEGF induces the expression of miR-17∼92 cluster members in endothelial cells.53 Our in vitro data have shown that upregulation of Bim mRNA in response to serum and growth factor withdrawal is accompanied by a corresponding decline in the levels of mir-17∼92 cluster member miR-17-5p and to a lesser extent miR-92a-3p (∼60% and ∼25%, respectively). Given our finding that Bim mRNA levels were elevated in miR-17∼92+/− endothelial cells, and mir-17∼92 +/− animals express 50% less miR-17∼92 miRNAs,37 the 60% reduction in miR-17-5p observed following serum and growth factor starvation would appear sufficient to promote an increase in Bim mRNA expression. The level of miR-92a reduction is less convincing in this regard. Levels of other miR-17∼92 cluster members following serum and growth factor starvation were not investigated. The lack of effect of mir-17∼92 heterozygosity on Bim levels following 24 h of serum and growth factor starvation is likely due to the advanced stage of cell death commitment at this time point (70% of cells are normally dead by 24 h), and maximal induction of Bim will likely have occurred. The role for miR-17∼92 miRNAs in Bim-dependent endothelial cell death may be stimulus specific, as only a small reduction in death was observed in endothelial cells overexpressing miR-17 in response to H2O2.54 As discussed above, it is possible that FOXO-dependent transcription may mediate the apoptotic response of endothelial cells to oxidative stress and the impact of miR-17 overexpression on serum and growth factor withdrawal was not investigated in that study.54
Given heterozygous loss of the miR-17∼92 cluster did not induce Bim to the same extent as serum and growth factor withdrawal, we consider it highly unlikely that it acts alone or is the predominant regulator of Bim expression in these cells. The transcription factor E2F1 has been reported to directly induce Bim transcription in endothelial cells in response to angiogensin II, and this activity was suppressed by PI3K/Akt.55 E2F1 may therefore be a critical transcriptional inducer of Bim following growth factor withdrawal and be a target for Akt-mediated repression of Bim transcription in endothelial cells. Interestingly, E2F1 is also a target of the miR-17∼92 cluster,56 and this may provide an additional conduit for miR-17∼92-mediated suppression of Bim induction. E2F1 is only one of eight E2F family members,57 therefore discerning its involvement in endothelial apoptosis may be complicated by functional redundancy.
Our results demonstrate a cell-autonomous role of BIM in endothelial cell apoptosis, confirming that the intrinsic apoptotic pathway is active within endothelial cells. This finding supports the likelihood that small-molecule death agonists that mimic the activity of BH3-only proteins like BIM (so called ‘BH3-mimetics’) may be capable of directly targeting endothelial cells and may find use in an anti-angiogenic context.
Materials and Methods
Mice
All experiments involving animals were performed with procedures approved by the Walter and Eliza Hall Institute of Medical Research Animal Ethics Committee. Bim−/−,42 BimlacZ/lacZ,32 Foxo3−/−,58 BimΔFoxo/ΔFoxo34 and miR-17∼92fl/+ mice37 have been described previously. Animals were maintained on an inbred C57BL/6 background. Bim mutants and littermate controls were generated through heterozygote intercrosses, Foxo3 mutants and littermate controls were generated through either heterozygote intercross or heterozygote × homozygote intercross. BimΔFoxo-mutant mice were generated through homozygote intercrosses and compared with age-matched C57BL/6 control mice. miR-17∼92+/− mutants were generated by crossing miR-17∼92fl/+ mice with a ubiquitous cre deleter,59 then mutants generated by heterozygote intercross.
Endothelial cell culture and viability assay
E9.5 embryos were dissected free of extra-embryonic membranes. Each embryo was dissociated individually in 1 ml of PBS containing 0.25 WU/ml Liberase TM (Roche, Dee Why, NSW, Australia), 10 μg/ml DNaseI and 5 mM MgCl2 for 45 min at 37 °C with constant agitation (800 r.p.m.). Dissociated cells were stained using anti-PECAM1-APC (clone 390, eBiosciences, San Diego, CA, USA), anti-ICAM2-FITC (BD Bioscience, North Ryde, NSW, Australia), anti-CD41-biotin (clone MWReg30, prepared in-house) and anti-CD45.2-biotin (clone S450, prepared in-house) antibodies, and streptavidin–PE. Endothelial cells were FACS sorted using an Influx cell sorter (BD Bioscience) and defined as PECAM1Hi ICAM2Hi CD45-CD41-cells. Dead cells were excluded based on DAPI uptake. Endothelial cells were directly deposited into bovine fibronectin-coated (2.5 μg/cm2) 96-well optical bottom plates (Nunc, Scoresby, VIC, Australia). Between 5700 and 7000 endothelial cells were plated per well. Cells were cultured for 3 days in complete media (MCDB131 media containing 5% serum, 100 U/ml penicillin, 100 μg/ml streptomycin, 10 mM HEPES, 2 mM glutamax, 5.5 μM 2-mercaptoethanol, 75 μg/ml ascorbic acid, 1 μg/ml hydrocortisone, 10 ng/ml rhVEGF-A, 5 ng/ml rhFGF2, 15 ng/ml rhIGF1 and 5 ng/ml rhEGF) at 37 °C in a 5% CO2-humidified incubator. To assess endothelial purity and viability, unfixed, live cells were incubated for 15 min with biotinylated anti-PECAM1 antibody in complete media under normal culture conditions, washed and then incubated for 15 min with streptavidin-Alexa-Fluor488 in complete media under normal culture conditions. Cells were then deprived of serum and growth factors by rinsing each well twice with MCDB131 media containing 0.1% serum and once with starvation media (complete media containing 0.1% serum and no growth factors). Cells were then cultured in starvation media containing 10 μM MitoTracker Red (Invitrogen, Mulgrave, VIC, Australia) and 0.1 μg/ml DAPI for 1 h before initiating time-lapse imaging. This was considered to be t=0. Time-lapse imaging was performed on a Nikon TiE inverted microscope equipped with a Spot Pursuit cooled CCD camera (Spot Imaging, Sterling Heights, MI, USA) and operated using MetaMorph software (Sunnyvale, CA, USA). Images were acquired hourly using a Plan Fluor ELWD × 20/0.45 NA objective (Nikon, Hilton, SA, Australia) for 36 h and binned 2 × 2 to minimize exposure time. Cell number per well was determined by manually counting the number of cells per field of view at t=0 based on the MitoTracker Red signal. Endothelial purity was determined at t=0 by determining the number of cells that were positive for PECAM1 in each field of view. As only dying cells take up DAPI in unfixed samples, viability was determined by counting the number of DAPI signals at each time point per field of view using the ‘count nuclei’ function in MetaMorph and expressed as a percentage of the number of viable cells observed in that field of view at t=0. Viability per well was taken as the average of two randomly selected fields of view. For RNA analysis, endothelial cells were cultured for 24 h with or without serum and growth factors in the presence of 25 μM QVD-OPH caspase inhibitor (MP Biomedicals, Seven Hills, NSW, Australia) to prevent cell death and consequent degradation of proteins and mRNA.
Immunohistochemical analysis of hyaloid and retinal vasculature
Hyaloid vessel dissection and quantitation was performed essentially as described.31 Briefly, eyes were enucleated, fixed in 4% paraformaldehyde for 4 h, injected with 5% gelatin at 40 °C then allowed to set at 4 °C overnight before dissection and mounting on polysine-coated slides (Thermo Scientific, Scoresby, VIC, Australia). Hyaloid vessels were stained for PECAM1 (clone MEC13.3, BD Pharmingen, North Ryde, NSW, Australia), which was detected using fluorescently conjugated anti-rat secondary antibodies (Jackson ImmunoResearch, West Grove, PA, USA). Vessels were mounted in ProLong Gold antifade reagent containing DAPI (Invitrogen). Terminal deoxynucleotidyl transferase dUTP nick-end labeling (TUNEL) assay was performed using the ApopTag Red in situ apoptosis detection kit as per the manufacturer’s instructions (Millipore, Kilsyth, VIC, Australia) with the exception that the reaction was conducted at room temperature for 2 h. Samples were then stained with PECAM1 antibody and DAPI as described above. All hyaloid samples were imaged by wide-field microscopy using a Zeiss Axiovert 200M (Zeiss, North Ryde, NSW, Australia) equipped with a Zeiss MRm Axiocam camera, × 0.63 C-mount, and either a Zeiss Fluar × 5/0.25 NA or Plan Apochromat × 10/0.45 NA objective. Post acquisition, tiled images were merged and minor adjustments to brightness and contrast were made using ImageJ software (NIH, Bethesda, MD, USA). Only non-regressing vessels were considered for quantitation. Regressing vessels were defined based on the presence of TUNEL+ cells or a discontinuous PECAM1 signal. All hyaloid data are presented as mean±S.E. of the mean with n values representing the number of individual animals studied per genotype (a minimum of three eyes were examined for each genotype in each experiment).
For retina analysis, eyes were enucleated and fixed in 4% paraformaldehyde for 2 h at 4 °C. Retinas were then dissected, incubated in blocking buffer (2% normal donkey serum, 1% Triton X-100 in HBSS) then stained with biotinylated isolectin B4 (Vector Laboratories, Burlingame, CA, USA), goat anti-collagen IV antibody (Millipore) and rabbit anti-cleaved (activated) caspase-3 antibody (Cell Signaling Technologies, Beverly, MA, USA; or Promega, Alexandria, NSW, Australia). Retinas were washed then incubated with secondary antibodies: Streptavidin-Alexa-Fluor488 (Jackson ImmunoResearch), anti-goat IgG Alexa-Fluor 647 (Jackson ImmunoResearch) and anti-rabbit IgG Cy3 (Jackson ImmunoResearch). Retinas were then washed before being mounted on slides in ProLong Gold antifade reagent (Invitrogen). Retinas were imaged using a Zeiss LSM 780 confocal microscope with a × 20/0.8 NA objective. Apoptotic endothelial cells were identified by the presence of a cleaved caspase-3 signal within an Isolectin B4-positive vessel. Quantification of cleaved caspase-3-positive endothelial cells was performed manually using ImageJ.
Expression analysis using sorted hyaloid endothelial cells
Hyaloid vessels were dissociated from P1 or P5 C57BL/6 mice. Vessels from multiple animals at each age were pooled and dissociated in PBS containing 0.25 WU/ml Liberase TM (Roche) and 0.1 μg/ml DNaseI. Dissociated cells were then stained using the following antibodies: anti-PECAM1-APC (clone MEC13.3, BD Bioscience) and anti-Mac1-FITC (clone M1/70). Endothelial cells were FACS sorted as PECAM1+ Mac1− cells that were viable based on propidium iodide exclusion. For QPCR analysis, total RNA from P1 and P5 hyaloid endothelial cells was extracted using Trizol Reagent (Invitrogen) and reverse transcribed using QuantiTect Reverse Transcription Kit (Qiagen, Chadstone, VIC, Australia) as per the manufacturer’s instructions. Quantitation of gene expression was performed by real-time PCR with 2 × SYBR mix (Roche) using a LightCycler 480 (Roche). Each reaction was performed in technical triplicate and expression calculated using the ΔΔct method. Oligonucleotides used were as follows: Pgk1 (5′-tacctgctggctggatgg-3′ and 5′-cacagcctcggcatatttct-3′) and Bim (5′-cgtccacccaatgtctgact-3′ and 5′-gctgcaattgtccaccttct-3′). Data are presented as mean±S.E. of the mean of three separate cell sorts at P1 and three separate cell sorts at P5. For semi-quantitative RT-PCR, hyaloid endothelial cells from P5 C57BL/6 mice were dissociated essentially as described above. Cells were stained with the following antibodies: anti-PECAM1-APC, anti-CD45.2-FITC (BD Bioscience) and anti-Ter119-PE (BD Pharmingen), then FACS sorted as PECAM1+ CD45.2− Ter119− cells that were viable based on propidium iodide exclusion. RNA was extracted using Trizol Reagent (Invitrogen) and reverse transcribed using Superscript III reverse transcriptase (Invitrogen) and PCR performed on serial 1/5 dilutions of cDNA using the JumpStart REDTaq ReadyMix (Sigma, Sydney, NSW, Australia). Oligonucleotide sequences used were as follows: Foxo1 (5′-aagagcgtgccctacttcaa-3′ and 5′-tgctgtgaagggacagattg-3′), Foxo3 (5′-cttcaaggataagggcgaca-3′ and 5′-ctgtgcagggacaggttgt-3′), Foxo4 (5′-ggtgccctacttcaaggaca-3′ and 5′-ctgtgcaaggacaggttgtg-3′) and Hprt (5′-gatggccacaggactagaaca-3′ and 5′-tccctggttaagcagtacagc-3′).
RNA isolation and qRT-PCR analysis of Bim and microRNA expression
Total RNA was isolated using TRIzol LS (Life Technologies, Mulgrave, VIC, Australia). For analysis of mRNA, 300 ng of total RNA was reverse transcribed using the Superscript III First Strand Synthesis System for RT-PCR (Life Technologies) following the manufacturer’s instructions. For quantitative PCR, 1 μl of the cDNA synthesis product was used. For the analysis of microRNA, 10 ng of Trizol-isolated RNA was reverse transcribed with microRNA-specific primers using the TaqMan Micro RNA assay kit (Life Technologies). TaqMan PCR was performed using a mouse Bim probe (Life Technologies Cat# MM00437796), a mouse HMBS probe (Life Technologies Cat# MM01143545), a U6 snRNA probe (Life Technologies Cat# 001973; 4427975), a miR-17-5p probe (Life Technologies Cat# 002308: 4427975) and a miR-92a-3p probe (Life Technologies Cat# 000430; 4427975) following the manufacturer’s protocols. Data were calculated using the 2−ΔΔct method.
In situ hybridization
Radioactive in situ hybridization was performed in order to locate Bim transcripts essentially as described previously.60 Briefly, eyes from postnatal mice were dissected and fixed in 4% paraformaldehyde, then embedded in 1% low-melting point agarose and then embedded in paraffin before 8-μm sections were cut. To localize Bim, mRNA sections were dewaxed, rehydrated through graded concentrations of alcohol and then treated with proteinase K (10 μg/ml) for 10 min, fixed in 4% paraformaldehyde for a further 10 min then dehydrated through a graded alcohol series. Sections were then air-dried. Bim sense and antisense cRNA probes were generated by in vitro transcription of a Bim cDNA using T3 and T7 RNA polymerases incorporating [35S]-labeled nucleotides. After purification, cRNA probes were partially hydrolyzed to allow efficient penetration and then incubated overnight at 56 °C on the tissue sections. Slides were washed at 65 °C including an RNaseA step to remove single-stranded RNA molecules. Finally, slides were dipped in liquid film emulsion (Ilford, Cheshire, UK), developed after 1–2 weeks at 4 °C and then counterstained with hematoxylin. Images were acquired on a Zeiss Axioplan 2 microscope equipped with an Axiocam HRc camera using a × 10/0.3 NA Plan Neofluar objective. For clarity, dark-field images were converted to grayscale using ImageJ software.
Statistical analysis
All statistical analysis was performed using a two-tailed Student’s t-test.
Abbreviations
- BIM:
-
BCL2-interacting mediator of cell death
- FOXO:
-
forkhead box O
- VEGF-A:
-
vascular endothelial growth factor A
- PI3K:
-
phosphoinositide-3 kinase
- BH3:
-
BCL2-homology region 3
- FACS:
-
fluorescence-activated cell sorting
- PECAM1:
-
platelet/endothelial cell adhesion molecule 1
- QVD-OPH:
-
quinoline-Val-Asp-CH2-O-Ph
References
Carmeliet P . Angiogenesis in life, disease and medicine. Nature 2005; 438: 932–936.
Coultas L, Chawengsaksophak K, Rossant J . Endothelial cells and VEGF in vascular development. Nature 2005; 438: 937–945.
Ebos JM, Kerbel RS . Antiangiogenic therapy: impact on invasion, disease progression, and metastasis. Nat Rev Clin Oncol 2011; 8: 210–221.
Miller JW, Le Couter J, Strauss EC, Ferrara N . Vascular endothelial growth factor a in intraocular vascular disease. Ophthalmology 2013; 120: 106–114.
Bergers G, Hanahan D . Modes of resistance to anti-angiogenic therapy. Nat Rev Cancer 2008; 8: 592–603.
Baffert F, Le T, Sennino B, Thurston G, Kuo CJ, Hu-Lowe D et al. Cellular changes in normal blood capillaries undergoing regression after inhibition of VEGF signaling. Am J Physiol Heart Circ Physiol 2006; 290: H547–H559.
Lee S, Chen TT, Barber CL, Jordan MC, Murdock J, Desai S et al. Autocrine VEGF signaling is required for vascular homeostasis. Cell 2007; 130: 691–703.
Huang J, Frischer JS, Serur A, Kadenhe A, Yokoi A, McCrudden KW et al. Regression of established tumors and metastases by potent vascular endothelial growth factor blockade. Proc Natl Acad Sci USA 2003; 100: 7785–7790.
Naik E, O'Reilly LA, Asselin-Labat ML, Merino D, Lin A, Cook M et al. Destruction of tumor vasculature and abated tumor growth upon VEGF blockade is driven by proapoptotic protein Bim in endothelial cells. J Exp Med 2011; 208: 1351–1358.
Gerber HP, McMurtrey A, Kowalski J, Yan M, Keyt BA, Dixit V et al. Vascular endothelial growth factor regulates endothelial cell survival through the phosphatidylinositol 3'-kinase/Akt signal transduction pathway. Requirement for Flk-1/KDR activation. J Biol Chem. 1998; 273: 30336–30343.
Alon T, Hemo I, Itin A, Pe’er J, Stone J, Keshet E . Vascular endothelial growth factor acts as a survival factor for newly formed retinal vessels and has implications for retinopathy of prematurity. Nat Med 1995; 1: 1024–1028.
Strasser A, Cory S, Adams JM . Deciphering the rules of programmed cell death to improve therapy of cancer and other diseases. EMBO J 2011; 30: 3667–3683.
Gerber HP, Dixit V, Ferrara N . Vascular endothelial growth factor induces expression of the antiapoptotic proteins Bcl-2 and A1 in vascular endothelial cells. J Biol Chem 1998; 273: 13313–13316.
Nor JE, Christensen J, Mooney DJ, Polverini PJ . Vascular endothelial growth factor (VEGF)-mediated angiogenesis is associated with enhanced endothelial cell survival and induction of Bcl-2 expression. Am J Pathol 1999; 154: 375–384.
Wang S, Sorenson CM, Sheibani N . Attenuation of retinal vascular development and neovascularization during oxygen-induced ischemic retinopathy in Bcl-2-/- mice. Dev Biol 2005; 279: 205–219.
Hahn P, Lindsten T, Tolentino M, Thompson CB, Bennett J, Dunaief JL . Persistent fetal ocular vasculature in mice deficient in bax and bak. Arch Ophthalmol 2005; 123: 797–802.
Wang S, Park S, Fei P, Sorenson CM . Bim is responsible for the inherent sensitivity of the developing retinal vasculature to hyperoxia. Dev Biol 2011; 349: 296–309.
Youle RJ, Strasser A . The BCL-2 protein family: opposing activities that mediate cell death. Nat Rev Mol Cell Biol 2008; 9: 47–59.
Kim I, Kim HG, So JN, Kim JH, Kwak HJ, Koh GY . Angiopoietin-1 regulates endothelial cell survival through the phosphatidylinositol 3'-Kinase/Akt signal transduction pathway. Circ Res 2000; 86: 24–29.
Dimmeler S, Assmus B, Hermann C, Haendeler J, Zeiher AM . Fluid shear stress stimulates phosphorylation of Akt in human endothelial cells: involvement in suppression of apoptosis. Circ Res 1998; 83: 334–341.
Fu Z, Tindall DJ . FOXOs, cancer and regulation of apoptosis. Oncogene 2008; 27: 2312–2319.
Dijkers PF, Medema RH, Lammers JW, Koenderman L, Coffer PJ . Expression of the pro-apoptotic Bcl-2 family member Bim is regulated by the forkhead transcription factor FKHR-L1. Curr Biol 2000; 10: 1201–1204.
Moller C, Alfredsson J, Engstrom M, Wootz H, Xiang Z, Lennartsson J et al. Stem cell factor promotes mast cell survival via inactivation of FOXO3a-mediated transcriptional induction and MEK-regulated phosphorylation of the proapoptotic protein Bim. Blood 2005; 106: 1330–1336.
Gilley J, Coffer PJ, Ham J . FOXO transcription factors directly activate bim gene expression and promote apoptosis in sympathetic neurons. J Cell Biol 2003; 162: 613–622.
Stahl M, Dijkers PF, Kops GJ, Lens SM, Coffer PJ, Burgering BM et al. The forkhead transcription factor FoxO regulates transcription of p27Kip1 and Bim in response to IL-2. J Immunol 2002; 168: 5024–5031.
Abid MR, Guo S, Minami T, Spokes KC, Ueki K, Skurk C et al. Vascular endothelial growth factor activates PI3K/Akt/forkhead signaling in endothelial cells. Arterioscler Thromb Vasc Biol 2004; 24: 294–300.
Potente M, Urbich C, Sasaki K, Hofmann WK, Heeschen C, Aicher A et al. Involvement of Foxo transcription factors in angiogenesis and postnatal neovascularization. J Clin Invest 2005; 115: 2382–2392.
Daly C, Wong V, Burova E, Wei Y, Zabski S, Griffiths J et al. Angiopoietin-1 modulates endothelial cell function and gene expression via the transcription factor FKHR (FOXO1). Genes Dev 2004; 18: 1060–1071.
Czymai T, Viemann D, Sticht C, Molema G, Goebeler M, Schmidt M . FOXO3 modulates endothelial gene expression and function by classical and alternative mechanisms. J Biol Chem 2010; 285: 10163–10178.
Rao S, Lobov IB, Vallance JE, Tsujikawa K, Shiojima I, Akunuru S et al. Obligatory participation of macrophages in an angiopoietin 2-mediated cell death switch. Development 2007; 134: 4449–4458.
Lobov IB, Rao S, Carroll TJ, Vallance JE, Ito M, Ondr JK et al. WNT7b mediates macrophage-induced programmed cell death in patterning of the vasculature. Nature 2005; 437: 417–421.
Bouillet P, Cory S, Zhang LC, Strasser A, Adams JM . Degenerative disorders caused by Bcl-2 deficiency prevented by loss of its BH3-only antagonist Bim. Dev Cell 2001; 1: 645–653.
Furuyama T, Nakazawa T, Nakano I, Mori N . Identification of the differential distribution patterns of mRNAs and consensus binding sequences for mouse DAF-16 homologues. Biochem J 2000; 349 (Pt 2): 629–634.
Herold MJ, Rohrbeck L, Lang MJ, Grumont R, Gerondakis S, Tai L et al. Foxo-mediated Bim transcription is dispensable for the apoptosis of hematopoietic cells that is mediated by this BH3-only protein. EMBO Rep 2013; 14: 992–998.
Simonavicius N, Ashenden M, van Weverwijk A, Lax S, Huso DL, Buckley CD et al. Pericytes promote selective vessel regression to regulate vascular patterning. Blood 2012; 120: 1516–1527.
Claxton S, Fruttiger M . Role of arteries in oxygen induced vaso-obliteration. Exp Eye Res 2003; 77: 305–311.
Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell 2008; 132: 875–886.
Xiao C, Srinivasan L, Calado DP, Patterson HC, Zhang B, Wang J et al. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat Immunol 2008; 9: 405–414.
Koralov SB, Muljo SA, Galler GR, Krek A, Chakraborty T, Kanellopoulou C et al. Dicer ablation affects antibody diversity and cell survival in the B lymphocyte lineage. Cell 2008; 132: 860–874.
Gaengel K, Genove G, Armulik A, Betsholtz C . Endothelial-mural cell signaling in vascular development and angiogenesis. Arterioscler Thromb Vasc Biol 2009; 29: 630–638.
Franco M, Roswall P, Cortez E, Hanahan D, Pietras K . Pericytes promote endothelial cell survival through induction of autocrine VEGF-A signaling and Bcl-w expression. Blood 2011; 118: 2906–2917.
Bouillet P, Metcalf D, Huang DC, Tarlinton DM, Kay TW, Kontgen F et al. Proapoptotic Bcl-2 relative Bim required for certain apoptotic responses, leukocyte homeostasis, and to preclude autoimmunity. Science 1999; 286: 1735–1738.
Lugus JJ, Park C, Ma YD, Choi K . Both primitive and definitive blood cells are derived from Flk-1+ mesoderm. Blood 2009; 113: 563–566.
Chen MJ, Yokomizo T, Zeigler BM, Dzierzak E, Speck NA . Runx1 is required for the endothelial to haematopoietic cell transition but not thereafter. Nature 2009; 457: 887–891.
Sheibani N, Morrison ME, Gurel Z, Park S, Sorenson CM . BIM deficiency differentially impacts the function of kidney endothelial and epithelial cells through modulation of their local microenvironment. Am J Physiol Renal Physiol 2012; 302: F809–F819.
Coultas L, Bouillet P, Stanley EG, Brodnicki TC, Adams JM, Strasser A . Proapoptotic BH3-only Bcl-2 family member Bik/Blk/Nbk is expressed in hemopoietic and endothelial cells but is redundant for their programmed death. Mol Cell Biol 2004; 24: 1570–1581.
Zhang F, Li Y, Tang Z, Kumar A, Lee C, Zhang L et al. Proliferative and survival effects of PUMA promote angiogenesis. Cell Rep 2012; 2: 1272–1285.
Villunger A, Michalak EM, Coultas L, Mullauer F, Bock G, Ausserlechner MJ et al. p53- and drug-induced apoptotic responses mediated by BH3-only proteins puma and noxa. Science 2003; 302: 1036–1038.
Coultas L, Bouillet P, Loveland KL, Meachem S, Perlman H, Adams JM et al. Concomitant loss of proapoptotic BH3-only Bcl-2 antagonists Bik and Bim arrests spermatogenesis. Embo J 2005; 24: 3963–3973.
Urbich C, Knau A, Fichtlscherer S, Walter DH, Bruhl T, Potente M et al. FOXO-dependent expression of the proapoptotic protein Bim: pivotal role for apoptosis signaling in endothelial progenitor cells. FASEB J 2005; 19: 974–976.
Shen B, Gao L, Hsu YT, Bledsoe G, Hagiwara M, Chao L et al. Kallistatin attenuates endothelial apoptosis through inhibition of oxidative stress and activation of Akt-eNOS signaling. Am J Physiol Heart Circ Physiol 2010; 299: H1419–H1427.
Huntzinger E, Izaurralde E . Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet 2011; 12: 99–110.
Suarez Y, Fernandez-Hernando C, Yu J, Gerber SA, Harrison KD, Pober JS et al. Dicer-dependent endothelial microRNAs are necessary for postnatal angiogenesis. Proc Natl Acad Sci USA 2008; 105: 14082–14087.
Doebele C, Bonauer A, Fischer A, Scholz A, Reiss Y, Urbich C et al. Members of the microRNA-17-92 cluster exhibit a cell-intrinsic antiangiogenic function in endothelial cells. Blood 2010; 115: 4944–4950.
Kim YC, Day RM . Angiotensin II regulates activation of Bim via Rb/E2F1 during apoptosis: involvement of interaction between AMPKbeta1/2 and Cdk4. Am J Physiol Lung Cell Mol Physiol 2012; 303: L228–L238.
O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT . c-Myc-regulated microRNAs modulate E2F1 expression. Nature 2005; 435: 839–843.
Biswas AK, Johnson DG . Transcriptional and nontranscriptional functions of E2F1 in response to DNA damage. Cancer Res 2012; 72: 13–17.
Miyamoto K, Araki KY, Naka K, Arai F, Takubo K, Yamazaki S et al. Foxo3a is essential for maintenance of the hematopoietic stem cell pool. Cell Stem Cell 2007; 1: 101–112.
Schwenk F, Baron U, Rajewsky K . A cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells. Nucleic Acids Res 1995; 23: 5080–5081.
Thomas T, Voss AK, Chowdhury K, Gruss P . Querkopf, a MYST family histone acetyltransferase, is required for normal cerebral cortex development. Development 2000; 127: 2537–2548.
Acknowledgements
We thank Lachlan Whitehead, Cameron Nowell and Kelly Rogers for expert imaging assistance, Lisa Sampurno for technical assistance, Merle Dayton, Keti Stoev, Chrystal Smith and Emma Lanera for expert animal care and members of the Development and Cancer Division (WEHI) for informative discussion. This work was made possible through Victorian State Government Operational Infrastructure Support and Australian Government NHMRC IRIISS. This work was supported by the National Health and Medical Research Council, Australia (project grants: 1010638, 1046010 and 1049720, program grants: 1016701, and fellowships to TT (1003435), AKV (575512) and AS (1020363)) and Leukemia & Lymphoma Society (Special Center of Research 7001-13). LC is supported by an Australian Research Council Future Fellowship (110100891).
Author contributions
MNK, EN, LR, MJH, TT, AKV conducted experiments and interpreted data. ET conducted experiments. PB generated essential reagents. AS planned experiments and interpreted data. LC conceived the study, planned experiments, conducted experiments, interpreted data and wrote the manuscript. All authors proof-read the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by S Kaufmann
Rights and permissions
About this article
Cite this article
Koenig, M., Naik, E., Rohrbeck, L. et al. Pro-apoptotic BIM is an essential initiator of physiological endothelial cell death independent of regulation by FOXO3. Cell Death Differ 21, 1687–1695 (2014). https://doi.org/10.1038/cdd.2014.90
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2014.90
This article is cited by
-
Contribution of cell death signaling to blood vessel formation
Cellular and Molecular Life Sciences (2021)
-
Idelalisib promotes Bim-dependent apoptosis through AKT/FoxO3a in hepatocellular carcinoma
Cell Death & Disease (2018)
-
A miRNA-101-3p/Bim axis as a determinant of serum deprivation-induced endothelial cell apoptosis
Cell Death & Disease (2017)
-
Endothelial cell apoptosis in angiogenesis and vessel regression
Cellular and Molecular Life Sciences (2017)
-
Endothelial cell survival during angiogenesis requires the pro-survival protein MCL1
Cell Death & Differentiation (2016)