Abstract
The programmed cell death 4 (Pdcd4), a translation inhibitor, plays an essential role in tumor suppression, but its role in apoptosis remains unclear. Here we show that Pdcd4 is a critical suppressor of apoptosis by inhibiting the translation of procaspase-3 mRNA. Pdcd4 protein decreased more rapidly through microRNA-mediated translational repression following apoptotic stimuli than did the activation of procaspase-3, cleavage of poly(ADP)ribose polymerase (PARP) by active caspase-3, and nuclear fragmentation. Strikingly, the loss of Pdcd4 by the specific RNA interference increased procaspase-3 expression, leading to its activation and PARP cleavage even without apoptotic stimuli, and sensitized the cells to apoptosis. Thus, our findings provide insight into a novel mechanism for Pdcd4 as a regulator of apoptosis.
Similar content being viewed by others
Main
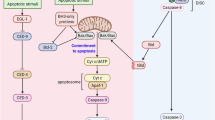
Apoptosis is a fundamental process controlling cell death and plays a critical role in normal development of multicellular organisms. When abnormalities occur in apoptosis, a variety of diseases are caused, including cancer,1, 2 autoimmunity,3, 4 and degenerative disorders.5, 6 Apoptosis is initiated through multiple independent pathways that trigger either activation of death receptors, DNA damage, mitochondrial stress, inhibition of protein synthesis, or inhibition of protein kinases.7 In contrast, all the pathways converge on the common machinery executing apoptosis, which consists mainly of a family of cysteine proteases (caspases).7 They are expressed as zymogens (procaspases) that have low intrinsic enzymatic activity, and once apoptosis is induced, they are activated by undergoing proteolysis and cleave protein substrates at their aspartate residues. It is well known that active caspase-3, the final executioner, cleaves poly(ADP)ribose polymerase (PARP), one of the substrates.8 Apoptosis is thus accompanied by proteolysis of cellular constituents, nuclear fragmentation, and DNA degradation;7 however, the complete molecular mechanisms specifying the transition of cellular fate from survival to death require elucidation.
Programmed cell death 4 (Pdcd4), also known as MA3,9 TIS,10 197/15a,11 H731,12 and DUG,13 was originally identified as a transcript upregulated under some conditions that induce apoptosis.9, 13 However, contradictory cases were reported, in which the mRNA levels are barely affected or even downregulated by different apoptotic stimuli.10, 14 Therefore, a role of Pdcd4 in apoptosis remains elusive. Further studies revealed that Pdcd4 functions as a tumor suppressor and a translation inhibitor (reviewed in Lankat-Buttgereit and Göke15; Allgayer16): Pdcd4 suppresses tumorigenesis and tumor progression and invasion and inhibits cap-dependent but not internal ribosomal entry site-dependent global and specific mRNA translation by binding eukaryotic initiation factor 4A and mRNA molecules selectively; the expression of Pdcd4 is lost or suppressed in some tumors, but induced or stimulated in the others.15 Yet, little is known concerning the expression and function of Pdcd4 protein during apoptosis and its downstream targets as well as upstream regulators. We aimed to elucidate the Pdcd4-mediated common mechanism controlling apoptosis with multiple types of non-cancerous or normal and cancer cells derived from several organs under various apoptotic conditions. Here we identify Pdcd4 as a critical regulator of apoptosis by inhibiting the procaspase-3 mRNA translation. We propose a novel microRNA-operated translational repression system for the disappearance of Pdcd4 protein by apoptotic stimuli and implicate the Pdcd4-dependent translational regulation of procaspae-3 mRNA in the cellular susceptibility to apoptosis.
Results and Discussion
Pdcd4 decreases following apoptotic stimuli
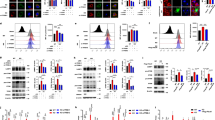
To reveal changes in the Pdcd4 protein levels during apoptosis, human cervical carcinoma HeLa cells cultured in the presence of serum were treated for 2 h without or with several kinds of apoptotic stimuli such as staurosporine (STS) that induces inhibition of protein kinases, an antagonizing antibody to Fas (CH11) that induces activation of the death receptor, etoposide (VP16) that induces DNA damage, and cycloheximide (CHX) that induces inhibition of protein synthesis, followed by western blot analyses. Pdcd4 protein decreased in cells treated with any apoptosis inducers, where PARP cleavage indicative of apoptosis occurred (Figure 1a). We also observed decreased Pdcd4 protein levels during the prolactin-dependent apoptosis in newt spermatogonia17 and during the STS-stimulated apoptosis in various cell types, such as human neuronal cells (SH-SY5Y), murine Sertoli cells (Sertoli B),18 murine spermatogonia (GC-1), and murine myoblasts (C2C12), where PARP was cleaved and thereby apoptosis progressed (Figure 1b). Next, the time course of Pdcd4 decrease was compared by western blotting in HeLa cells with that of procaspase-3 expression, its activation to active caspase-3, and PARP cleavage following STS treatment. Pdcd4 protein rapidly decreased within 5 min and disappeared in a time-dependent manner (Figure 1c, 1st panel). The procaspase-3 level was faintly detectable in the presence of Pdcd4, but significantly increased when the Pdcd4 level decreased (Figure 1c, 2nd panel), raising a possibility that the downregulation of Pdcd4 is associated with the upregulation of procaspase-3. Active caspase-3 apparently appeared within 1 h and its amount drastically increased 2 h after STS treatment (Figure 1c, 3rd panel). Accordingly, PARP (116 kDa) was slightly cleaved within 1 h and thereafter the 85-kDa product was remarkably detected (Figure 1c, 4th panel). Interestingly, 2 h after cells were treated with STS, the procaspase-3 level considerably decreased, whereas active caspase-3 and the cleaved PARP product were easily detectable. Thus, the decrease of Pdcd4 occurred much earlier than procaspase-3 expression, its activation, and PARP cleavage by active caspase-3 during apoptosis. Similar results were obtained in not only HeLa cells but also other cells used here under apoptotic conditions induced by not only STS but also other stimuli used here. Taken together, these results suggest that the loss of Pdcd4 protein by apoptotic stimuli is a critical cause that induces apoptosis in several cells.
Rapid decrease of Pdcd4 protein in several cells following various apoptotic stimuli. (a) Human cervical carcinoma HeLa cells were treated for 2 h without (−) or with a variety of apoptosis inducers at the respective dose as indicated and analyzed by western blotting for Pdcd4, PARP, and β-actin. PARP is an apoptotic marker: an arrow, full-length PARP (116 kDa); an arrowhead, the cleaved product (85 kDa). β-Actin is the loading control. (b) Not only HeLa cells but also human neuroblastoma SH-SY5Y, murine testicular somatic Sertoli B, murine spermatogonial GC-1, and murine myoblast C2C12 cells were treated for 2 h with dimethyl sulfoxide (DMSO) (−) or STS (+), followed by western blotting for the indicated proteins. (c) HeLa cells were treated for 2 h with DMSO (0) or for the indicated periods with STS and subjected to western blotting for the indicated proteins. (d) The experiment was performed as in (c), except that cells were subjected to RNA extraction and RT-PCR (30 cycles) for Pdcd4 (350 and 270 bp) and GAPDH as the internal control (212 bp)
As the disappearance of Pdcd4 protein was so rapid, the transcriptional repression of Pdcd4 gene seemed unlikely. To rule this possibility out, we treated HeLa cells under the same conditions as above and performed RT-PCR analysis for Pdcd4; the two bands are shown: an upper band (350 bp) detected on agarose gel electrophoresis for the variant 2 of Pdcd4 mRNA that includes an alternate in-frame exon in the 5′ coding region and utilizes a downstream start codon, generating the isoform 2 of Pdcd4 protein with a little faster electrophoretic mobility on SDS-PAGE that has a shorter N terminus, compared with a lower band (270 bp) for the variant 1, generating the isoform 1. The Pdcd4 mRNA expression remained barely changed following activation of apoptotic pathways (Figure 1d). Overall, we speculated that Pdcd4 undergoes the rapid post-transcriptional downregulation such as proteolysis and microRNA-mediated translational repression at the early phase of apoptosis.
Pdcd4 is downregulated by microRNA-mediated translational repression following apoptotic stimuli
Pdcd4 undergoes mitogen-dependent proteolysis by proteasome during cell growth.19 We found in cells used that serum starvation lowered the Pdcd4 protein level, but induced rather growth suppression than apoptosis (data not shown), as reported.19 Because here we have cultured cells in the presence of serum, the Pdcd4 protein level should be maintained low; we demonstrated that it lowered more after apoptosis was induced in the presence of serum (Figure 1). To determine whether Pdcd4 is degraded by proteasome in response to apoptotic stimuli, HeLa cells were treated in the presence of the serum for 2 h without or with the proteasome inhibitor MG132 before 0.5-h stimulation with STS and analyzed by western blotting. Although more Pdcd4 protein likely accumulated in cells treated with MG132 than without MG132, consistent with the previous report,19 the STS-stimulated Pdcd4 protein disappearance was not suppressed by MG132 (Figure 2a, top panels). Accordingly, the STS-induced PARP cleavage was not inhibited by MG132 (Figure 2a, middle panels). On the other hand, treatment of cells with MG132 alone resulted in the appearance of the cleaved PARP product even in the absence of apoptotic stimuli, supporting a lot of recent reports demonstrating that inhibition of proteasome induces apoptosis (reviewed in Giuliano et al.20). In addition, inhibitors of calpain (calpeptin), lysosome (E64d), caspase-like proteases (VAD-fmk), and serine proteases (methylamine, N-p-tosyl-L-phenylalanyl chloromethyl ketone) had little effects on the Pdcd4 disappearance in cells treated with apoptosis inducers (data not shown). These results suggest that the loss of Pdcd4 protein in the presence of serum by apoptotic stimuli is not only independent of the proteasome pathway but also independent of other well-known protein degradation systems.
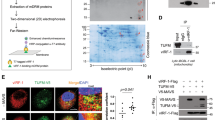
miR-199a-5p-mediated translational downregulation of Pdcd4 in apoptosis. (a) Cells were treated for 2 h with DMSO (−), MG132, or LY294002, and then for 0.5 h with DMSO (−) or STS (+), followed by western blotting for the indicated proteins. An arrow indicates full-length PARP (116 kDa); an arrowhead indicates the cleaved product (85 kDa); and asterisk indicates a nonspecific band. (b and c) Cells were treated for 0.5 h with DMSO (−) or STS (+) and subjected to small RNA extraction and RT-PCR for the indicated microRNA molecules; results were representative of three separate experiments (b) and quantified by densitometry as a fold of miR-199a-5p and miR-564 expression, normalized to let-7 expression, in cells treated with STS, relative to those in the control, respectively (c). *P<0.01 and **P<0.05 versus each of the controls. (d and e) Cells were transfected with 8 nM of control microRNA (Cont.) or the indicated doses of miR-199a-5p and cultured for 8 h, followed by western blotting for Pdcd4 and β-actin; results were representative of three independent experiments (d) and quantified by densitometry as a fold of Pdcd4 expression, normalized to β-actin expression, in cells transfected with miR-199a-5p, relative to that in the control (e). *P<0.01 versus the control. (f and g) The experiment was performed as in (d), except that cells were cultured for the indicated periods after transfection with 8 nM of control microRNA (−) or miR-199a-5p (+), and results were presented as in (e). *P<0.01 versus each of the controls. (h) The experiment was performed as in (f), except that the transfectants were subjected to RT-PCR (30 cycles) for Pdcd4 and GAPDH. (i–o) The experiment was performed as in (f and g), except that the transfectants were analyzed by DAPI staining (i–m) and TUNEL assay (n); a minimum of 300 cells (100 cells a microscopic field) was scored in the transfectants under each condition and results were presented as a percentage (means±S.E.) of apoptotic cells with nuclear fragmentation among the total ones derived from three independent experiments (o). 12-hour post-transfection with control microRNA (i); 4 (j), 8 (k), or 12 h (l) post-transfection with miR-199a-5p. *Representative apoptotic cells (i–n) and P<0.01 versus each of the controls (o)
To gain insight into the phosphoinositide 3-kinase (PI3K) pathway, which induces activation of the protein kinase S6K1 and degradation of Pdcd4 by proteasome19 in the disappearance of Pdcd4 protein following apoptotic stimuli, HeLa cells were treated for 2 h with the PI3K inhibitor LY294002 and then for 0.5 h without or with STS. The inhibitor did not prevent the STS-stimulated Pdcd4 decrease, but enhanced PARP cleavage independent of treatment with apoptosis inducers (Figure 2a). These results suggest that the PI3K pathway plays a role in cell survival, but not in the decrease of Pdcd4 protein during apoptosis, which is distinct from the Pdcd4 proteolysis during cell growth.
To test whether Pdcd4 decreases by microRNA-mediated translational repression during apoptosis, we searched for a microRNA molecule(s) that is capable of binding to the 3′-untranslated region (UTR) of human Pdcd4 by computational analyses using the Microinspector software (Institute of Molecular Biology and Biotechnology, Heraklion, Greece and University of Plovdiv, Bulgaria) and examined changes in the expression levels by RT-PCR of small RNA fractions prepared from HeLa cells treated for 0.5 h without or with STS.21, 22 Computational analyses revealed 14 microRNA candidates, which might interact possibly with the putative 3′-UTR from the translation stop codon to the poly(A) signal sequence (639 bp). Among them, screened by RT-PCR, we identified two microRNA molecules, miR-564 and miR-199a-5p, that were upregulated by STS treatment; compared with miR-564, miR-199a-5p increased greatly at threefold (Figures 2b and c). The increase of miR-199a-5p was observed in not only HeLa cells but also other cells by treatment with not only STS but also other apoptotic stimuli. We speculated that this is due to the activation of the microRNA precursor transcription or/and enzymes, including Drosha and Dicer, producing the mature microRNA under apoptotic conditions. Meanwhile, miR-21, a microRNA that is known to downregulate Pdcd4 expression,16, 23 was contained in the candidates, but no change in the miR-21 levels was found during apoptosis (data not shown). Next, we transfected HeLa cells with miR-199a-5p and analyzed its effect in the absence of apoptotic stimuli on changes in the Pdcd4 expression by western blotting and RT-PCR and on the incidence of apoptosis by 4′, 6-diamidino-2-phenylindole (DAPI) staining and terminal deoxynucleotidyl transferase-mediated dUTP-biotin nick-end labeling (TUNEL) assay. The Pdcd4 protein levels significantly decreased 8 h after cells were transfected with 2, 4, or 8 nM of miR-199a-5p in a dose-dependent manner (Figures 2d and e), and in those transfected with 8 nM of miR-199a-5p with longer incubation periods and by a half to the control after 8 h post-transfection (Figures 2f and g). On the other hand, transfection with the microRNA had little effect on the Pdcd4 mRNA expression (Figure 2h). The influence of the Pdcd4-targeted microRNA on the expression of Pdcd4 mRNA and protein was consistent with that of apoptotic stimuli (Figure 1). DAPI staining showed that the nuclear fragmentation indicative of apoptosis occurred significantly 12 h after transfection with 8 nM of miR-199a-5p (Figures 2l and o), compared with the control (Figures 2i and o), which was confirmed by TUNEL assay demonstrating apoptotic DNA degradation (Figures 2m and n). Similar results were obtained in other cells than HeLa cells. These results indicate that the decrease of Pdcd4 is followed by the induction of apoptosis and suggest that the loss of Pdcd4 protein by miR-199a-5p causes apoptosis. Therefore, we concluded that apoptotic Pdcd4 disappearance is dependent on the microRNA-operated translational downregulation system, which may not enhance degradation of Pdcd4 mRNA but inhibit its translation and may be a difference in the way for Pdcd4 protein to disappear between during apoptosis and cell growth.
Pdcd4 regulates the sensitivity of cells to apoptosis
To determine the functional role of Pdcd4 and the significance of its disappearance in apoptosis, HeLa cells were transfected with the empty vector (mock) or a construct expressing either control small interfering (si) RNA or Pdcd4-targeted siRNA.24 At 40 h after transfection, cells were treated for the indicated periods without or with STS and then stained with DAPI to detect apoptotic nuclear fragmentation. Western blotting verified that Pdcd4 decreased in cells transfected with the Pdcd4-targeted siRNA expression construct in an amount-dependent manner, but not in those with the control siRNA expression construct or with mock (Figures 3a and b), suggesting that Pdcd4 is specifically targeted by the specific siRNA to disappear. DAPI staining demonstrated that apoptotic nuclear fragmentation in the control and mock cells containing normal amounts of Pdcd4 slightly occurred 60 min after STS treatment (Figures 3d, f, and i). In contrast, cells deficient in Pdcd4 significantly became apoptotic as earlier as 10 min and displayed more nuclear fragmentation even 60 min after STS treatment (Figures 3h and i), indicating that they were sensitized to apoptosis. Similar results were obtained in not only HeLa cells but also other cells by treatment with not only STS but also other apoptotic stimuli; Pdcd4 deficiency resulted in enhanced sensitivity of C2C12 cells treated with CH11 to apoptosis (Figure 3j). Thus, the presence or absence of Pdcd4 protein may decide whether cells are committed to survival or death by affecting the cellular susceptibility to apoptosis.
Susceptibility of cells deficient in Pdcd4 to apoptosis. (a) HeLa cells were transfected with 0.4 μg of the empty vector (mock), the control siRNA expression construct, or the Pdcd4-targeted siRNA expression construct and 40 h later subjected to western blotting for Pdcd4 and β-actin. (b) The experiments were performed as in (a), except that the indicated amounts of the Pdcd4-targeted siRNA expression construct were used for transfection. (c–h) Each transfectant with mock (c and d), the control siRNA expression construct (e and f), or the Pdcd4-targeted siRNA expression construct (g and h) was treated for 60 min with DMSO (−; c, e and g) or STS (+; d, f and h), followed by DAPI staining. Asterisk and an inset, representative apoptotic cells with nuclear fragmentation. (i) The experiment was performed as in (c–h), except that transfectants were treated for 60 min with DMSO (0) or for the indicated periods with STS; a minimum of 300 cells (100 cells a microscopic field) was scored in the respective transfectant under each condition and results are presented as a percentage (means±S.E.) of apoptotic cells with nuclear fragmentation among the total ones derived from three independent experiments. *P<0.01 versus the control cells treated for each period with DMSO or STS. (j) The experiment was performed as in (a–h), except that C2C12 cells transfected with 0.4 μg of the expression construct for control siRNA (−) or Pdcd4-targeted siRNA (+) were treated for 60 min with PBS containing 50% glycerol (0) or for the indicated periods with CH11, and results were presented as in (i). Insets, western blotting of the transfectants for Pdcd4 and β-actin; *P<0.01 versus the control cells treated for each period with CH11. (k and l) The experiment was performed as in (a–h), except that HeLa cells transfected with the expression construct for control siRNA (−) or Pdcd4-targeted siRNA (+) were cultured for the indicated periods in the absence of apoptotic stimuli, and results were presented as in (i). *P<0.01 and **P<0.05 versus 16 h after cells were transfected with the Pdcd4-targeted siRNA expression construct. (m and n) The experiment was performed as in (k), except that C2C12 cells were used, and results were presented as in (l)
To determine whether the loss of Pdcd4 alone causes apoptosis, we examined the expression of Pdcd4 protein by western blotting and the numbers of apoptotic nuclei by DAPI staining in HeLa cells transfected with the expression construct for control siRNA or Pdcd4-targeted siRNA in the absence of apoptotic stimuli. Apoptotic nuclei significantly increased when the Pdcd4 protein levels decreased depending on the time after transfection (Figures 3k and l). Similar results were obtained in other cells beyond HeLa cells; Pdcd4 deficiency resulted in increased numbers of apoptotic nuclei with longer culture periods in C2C12 cells (Figures 3m and n). There seemed to be a time lag until apoptotic nuclei increased significantly after the Pdcd4 protein levels decreased, but it may be necessary for the progress of apoptosis. That is probably because the microRNA/siRNA-mediated inhibition of Pdcd4 mRNA translation induces activation of only one apoptosis signaling pathway comprising the loss of Pdcd4 protein, although apoptotic stimuli induce full activation of all apoptosis signaling pathways, including the loss of Pdcd4 protein. Therefore, the loss of Pdcd4 protein may not be sufficient but may be required for inducing apoptosis, and we propose that the loss of Pdcd4 protein is a novel hallmark of apoptosis in multiple types of cells.
Pdcd4 is a direct suppressor of the procaspase-3mRNA translation
To determine how Pdcd4 regulates the cellular susceptibility to apoptosis, we have screened by western blotting for a factor(s) involved in apoptosis, whose expression is upregulated in cells lacking Pdcd4 with the specific siRNA. We investigated procaspase-3 expression in the cells because it increased in HeLa cells containing reduced Pdcd4 by STS treatment as shown in Figure 1c. Notably, Pdcd4-deficient HeLa and C2C12 cells displayed increased expression of procaspase-3 independent of apoptotic stimuli compared with the control cells (Figure 4a, 1st and 2nd panels), suggesting that the loss of Pdcd4 protein results in the increase of procaspase-3 mRNA translation. This was paralleled with the previous report demonstrating that procasapase-3, -7, and -10 are downregulated by overexpression of Pdcd4 in a neuroendocrine cell line Bon-1.25 Strikingly, in Pdcd4-deficient cells, the procaspase-3 activation generating the active form and the PARP cleavage generating the 85-kDa product occurred even without apoptotic stimuli (Figure 4a, 3rd and 4th panels). These findings suggest that increased translation of procaspase-3 mRNA causes spontaneous activation of the protein by self-proteolysis in Pdcd4-deficient cells, leading to enhanced cleavage of PARP, even in the absence of apoptotic stimuli. This was consistent with the previous report demonstrating that overexpression of procaspase-3 in insect Sf9 cells resulted in DNA fragmentation even in the absence of apoptotic stimuli.26 Similar results were obtained by Pdcd4 deficiency in not only HeLa and C2C12 cells but also other cells. Thus, the loss of Pdcd4 protein may be a widespread event that causes apoptosis by releasing the procaspase-3 mRNA translation from the Pdcd4-mediated inhibition at least in several cells.
Pdcd4-regulated procaspase-3 mRNA translation and apoptosis. (a) HeLa and C2C12 cells were transfected with the expression construct for control siRNA or Pdcd4-targeted siRNA and 40 h later treated for 60 min with DMSO (0) and PBS containing 50% glycerol (0) or for the indicated periods with STS and CH11, respectively, followed by western blotting for the indicated proteins. Arrows indicate full-length PARP (116 kDa); arrowheads indicate the cleaved product (85 kDa). (b and c) Either control template (Cont.), the HA-tagged procaspase-3 template, or the HA-tagged β-actin template as the specificity control was incubated in a cell-free protein synthesis system in the absence (0) or presence of the indicated amounts of Pdcd4 protein, followed by western blotting with the specific and the HA tag antibodies to detect procaspase-3 and β-actin. CBB indicates loading controls by staining the western blots with CBB; western blotting for Pdcd4 indicates the amounts of Pdcd4 protein added. Arrows indicate the indicated proteins translated from each template; an arrowhead indicates the translated protein derived from control template; and an asterisk indicates nonspecific bands. Results were representative of three separate experiments (b) and quantified by densitometry as a fold of procaspase-3 and β-actin expression in the presence of Pdcd4 protein, relative to those in its absence (c). *P<0.01 versus procaspase-3 or β-actin expression in the absence of Pdcd4 protein. (d) Extracts prepared from cells were subjected to immunoprecipitation (IP) with normal rabbit immunoglobulins (NRI) or an antibody to Pdcd4, followed by RT-PCR (45 cycles) for procaspase-3 (323 bp) (top panels) and GAPDH (212 bp) as the specificity control (middle panels) and western blotting for Pdcd4 (bottom panels). An arrow indicates Pdcd4; an asterisk indicates heavy chains of immunoglobulins
Conversely, to clarify whether Pdcd4 suppresses the procaspase-3 mRNA translation directly in normal cells, we utilized a cell-free protein synthesis system. A cDNA encoding human procaspase-3 or human β-actin as the control, which was tagged 3′ terminally with the hemagglutinin (HA) sequence, was added with the SP6 promoter sequence by two steps of PCR to generate the respective template for cell-free protein synthesis. After the template was subjected to the SP6 RNA polymerase-driven transcription reaction, the resulting reaction mixture was divided into several aliquots and each of them was processed for the translation reaction in the absence or presence of Pdcd4 protein, followed by western blotting for procaspase-3 and β-actin with antibodies specific for them and against the HA tag (Figures 4b and c). The translated procaspase-3 protein was detected in the absence of Pdcd4, but decreased when the amounts added increased, whereas no effect of Pdcd4 protein added on the translation of β-actin mRNA was found, suggesting that Pdcd4 suppresses the translation of at least procaspase-3 mRNA specifically. Moreover, we performed immunoprecipitation with an antibody to Pdcd4 from HeLa cells, followed by RT-PCR for procaspase-3 and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) as the control (Figure 4d). Procaspase-3 mRNA co-precipitated in the immunoprecipitates of Pdcd4, indicating the interaction between Pdcd4 protein and procaspase-3 mRNA. By contrast, no mRNA for GAPDH was detected in the immunoprecipitates. In addition, control immunoprecipitation with normal immunoglobulins gave no enrichment in procaspase-3 mRNA. These results suggest that Pdcd4 binds procaspase-3 mRNA specifically and inhibits its translation directly in normal cells. Overall, the loss of Pdcd4 protein by apoptotic stimuli may stimulate the procaspase-3 mRNA translation, and then procaspase-3 is activated by upstream caspases and the resulting active form cleaves PARP and induces nuclear fragmentation during apoptosis. Therefore, Pdcd4 might be a threshold in determining cellular fate: cells commit to survival when Pdcd4 protein is expressed in cells; even though Pdcd4 protein decreases to some extent and because it is still present in the presence of serum, cells survive and proliferate;19 however, when Pdcd4 protein decreases more and disappears following apoptotic stimuli in the presence of serum, cells commit to death ultimately.
We demonstrated that Pdcd4 protein is downregulated in several cells by various apoptotic stimuli, although the mRNA remains largely unchanged as reported.10, 14 There are contradictory reports concerning the Pdcd4 mRNA expression during apoptosis: it increases,9, 13 but remains barely affected or even decreases.10, 14 This discrepancy suggests the diverse regulation, which could vary the Pdcd4 mRNA levels depending on various factors such as genetic backgrounds, responsiveness to apoptotic stimuli, and apoptotic pathways in different cell types, although they are not fully understood yet. On the other hand, very little has been addressed about the Pdcd4 protein expression in apoptosis. To our knowledge, this is the first report determining changes in the Pdcd4 protein levels under apoptotic conditions in mammals. The difference between changes in the Pdcd4 mRNA and protein expression during apoptosis is consistent with the previous reports suggesting that changes in the mRNA levels are not always paralleled by concomitant changes in the protein levels.17, 27 This observation may be explained by our data implicating miR-199a-5p-operated translational repression in the apoptotic downregulation of the Pdcd4 protein expression independent of the mRNA expression: even though Pdcd4 mRNA is continuously expressed, its translation is inhibited by action of miR-199a-5p, resulting in the decrease of the protein. In addition, this observation may be supported by the previous report describing the involvement of an RNA-binding protein in the Pdcd4 mRNA stability and translation.17
Likewise, there are conflicting results concerning Pdcd4 function in tumor suppression and apoptosis. Although the loss of Pdcd4 is correlated with tumor progression in lung cancer,28 high expression of Pdcd4 is found in bladder and breast carcinoma;29 overexpression of Pdcd4 induces apoptosis in human breast cancer cells30 and inhibits proliferation in human endocrine tumor cells,31 but does not alter cell cycle or induce apoptosis in colon carcinoma cells.32 The TGF-β signaling increases Pdcd4 expression and apoptotic death in hepatocellular carcinoma Huh7 cells,33 but increases mature miR-21, which decreases Pdcd4 expression,16, 23 in vascular smooth muscle cells.34 In addition, Pdcd4 is implicated in conferring the sensitivity of gastric cancer cells to apoptosis induced by a cytotoxic tumor necrosis factor (TNF) family member TNF-related apoptosis induced ligands (TRAIL) by inhibiting the expression of FLICE-inhibiting protein (FLIP).35 However, expression levels of Pdcd4 are not correlated in several cells with those of various proteins involved in cell cycle or apoptosis or with cell growth or the sensitivity to TRAIL.25 We demonstrated that the loss of Pdcd4 protein induces apoptosis through miR-199a-5p-operated translation repression in response to various stimuli in multiple types of cells, indicating that Pdcd4 plays a role in lowering the sensitivity to apoptosis by inhibiting the expression of procaspase-3. These findings suggest that Pdcd4 affects multiple mechanisms including signal transduction pathways and mediates diverse responses to various factors including genetic backgrounds, pathophysiological conditions such as tumor grade, and apoptosis inducers in different cell types,15, 25 although they are not fully clarified yet. Overall, alterations in the Pdcd4 protein expression could be governed by interactions of the RNA-binding protein,17 microRNA molecules such as miR-199a-5p and miR-21,16, 23 and Pdcd4 mRNA, leading to divergent functions of Pdcd4.
In conclusion, the current study reports an antiapoptotic role of Pdcd4 and some features of the molecular mechanisms underlying the control of translation during apoptosis: Pdcd4 protein decreases following apoptotic stimuli; this decrease is mediated by miR-199a-5p-dependent translational repression; the loss of Pdcd4 protein increases the sensitivity of cells to apoptosis by promoting the translation of procaspase-3 mRNA; however, normally Pdcd4 lowers or/and suppresses the translation of procaspase-3 mRNA and thereby protects cells from apoptosis. Thus, Pdcd4 appears to be an antiapoptotic regulator that inhibits the translation of at least procaspase-3 mRNA in cells and may be an eventual translation organizer (ETO) that tunes the gene expression of caspase-3, the final executioner of apoptosis.
Materials and Methods
Cell culture, treatments, and transfection
The Pdcd4 mRNA levels increase by apoptotic stimuli such as dexamethasone and serum starvation in several cells including hematopoietic and insulinoma cells,9, 13 but remain largely unchanged or even decrease by those like topoisomerase inhibitors in lymphoma cells.10, 14 To investigate the expression and function of Pdcd4 protein during apoptosis, we performed experiments under apoptotic conditions induced by various stimuli with multiple types of cells originated from some organs as follows: cancer HeLa and SH-SY5Y, and non-cancerous GC-1 and C2C12 cells were purchased from the American Type Culture Collection (Manassas, VA, USA) and non-cancerous Sertoli B cells were kindly provided by Drs. K Tanaka and T Hara.18 HeLa is the most commonly used cell line established from a human epithelial cervical carcinoma; SH-SY5Y is neuronal cells derived from a human neuroblastoma; Sertoli B is a somatic Sertoli cell line established from murine testes; GC-1 is germ cells at the spermatogonial stage established from murine testes; and C2C12 is myoblasts derived from murine skeletal muscles after a crush injury. Cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. At 1 day after cells were plated, they were treated for 2 h with each of the vehicle or the inhibitor for proteasome (MG132, 100 μM; Peptide Institute Inc., Osaka, Japan) and PI3K (LY294002, 100 μM; Cell Signaling Technology, Danvers, MA, USA), and then stimulated for the indicated periods with each of the vehicle or several apoptosis inducers (STS, 1 μM, Sigma, St. Louis, MO, USA; CH11, 1.6 mg/ml, MBL, Nagoya, Japan; etoposide, 50 μg/ml, Sigma; CHX, 100 μM, Sigma), followed by DAPI staining, TUNEL assay, RT-PCR, and western blotting. Alternately, cells were transfected in OPTI-MEM (Invitrogen, Carlsbad, CA, USA) with the indicated doses of control microRNA (Life Technologies, Carlsbad, CA, USA) or miR-199a-5p (Life Technologies) using INTERFERin (Invitrogen) and with the indicated amounts of the empty pTER vector (mock),24 the pTER expression construct for Pdcd4-targeted siRNA (the target sequence GUGUUGGCAGUAUCCUUA) used in Bitomsky et al.24 or that for green fluorescent protein (GFP)-targeted siRNA (the target sequence GGCUACGUCCAGGAGCGCACC) as the control using FuGene HG (Roche, Basel, Switzerland) according to the manufacturer's instructions, respectively, cultured for the indicated periods, and treated without or with apoptotic stimuli. The GFP-targeted siRNA expression plasmid was constructed using the target sequence as follows:36 oligonucleotides 5′-GATCCCGGCTACGTCCAGGAGCGCACCTTCAAGAGAGGTGCGCTCCTGGACGTAGCCTTTTTGGAAA-3′ and 5′-AGCTTTTCCAAAAAGGCTACGTCCAGGAGCGCACCTCTCTTGAAGGTGCGCTCCTGGACGTAGCCGG-3′ (100 pmol, each) were separately treated for 1 h with T4 polynucleotide kinase (Takara, Shiga, Japan) in a total volume of 10 μl for their phosphorylation. To anneal the oligonucleotides, they were mixed, boiled for 2 min in the presence of 200 mM NaCl, incubated at 57°C for 2 h, and cooled slowly. A measure of 1 μl of the mixture was used for ligation with pTER vector that had been digested with BglII (Takara) and HindIII (Takara) and then treated with calf intestinal phosphatase (Takara).
RT-PCR
Total RNA and co-precipitated RNA were extracted from cells and immunoprecipitates of Pdcd4, respectively, by using ISOGEN (Nippon Gene, Tokyo, Japan), treated with RNase-free DNase I (Takara), and reverse-transcribed using random hexamers with a reverse transcriptase Superscript III (Invitrogen). In all, 10% of the reaction mixture was subjected as the template to PCR with Ex Taq DNA polymerase (Takara) as follows: for 30 or 45 cycles at 95°C for 30 s, at 56°C for 30 s, and at 72°C for 30 s using forward and reverse primers 5′-AGAGACTCTCCGAGGCGGCG-3′ and 5′-CCACTTCTAAGGGCGTCACT for human Pdcd4 and 5′-GTCAGTGGTGGACCTGACCT-3′ and 5′-TGAGCTTGACAAAGTGGTCG-3′ for human GAPDH; for 45 cycles at 95°C for 30 s, at 58°C for 30 s, and at 72°C for 30 s using forward and reverse primers 5′-CTGGAATATCCCTGGACAAC-3′ and 5′-CAGGTCAACAGGTCCATTTG-3′ for human procaspase-3. The PCR products were separated by 1% agarose gel electrophoresis and detected by ethidium bromide staining. They were ligated with pT7Blue vector (Merck, Darmstadt, Germany) and their nucleotide sequences were verified. For RT-PCR of microRNA molecules, small RNA pools were fractionated from total RNA (7.2 μg) by incubating on ice for 1 h with 13% polyethylene glycol 6000 and 0.8 M NaCl, added with the poly(A) sequence by incubating at 37°C for 1 h with 0.04 U/μl poly(A) polymerase (Takara) in the presence of 1 mM ATP and 2.5 mM MnCl2,21 and after the enzyme was denatured at 85°C for 5 min, reverse-transcribed using 5 μM poly(T) adapter 5′-GCGAGCACAGAATTAATACGACTCACTATAGG(T)25-3′.22 In all, 10% of the reaction mixture was subjected to PCR for 45 cycles at 95°C for 30 s, at 55°C for 30 s, and at 72°C for 30 s using a forward primer 5′-CCCAGTGTTCAGACTACCTGTTC-3′ for miR-199a-5p, 5′-AGGCACGGTGTCAGCAGG C for miR-564, 5′-TAGCTTATCAGACTGATGTTGA-3′ for miR-21, or 5′-TGAGGTAGTAGGTTGTATAGTT for let-7 and a reverse primer 5′-GCG AGCACAGAATTAATACGAC-3′. The PCR products were separated by 12% polyacrylamide gel electrophoresis and detected by ethidium bromide staining. They were ligated with pT7Blue vector and their nucleotide sequences were verified.
Western blotting
Whole protein extracts and precipitated proteins were eluted from cells and immunoprecipitates of Pdcd4, respectively, by using SDS-PAGE sample buffer (62.5 mM Tris/HCl, pH 6.8, 25% glycerol, 2% SDS, 0.01% bromophenol blue, 0.5% β-mercaptoethanol) and subjected to western blot analyses with rabbit anti-Pdcd4 (1 : 2000; Rockland, Gilbertsville, PA, USA), mouse anti-procaspase-3 (1 : 1000; BioLegend, San Diego, CA, USA), rabbit anti-active caspase-3 (1 : 1000; Abcam, Cambridge, UK), mouse anti-PARP (1 : 2000; Enzo, Farmingdale, NY, USA), rabbit anti-HA (1 : 1000; Santa Cruz Biotechnology, Santa Cruz, CA, USA), and mouse anti-β-actin antibodies (1 : 3000; Millipore, Piscataway, NJ, USA), followed by chemiluminescence detection (Millipore). In some experiments, cells were immediately washed in phosphate-buffered saline (PBS) and lysed in RIPA buffer (10 mM Tris/HCl, pH 7.4, 150 mM NaCl, 5 mM EDTA, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, protease inhibitor cocktail (Roche), 1 mM phenylmethylsulfonyl fluoride, phosphatase inhibitor cocktail (Merck)), and then the extracts (5 μg, each) were analyzed by western blotting for Pdcd4 and β-actin.
DAPI staining and TUNEL assay
Cells were fixed at room temperature for 10 min with 4% paraformaldehyde in PBS, permealized at room temperature for 10 min with 0.2% Triton X-100 in PBS, stained at room temperature for 1 min with 0.5 μg/ml DAPI in Tris-buffered saline, and observed under a fluorescence microscopy (BX60; Olympus, Tokyo, Japan). TUNEL assay was performed with an in situ apoptosis detection kit (Takara) according to the manufacturer's instructions. For quantification of apoptotic cells, a minimum of 100 cells, each from at least three microscope fields, was examined for DAPI-stained cells with nuclear fragmentation among the total ones or TUNEL-positive nuclei among DAPI-stained ones. The frequency of apoptosis was expressed as a percentage (means±S.E.) of apoptotic cells obtained from three independent experiments.
Cell-free protein synthesis system
The human procaspase-3 cDNA (1.1 kbp) was cloned from the cDNA library prepared from HeLa cells and added 3′ terminally with the HA tag sequence by PCR for 45 cycles at 95°C for 1 min, at 52°C for 1 min, and at 72°C for 2.5 min with Ex Taq DNA polymerase using forward and reverse primers 5′-GGCGAATTCAGGGAGAACTGAGGTATTAA-3′ and 5′-GATCTCGAGTTAGGCATAGTCAGGGACGTCATAAGGATAGTGATAAAAATAGAGTTCTT-3′. The PCR product was ligated with pT7Blue vector and the nucleotide sequence was verified. The template for cell-free protein synthesis was constructed by two steps of PCR with Ex Taq DNA polymerase to add the SP6 promoter sequence as follows: the first PCR was performed for 35 cycles at 98°C for 10 s, at 53°C for 1 min, and at 72°C for 3 min using the cDNA clone and a forward gene-specific primer 5′-CCACCCACCACCACCAATGAGGGAGAACTGAGGTATTAA-3′ and a reverse pT7Blue primer 5′-CAAGGCGAGTTACATGATCC-3′ derived from the vector; the second PCR for 5 cycles at 98°C for 10 s, at 53°C for 1 min, and at 72°C for 1 min, followed by 35 cycles at 98°C for 10 s, at 58°C for 40 s, and at 72°C for 3 min using 1/10 volumes of the first PCR product and forward primers 5′-GCGTAGCATTTAGGTGACACT-3′ and 5′-GGTGACACTATGGAACTCACCTATCTCCCCAACACCTAATAACATTCAATCACTCTTTCCACTAACCACCTATCTACATCACCACCCACCACCACCAAT G-3′ and the reverse pT7Blue primer. Similarly, the HA-tagged human β-actin template was constructed using the cDNA (1.1 kbp) as described above: for cloning β-actin cDNA with the HA tag sequence, forward and reverse primers 5′-GGCGAATTCACCGCCGAGACCGCGTCCGC-3′ and 5′-GATCTCGAGCTAGGCATAGTCAGGGACGTCATAAGGATAGAAGCATTTGCGGTGGACGA-3′; for the first PCR, a forward gene-specific primer 5′-CCACCCACCACCACCAATGACCGCCGAGACCGCGTCCGC-3′ and the reverse pT7Blue primer; for the second PCR, the same forward primers as used above for constructing the procaspase-3 template and the reverse pT7Blue primer. The template (1 μg, each), which had been concentrated by ethanol precipitation, was added to the transcription mixture provided by ENDEXT Technology Premium Expression Kit, PCR (Cell Free Sciences Co., Ltd, Ehime, Japan), mixed gently, and incubated at 37°C for 6 h according to the manufacturer's instructions. An aliquot of the reaction mixture was analyzed by agarose gel electrophoresis to confirm cRNA synthesis. The remaining reaction mixture was added 10 μl each into the translation mixture provided by the same kit according to the manufacturer's instructions and then incubated at 15°C for 20 h without or with the indicated amounts of recombinant human Pdcd4 protein (Atgen, Montevideo, Uruguay), followed by western blotting with the respective antibody specific for procaspase-3 and β-actin and the antibody to the HA tag for the both proteins.
Immunoprecipitation
Extracts (1 mg of protein) were prepared from cells by lysis in cell extraction buffer (50 mM PIPES, pH 7.4, 50 mM KCl, 5 mM EGTA, 2 mM MgCl2, 1 mM DTT, 1 mM PMSF, protease inhibitor cocktail, RNase inhibitor (Promega, Madison, WI, USA)), followed by centrifugation at 4°C for 1 min at 18 000 × g, pre-cleared at 4°C for 8 h with protein G-sepharose beads (GE Healthcare), and incubated at 4°C for 12 h with protein G-sepharose beads and 4 μg of either an antibody to Pdcd4 or normal rabbit immunoglobulins (Dako, Glostrup, Denmark) under constant rotation. After centrifugation at 4°C for 1 min at 1000 × g, immunoprecipitates were washed five times with cell extraction buffer. Co-precipitated RNA was extracted from 4/5 of the immunoprecipitates and subjected to RT-PCR for procaspase-3 and GAPDH. The remaining immunoprecipitates were eluted in SDS-PAGE sample buffer, followed by western blotting for Pdcd4.
Statistics
Data were presented as the means±S.E. For statistical comparison, Student's t-test was used. P-values <0.05 were considered to be statically significant.
Others
Protein concentrations were estimated by Coomassie dye binding (Bio-Rad, Hercules, CA, USA) using bovine serum albumin as a standard. For some experiments, signals detected in the western blotting and RT-PCRs were quantified by densitometry using the ImageJ software that had been developed by NIH (Bethesda, MD, USA).
Abbreviations
- Pdcd4:
-
programmed cell death 4
- PARP:
-
poly(ADP)ribose polymerase
- STS:
-
staurosporine
- CHX:
-
cycloheximide
- PI3K:
-
phosphoinositide 3-kinase
- siRNA:
-
small interfering RNA
- HA:
-
hemagglutinin
- GAPDH:
-
glyceraldehyde 3-phosphate dehydrogenase
- DMSO:
-
dimethyl sulfoxide
References
Strasser A, Harris AW, Bath ML, Cory S . Novel primitive lymphoid tumors induced in transgenic mice by cooperation between myc and bcl-2. Nature 1990; 348: 331–333.
McDonnell TJ, Korsmeyer SJ . Progression from lymphoid hyperplasia to high grade malignant lymphoma in mice transgenic for the t(14; 18). Nature 1991; 349: 254–256.
Strasser A, Whittingham S, Vaux DL, Bath ML, Adams JM, Cory S et al. Enforced BCL2 expression in B lymphoid cells prolongs antibody responses and elicits autoimmune disease. Proc Natl Acad Sci USA 1991; 88: 8661–8665.
Watanabe-Fukunaga R, Brannan CL, Copeland NG, Jenkins NA, Nagata S . Lymphoproliferation disorder in mice explained by defects in Fas antigen that mediates apoptosis. Nature 1992; 356: 314–317.
Barr PJ, Tomei LD . Apoptosis and its role in human disease. Biotechnology 1994; 12: 487–493.
Thompson CB . Apoptosis in the pathogenesis and treatment of disease. Science 1995; 267: 1456–1462.
Strasser A, O’Connor L, Dixit VM . Apoptosis signaling. Annu Rev Biochem 2000; 69: 217–245.
Lazebnik YA, Kaufmann SH, Desnoyers S, Poirier GG, Eamshaw WC . Cleavage of poly(ADP-ribose) polymerase by a proteinase with properties like ICE. Nature 1994; 371: 346–347.
Shibahara K, Asano M, Ishida Y, Aoki T, Koike T, Honjyo T . Isolation of a novel mouse gene MA-3 that is induced upon programmed cell death. Gene 1995; 166: 297–301.
Onishi Y, Hashimoto S, Kizaki H . Cloning of the TIS gene suppressed by topoisomerase inhibitors. Gene 1998; 215: 453–459.
Azzoni L, Zatsepina O, Abebe B, Bennett IM, Kanakaraj P, Perussia B . Differential transcriptional regulation of CD161 and a novel gene, 199/15a, by IL-2, IL-15, and IL-12 in NK and T cells. J Immunol 1998; 161: 3493–3500.
Matsuhashi S, Yoshinaga H, Yatsuki H, Tsugita A, Hori K . Isolation of a novel gene from a human cell line with Pr-28mAb which recognizes a nuclear antigen involved in the cell cycle. Res Commun Biochem Cell Mol Biol 1997; 1: 109–120.
Göke A, Göke R, Knolle A, Trusheim H, Schmidt H, Wilmen A et al. DUG is a novel homologue of translation initiation factor 4G that binds eIF4A. Biochem Biophys Res Commun 2002; 297: 78–82.
Onishi Y, Kizaki H . Molecular cloning of the genes suppressed in RVC lymphoma cells by topoisomerase inhibitors. Biochem Biophys Res Commun 1996; 228: 7–13.
Lankat-Buttgereit B, Göke R . The tumour suppressor Pdcd4: recent advances in the elucidation of function and regulation. Biol Cell 2009; 101: 309–317.
Allgayer H . Pdcd4, a colon cancer prognostic that is regulated by a microRNA. Crit Rev Oncol Hematol 2010; 73: 185–191.
Eto K, Eda K, Hayano M, Goto S, Nagao K, Kawasaki T et al. Reduced expression of an RNA-binding protein prolactin leads to translational silencing of programmed cell death protein 4 and apoptosis in newt spermatogonia. J Biol Chem 2009; 284: 23260–23271.
Fujino R, Tanaka K, Morimatsu M, Tamura K, Kogo H, Hara T . Spermatogonial cell-mediated activation of an IκBζ-independent nuclear factor-κB pathway in Sertoli cells induces transcription of the lipocalin-2 gene. Mol Endocrinol 2006; 20: 904–915.
Dorrello NV, Peschiaroli A, Guardavaccaro D, Colburn NH, Sherman NE, Pagano M . S6K1- and βTRCP-mediated degradation of PDCD4 promotes protein translation and cell growth. Science 2006; 314: 467–471.
Giuliano M, D’Anneo A, DeBlasio A, Vento R, Tesoriere G . Apoptosis meets proteasome, an invaluable therapeutic target of anticancer drugs. Ital J Biochem 2003; 52: 112–121.
Watanabe T, Takeda A, Mise K, Okuno K, Suzuki T, Minami N et al. Stage-specific expression of microRNAs during Xenopus development. FEBS Lett 2005; 579: 318–324.
Shi R, Chiang VL . Facile means for quantifying microRNA expression by real-time PCR. BioTechniques 2005; 39: 519–525.
Young MR, Santhanam AN, Yoshikawa N, Colburn NH . Have tumor suppressor PDCD4 and its counteragent oncogenic miR-21 gone rogue? Mol Interv 2010; 10: 76–79.
Bitomsky N, Wethkamp N, Marikkannu R, Klempnauer K-H . siRNA-mediated knockdown of Pdcd4 expression causes upregulation of p21(Waf1/Cip1) expression. Oncogene 2008; 27: 4820–4829.
Lankat-Buttgereit B, Lenschen B, Schmidt H, Göke R . The action of Pdcd4 may be cell type specific: evidence that reduction of dUTPase levels might contribute to its tumor suppressor activity in Bon-1 cells. Apoptosis 2008; 13: 157–164.
Fernandes-Alnemri T, Litwack G, Alnemri ES . CPP32, a novel human apoptotic protein with homology to Caenorhabditis elegans cell death protein Ced-3 and mammalian interleukin-1β-converting enzyme. J Biol Chem 1994; 269: 30761–30764.
Kalinichenko SV, Kopantzev EP, Korobko EV, Palgova IV, Zavalishina LE, Bateva MV et al. Pdcd4 protein and mRNA level alternations do not correlate in human lung tumors. Lung Cancer 2008; 62: 173–180.
Chen Y, Knösel T, Kristiansen G, Pietas A, Garber ME, Matsuhashi S et al. Loss of PDCD4 expression in human lung cancer correlates with tumor progression and prognosis. J Pathol 2003; 200: 640–646.
Yoshinaga H, Matsuhashi S, Fujiyama C, Masaki Z . Novel human PDCD4 (H731) gene expressed in proliferative cells is expressed in small duct epithelial cells of the breast as revealed by an anti-H731 antibody. Pathol Int 1999; 49: 1067–1077.
Afonja O, Juste D, Das S, Matsuhashi S, Samuels HH . Induction of PDCD4 tumor suppressor gene expression by RAR agonists, antiestrogen and HER-2/neu antagonist in breast cancer cells. Evidence for a role in apoptosis. Oncogene 2004; 23: 8135–8145.
Göke R, Gregel C, Göke A, Arnold R, Schmidt H, Lankat-Buttgereit B . Programmed cell death 4 (PDCD4) acts as a tumor suppressor in neuroendocrine tumor cells. Ann N Y Acad Sci 2004; 1014: 220–221.
Yang HS, Matthews CP, Clair T, Wang Q, Baker AR, Li CC et al. Tumorigenesis suppressor Pdcd4 down-regulates mitogen-activated protein kinase kinase kinase kinase 1 expression to suppress colon carcinoma cell invasion. Mol Cell Biol 2006; 26: 1297–1306.
Zhang H, Ozaki I, Mizuta T, Hamajima H, Yasutake T, Eguchi Y et al. Involvement of programmed cell death 4 in transforming growth factor-β1-induced apoptosis in human hepatocellular carcinoma. Oncogene 2006; 25: 6101–6112.
Davis BN, Hiyard AC, Lagna G, Hata A . Smad proteins control DROSHA-mediated microRNA maturation. Nature 2008; 454: 56–61.
Wang W, Zhao J, Wang H, Sun Y, Peng Z, Zhou G et al. Programmed cell death 4 (PDCD4) mediates the sensitivity of gastric cancer cells to TRAIL-induced apoptosis by down-regulation of FLIP expression. Exp Cell Res 2010; 316: 2456–2464.
van de Wetering M, Oving I, Muncan V, Pon Fong MT, Brantjes H, van Leenen D et al. Specific inhibition of gene expression using a stably integrated, inducible small-interfering-RNA vector. EMBO Rep 2003; 4: 609–615.
Acknowledgements
We thank Dr. K-H Klempnauer (Institut für Biochemie, Westfälische Wilhelms-Universität Münster, Münster, Germany) for providing us with pTER plasmid and the expression construct for Pdcd4-targeted siRNA, and Professor emerita MA DiBerardino for editorial review of the manuscript. This work was funded in part by Grants-in-Aid for Scientific Research (No. 22247008) from the Ministry of Education, Science, Sports, and Culture of Japan.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by S Kumar
Rights and permissions
About this article
Cite this article
Eto, K., Goto, S., Nakashima, W. et al. Loss of programmed cell death 4 induces apoptosis by promoting the translation of procaspase-3 mRNA. Cell Death Differ 19, 573–581 (2012). https://doi.org/10.1038/cdd.2011.126
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2011.126
Keywords
This article is cited by
-
Atg7 senses ATP levels and regulates AKT1-PDCD4 phosphorylation-ubiquitination axis to promote survival during metabolic stress
Communications Biology (2023)
-
Noncanonical functions of Ku may underlie essentiality in human cells
Scientific Reports (2023)
-
Induction of premature senescence and a less-fibrogenic phenotype by programmed cell death 4 knockdown in the human hepatic stellate cell line Lieming Xu-2
Human Cell (2022)
-
The protective effect of microRNA-21 in neurons after spinal cord injury
Spinal Cord (2019)
-
Cytoplasmic localization of programmed cell death 4 contributes to its anti-apoptotic function
Molecular and Cellular Biochemistry (2018)