Abstract
Here we investigate the function of zebrafish Bcl-2 family proteins and demonstrate important conservation of function across zebrafish and mammalian systems. We have isolated a zebrafish ortholog of mammalian BIM and show that it is the most toxic of the zebrafish BH3-only genes examined, sharing this characteristic with the mammalian BIM gene. The zebrafish bad gene shows a complete lack of embryonic lethality, but like mammalian BAD, its pro-apoptotic activity is regulated through phosphorylation of critical serines. We also found that the pattern of mitochondrial dysfunction observed by zebrafish BH3 domain peptides in a mammalian cytochrome c release assay recapitulates the pattern of embryonic lethality induced by the respective mRNA injections in vivo. In contrast to zebrafish Bim, Bid exhibited only weak binding to zebrafish Bcl-2 and moderate-to-weak overall lethality in zebrafish embryos and isolated mitochondria. Given that zebrafish Bcl-2 binds strongly to mammalian BID and BIM peptides and proteins, the protein identified as the zebrafish Bid ortholog has different properties than mammalian BID. Overall, our results demonstrate the high degree of functional conservation between zebrafish and mammalian Bcl-2 family proteins, thus validating the zebrafish as a model system to further dissect the molecular mechanisms that regulate apoptosis in future forward genetic and chemical modifier screens.
Similar content being viewed by others
Main
Members of the BCL-2 family of proteins are key regulators of the intrinsic, or mitochondrial, apoptotic pathway. The BCL-2 family can be subdivided into three main functional classes based primarily on number of conserved α-helical BCL-2 homology (BH) domains. The pro-survival members contain all four BH domains and include BCL-2, BCL-XL, MCL-1, A1/BFL-1, and BCL-W. The multidomain pro-apoptotic members (BAX, BAK, and BOK) each contain BH domains 1, 2, and 3, but members of the pro-apoptotic BH3-only class (BID, BIM, BAD, NOXA, PUMA, HRK, BIK, BMF) show conservation only in the BH3 domain.
BCL-2 family proteins control the commitment to programmed cell death in the mitochondrial-mediated apoptosis pathway.1, 2 Exposure of the cell to a wide variety of pro-death signals leads to activation of the BH3-only proteins.3, 4 The pro-apoptotic activity of these proteins is dependent on a functional BH3 domain 5 and the presence of BAX and BAK.6, 7 BID and BIM are able to interact with and consequently induce the homo-oligomerization and activation of BAX and BAK, and are therefore called activator BH3-only proteins.8, 9 Activation of BAX and BAK causes mitochondrial outer membrane permeabilization followed by release of cytochrome c and other apoptosis-promoting factors into the cytosol.10, 11 Cytochrome c is then able to bind to APAF-1 and Caspase-9 to form a holoenzyme that activates Caspase-3 through proteolytic cleavage, thus culminating in cell-wide proteolysis.2
Pro-survival BCL-2 family members (like BCL-2) can bind and sequester the activators, thus preventing them from activating BAX and BAK.9, 12, 13 A class of pro-death BH3-only proteins called sensitizers (including BAD, NOXA, BIK, HRK, BMF) cannot activate BAX and BAK directly.9, 14 Rather, they are thought to act indirectly by displacing the activators from pro-survival member-binding pockets, thereby releasing the activators to trigger the activation of BAX and BAK. One notable characteristic of activators (including BID and BIM) is that they are capable of binding to all pro-survival BCL-2 family members, whereas sensitizers demonstrate more selective interaction patterns.8, 14, 15, 16
It has recently become evident that some of the historically most useful genetically tractable models of apoptosis differ significantly from mammals in the mechanism by which apoptosis is controlled. For instance, BAX and BAK are essential for apoptosis in mammals, but no ortholog has been identified in Caenorhabditis elegans. Thus, it would be of great utility to validate a genetically tractable vertebrate model that shares key features of apoptotic control with the human system. The zebrafish genome contains orthologs of nearly all of the known BCL-2 family genes.17, 18, 19 Overexpression and morpholino knockdown studies in zebrafish have demonstrated lethality by pro-apoptotic members and cytoprotection by antiapoptotic members of the bcl-2 family.20 Although the identification and cloning of the zebrafish bcl-2 family of genes is nearly complete, zebrafish bim was previously reported to be a pseudogene20 and has not yet been functionally analyzed. BIM plays such a critical role in numerous types of mammalian cell death; its absence would call into serious question the applicability of zebrafish models. Here we report the cloning and functional analysis of the zebrafish bim gene. We furthermore demonstrate the cross-species conservation of function of a number of zebrafish BH3-only genes as well as the antiapoptotic zebrafish gene bcl-2. This confirmation of the function of key members, like Bim, of the zebrafish bcl-2 family provides significant support for the use of this vertebrate to model mammalian cell death biology.
Results
Identification of a functional zebrafish ortholog of human BIM
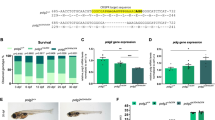
We identified predicted zebrafish bim exons by syntenic analysis. Specifically, predicted coding sequences surrounding human and mouse BIM were analyzed to identify highly conserved neighboring genes. The ACOXL (Acyl-Coenzyme A Oxidase-Like) gene is an ideal candidate, as it is located approximately 50 kb 5′ to the BIM gene in the mammalian genomes. To identify the corresponding zebrafish acoxl gene, we performed a BLASTX search in the Ensembl Zebrafish genome database using the human ACOXL protein sequence. We analyzed sequences 3′ to the zebrafish acoxl gene and identified GeneScan-predicted exonic sequences that encoded a putative protein containing a BH3-only domain. We confirmed that this predicted transcript was an expressed gene in zebrafish embryos by performing RT-PCR on RNA derived from 24 h post-fertilization (hpf) embryos and subcloning the expressed product into the pCS2+ plasmid. Wild-type cDNA coding sequence was then compared to the GeneScan-predicted transcript and found to be identical. An alignment of the predicted translation product revealed an overall 26% identity and 35% similarity with human BIM EL (Figure 1), with 100% conservation of the BH3 domain. Thus, we have identified a zebrafish ortholog of mammalian BIM (GenBank EF539840).
Alignment of zebrafish Bim and mouse and human BIM (EL) proteins. The zebrafish bim gene was identified and its protein product aligned with mouse and human BIM sequences (GenBank AF032459 and AF032457, respectively) using ClustalW and Boxshade from the EMBnet server. The domain specific to the EL isoform lies in the area between the arrows and the nine-amino-acid core BH3 domain is underlined. *Conservation of human serine 69, shown previously to be phosphorylated by ERK1/2.21, 22 #Possible conservation of human serine 87 (depending on where the zebrafish serine 68 (zS68) is aligned), shown previously to be phosphorylated by AKT.23 Also shown, the mammalian dynein light-chain (DLC) binding site24 does not appear to be conserved in zebrafish Bim. (b) Individual pairwise alignments were used to determine percent identity and overall similarity between protein sequences
An intact mitochondrial-mediated apoptosis pathway is maternally provided in zebrafish
Zygotic transcription in the zebrafish embryo begins at 2.5 hpf, at the mid-blastula transition, before which only maternally provided proteins are found within the embryo.25 Overexpression of zebrafish BH3-only genes leads to varying levels of lethality in developing embryos as measured at 8 h post-mRNA injection.20 As all mitochondria are maternally derived, it is an important question whether zygotic mitochondria already bear a maternal imprint of BCL-2 family proteins, or whether they arrive ‘empty’ and expression of BCL-2 family members is only established during embryogenesis. As such, this knowledge will prove useful in zebrafish forward genetic screens designed to interrogate the mitochondrial-mediated apoptosis pathway, as it will reveal the likelihood of identifying maternal effect mutations that affect cell death.
Thus, we asked whether zebrafish BH3-only proteins could induce apoptosis before the advent of zygotic transcription. To address this question, we injected zebrafish embryos with 100 ng/μl of mRNA encoding each gene or egfp as a negative control, at the one-cell stage of development (approximately 15 min post-fertilization), and followed their development during the first 2 hpf. Within the first hpf, embryos injected with mRNAs encoding bim, noxa, or puma ceased normal development and underwent a structural breakdown (Figure 2a). Conversely, development of embryos injected with egfp, bid, or bad mRNA proceeded normally during this time period. To determine if the structural breakdown involved activation of the mitochondrial-mediated apoptosis pathway, we fixed the injected embryos at the two- to four-cell stage and performed immunofluorescence with an antibody to activated-human Caspase-3 (from here on referred to as the Casp3 assay). Caspase-3 activation in embryos was an early indication of active apoptosis and predicted structural breakdown by the eight-cell stage (Figure 2a). At the 8- to 16-cell stage (1.25–1.5 hpf), embryos injected with bim, noxa, and puma exhibited similar phenotypes to each other, as did embryos injected with egfp, bid, and bad (Figure 2b).
Overexpression of the zebrafish BH3-only genes bim, noxa, and puma, but not bad or bid, causes widespread embryonic death via maternally provided apoptotic machinery. (a) One-cell stage embryos were injected with 100 ng/μl of mRNA encoding the indicated genes, and development of the embryos was evaluated by brightfield microscopy. One representative embryo from each group is shown. At the two- to four-cell stage, half of the embryos were fixed and analyzed for apoptosis by Casp3 assay. Fluorescence indicates the presence of activated caspase-3 in a representative embryo from each group. (b) Groups treated as in (a) are shown at the 8- to 16-cell stage to demonstrate that bim, noxa, and puma mRNA injections give rise to overall similar phenotypes. (c) One-cell stage embryos were injected with a range of doses of mRNA encoding the indicated genes, and embryos were scored for survival at 2 h post-injection. The concentration of wild-type mRNA in ng/μl is indicated for each gene. The total mRNA concentration of each injection was held constant at 100 ng/μl using egfp mRNA to make up the difference. BH3 mutant forms of each gene (bh3mut) described in (d) were injected at 100 ng/μl and included to demonstrate that the lethality is caused by the respective BH3-domain. Standard deviation is representative of three to six independent experiments. (d) BH3 domain mutations were created for each zebrafish BH3-only gene by changing the residue(s) denoted by an asterisk to alanine
To quantify the embryonic toxicity induced by each BH3-only gene, we injected a range of doses of mRNA and calculated embryo survival at 2 hpf (Figure 2c). As has been found in mammalian systems, bim and puma were found to be most toxic. Zebrafish bim mRNA proved the most toxic as most embryos were dead at an mRNA concentration of 3 ng/μl. Lethality caused by puma and noxa was nearly maximal at 10 and 30 ng/μl, respectively. At the maximal dose of 100 ng/μl, zebrafish bid showed moderate lethality whereas bad and egfp were nontoxic. Although higher amounts of bid mRNA may induce more toxicity (as demonstrated by 20Kratz et al.), we occasionally see nonspecific morphological defects arising from injection of greater than 100 ng/μl mRNA, and thus, this is our upper injection limit. By 24 hpf, about half of the 100 ng/μl bid-injected embryos had survived, whereas all of the bad- and egfp-injected embryos continued to develop normally (data not shown).
As the pro-apoptotic activity of mammalian BH3-only proteins requires an intact BH3 domain,5 we asked whether the embryonic apoptosis of each zebrafish BH3-only gene was specifically mediated by their respective BH3 domains, or by some other mechanism. Thus, we introduced mutations into the nine-amino-acid core BH3 domains of bid, bim, noxa, and puma (Figure 2d) and injected mRNA encoding these mutant proteins into embryos (Figure 2c). Mutation of the BH3 domain abrogated the lethality induced by each of the mRNAs indicating that embryonic death is occurring in a BH3-domain-dependent manner. Consistent with a specific requirement for the BH3 domain in inducing embryonic lethality, co-injection of zebrafish bcl-2 mRNA rescues the lethality induced by overexpression of bid, bim, puma, and noxa mRNA overexpression in this assay (data not shown). These experiments demonstrate that zygotes inherit maternal mitochondria that are fully competent to execute apoptotic signals.
Analogous to mammalian BAD, zebrafish Bad can radiosensitize neural tissue, and conserved serines are required for its pro-apoptotic activity
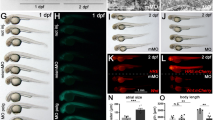
Expression of mammalian BAD has been shown to enhance radiation-induced apoptosis in cell culture.26, 27 To validate the use of the zebrafish to model mammalian apoptosis, we wanted to test whether this very specific function was shared by zebrafish Bad. As overexpression of zebrafish bad was nontoxic to zebrafish embryos, we asked whether it could enhance radiation-induced apoptosis in neural tissue. To verify that Bad protein was being expressed after mRNA injection, we created a fusion gene between bad and mcherry (a dsRED variant). Similar to unfused bad, mcherry-bad failed to induce embryonic lethality (data not shown) even though it was clearly expressed by 2 h post-injection (Figure 3a). We next injected embryos with 100 ng/μl of mRNA encoding mcherry-bad or mcherry as a negative control. At 24 hpf, half of the embryos in each group were exposed to 15 Gy γ-radiation (IRR), and all embryos were fixed 3 h later and analyzed by the Casp3 assay. Exposure of the control-injected embryos to IRR caused widespread apoptosis throughout the brain and spinal cord (Figure 3b). Overexpression of mcherry-bad in the absence of IRR induced a low level of apoptosis in the head at 27 hpf. However, upon exposure to IRR at 24 hpf, mcherry-bad-expressing embryos appeared to exhibit a greater level of activated Caspase-3 activity in both the brain and spinal cord. To capitalize on the difference in radiosensitivity between control- and mcherry-bad-injected embryos, we identified a dose of radiation (4 Gy) that led to a clear (Figure 3c) and quantifiable (Figure 3d) difference between control- and bad-injected embryos. The ability of mcherry-bad to enhance radiation-induced apoptosis required a functional BH3 domain because embryos injected with mRNA encoding a mutation in the zebrafish Bad BH3 domain (mcherry-bad bh3mut, Figure 2d) showed apoptotic levels of control-injected embryos.
Zebrafish Bad enhances radiation-induced apoptosis and its activity is likely regulated by serine phosphorylation. (a) Embryos were either uninjected or injected with 100 ng/μl of mcherry-bad and analyzed for mcherry expression by fluorescence microscopy 2 h after injection. One representative embryo from each group is shown. Asterisks denote the yolk region. Arrows point to the blastomeres of the animal pole. (b) Embryos were injected at the one-cell stage with mRNA encoding mcherry (as a negative control) or mcherry-bad. At 24 hpf, embryos were either left unexposed or exposed to 15 Gy γ-radiation and harvested 3 h later. The Casp3 assay was performed, and apoptotic cells were visualized by fluorescence microscopy. (c) Embryos were injected with mRNA encoding mcherry, mcherry-bad, mcherry-bad bh3mut (mcherry-bad L99A, see Figure 2c), or mcherry-bad-2SD and either left unexposed or exposed to 4 Gy γ-radiation and harvested and analyzed as in (b). In the whole embryo (above), the white box denotes the region of the spinal cord documented below. (d) Five to ten embryos from each group of embryos analyzed in (c) were blindly selected and apoptotic cells were documented by fluorescence microscopy. Fluorescence intensity from a representative region of the spinal cord from each embryo was quantified by Volocity software and displayed as arbitrary units of fluorescence (pixel) intensity. Student's t-tests were performed as indicated to measure statistical significance. **P<0.05. (e) Alignment of part of zebrafish Bad and mouse and human BAD proteins. Asterisks denote serines 84 and 103 in zebrafish Bad that show conservation with mouse serines 136 and 155 (and human serines 99 and 118), respectively. (f) Messenger RNA encoding mcherry (control), bad, and bad 2SA was injected into zebrafish embryos at the one-cell stage, and survival was measured at the indicated time points. (g) Embryos surviving at 5-h post-injection in the experiment described in (f) were analyzed by Casp3 assay. Asterisks denote the yolk region. Arrows point to the blastomeres of the animal pole
The activity of mammalian BAD is negatively regulated through phosphorylation of (mouse) serines 112, 136, and 155 and subsequent binding by 14-3-3 proteins.28 Zebrafish Bad shows conservation of mouse serines 136 and 155 (zebrafish Bad serines 84 and 103, respectively, shown in Figure 3e). To mimic phosphorylation at these serines and to assess whether the phosphoregulation of BAD activity is conserved in zebrafish, we mutated zebrafish serines 84 and 103 to aspartate (bad 2SD). Indeed, phosphomimicking at these serines prevented zebrafish Bad from radiosensitizing nervous tissue (Figure 3c and d and data not shown).
We next asked whether the lack of Bad toxicity in the early embryonic assay (Figure 1) was due to serine phosphorylation immediately following translation of the injected mRNA. To prevent phosphorylation at these sites, we mutated serines 84 and 103 to alanine (bad 2SA). Embryos injected with bad 2SA began to die by apoptosis by 4 h post-injection (Figure 3f and g). Most embryos failed to survive by 8 h post-injection. Thus, similar to mammalian BAD, the pro-apoptotic activity of zebrafish Bad appears to be regulated by serine phosphorylation.
Zebrafish Bcl-2 and BH3-only proteins function in mammalian systems
One of the strengths of the zebrafish is the ability to perform forward genetic screens in a vertebrate model system. However, it is important to test whether the genetic pathway in question maintains a high degree of functional conservation, such that information gained from these screens will be applicable to human biology. To establish the degree to which zebrafish Bcl-2 family proteins are functionally conserved, we asked whether they could function in mammalian systems similar to their mammalian counterparts. To this end, we tested the ability of zebrafish BH3 domain peptides to induce cytochrome c release in mitochondria isolated from mouse embryonic fibroblasts (MEFs). We designed zebrafish BH3 domain peptides that align in sequence to human or mouse peptides that have been analyzed previously (9Letai et al. and 14Certo et al.; Figure 4a), and results were compared to these mammalian peptides (Figure 4b). Among the zebrafish BH3 domain peptides, Bim was the most potent inducer of cytochrome c release, similar to mouse BIM and in keeping with its expected role as an ‘activator’.9 On the contrary, Bid was one of the least potent of the zebrafish peptides, similar to its modest activity in vivo (Figure 2c). Like the human PUMA peptide, the zebrafish Puma peptide had substantial activity in this assay. The zebrafish Noxa peptide, on the other hand, showed only moderate activity. Finally, the zebrafish Bad BH3 peptide was unable to induce cytochrome c release, consistent with the role of mammalian BAD as a sensitizer BH3-only protein.9 None of the peptides showed any activity in cells that lacked BAX and BAK (Figure 4c), indicating that the BH3 peptides function through the genetic pathway of mitochondrial-mediated apoptosis rather than by nonspecific damage to the mitochondria.
Zebrafish BH3-only peptides display a toxicity in isolated mouse mitochondria that is similar to lethality of the respective full-length proteins in vivo. (a) To design zebrafish BH3 domain peptides, zebrafish BH3-only protein sequences were aligned with human or mouse peptides as described previously.9, 14 (b) Mitochondria (0.5 mg/ml) isolated from wild-type MEFs were treated with zebrafish, mouse, or human BH3 domain peptides (10 μM). Percent cytochrome c release was measured by ELISA. (c) BH3 domain peptides were analyzed as in (b) but with mitochondria isolated from BAX−/−, BAK−/− MEFs (DKO). Standard deviations of at least three independent measurements are in parentheses
Zebrafish BH3 domains exhibit selective interaction with mammalian and zebrafish antiapoptotic BCL-2 proteins
A common characteristic of mammalian activator proteins like BID and BIM is that they are able to bind to all of the pro-survival Bcl-2 family proteins, whereas sensitizer BH3-only proteins tend to exhibit more selectivity in binding.8, 14, 15 For instance, the mammalian BAD BH3 peptide binds to BCL-2 and BCL-xL, but not to MCL-1, whereas the NOXA BH3 peptide shows the opposite binding pattern. To investigate the importance of this trend, we asked whether zebrafish BH3-only proteins exhibited protein–protein interaction patterns that were similar to those observed with mammalian BH3-only proteins. We generated full-length GST-tagged zebrafish Bcl-2 protein and tested the binding affinity of BH3-domain peptides by fluorescence polarization (Figure 5a and b). Among the mammalian peptides, the activators BID and BIM demonstrated the highest affinity for binding to zebrafish Bcl-2. Zebrafish Bim followed suit showing the highest affinity among the zebrafish peptides. However, zebrafish Bid showed no significant binding activity, again suggesting that the function of this protein has diverged in zebrafish. Interestingly, human PUMA had approximately 15 times stronger binding affinity than zebrafish Puma, whereas both mouse and zebrafish Bad and Noxa showed comparatively weak binding.
Zebrafish Bcl-2 has evolved to bind a distinct set of BH3-only proteins. Binding affinities between GST-zBcl-2 and BH3 domain peptides were analyzed by fluorescence polarization. (a) Representative binding curves show selective binding between antiapoptotic zebrafish Bcl-2 and zebrafish and mouse BH3-only family members. (b) Dissociation constants (in nM) were calculated for each interaction and then corrected for impurities in the protein (62.8% pure protein, 37.2% impurities). Standard deviations of at least three independent measurements are in parentheses. Yellow blocks signify high affinity binding, blue blocks and minus sign signify no observed binding (Kd>700 nM). Activators are in purple, sensitizers in green. n.a., not assayed. We previously published the GST-BCL-2 binding data,14 and permission was kindly granted by Cancer Cell to reprint the data here for the sake of comparison
As zebrafish Bcl-2 was proficient at binding to human BID and BIM peptides, we asked whether it could also bind full-length protein in the context of LY-1 cell lysates. We generated affinity-purified GST-tagged zebrafish Bcl-2 and performed a GST pull-down assay (Figure 6a). We found that GST-zebrafish Bcl-2, but not GST alone, was indeed able to bind to human BID and the long (L) and extra-long (EL) isoforms of BIM. As the ability to bind pro-death BID and/or BIM is a requisite for the pro-survival function of Bcl-2,29, 30, 31, 32 we next examined whether zebrafish Bcl-2 could promote survival in mammalian cells (Figure 6b). We transfected FL5.12 cells with either pCMV-3XFLAG-zebrafish Bcl-2 or vector alone. Upon IL-3 withdrawal, these cells normally undergo a mitochondrial-mediated apoptotic death. However, overexpression of zebrafish Bcl-2 reduced the Annexin V positivity in serum-starved cells by almost half, proving that the pro-survival function of zebrafish Bcl-2 is conserved across species. Finally, we reasoned that in order for zebrafish Bcl-2 to exert its pro-survival function in FL5.12 cells (which express undetectable levels of BID), it must be binding to BIM upon IL-3 withdrawal. Thus, we immunoprecipitated FLAG-tagged zebrafish Bcl-2 with an anti-FLAG antibody and analyzed its binding partners in the presence and absence of IL-3 (Figure 6c). Upon withdrawal of IL-3, FLAG-zebrafish Bcl-2 bound to substantially more BIM EL and BIM L, whereas the opposite binding pattern was observed for BAX. Finally, zebrafish Bcl-2 bound well to PUMA before and after IL-3 withdrawal.
Zebrafish Bcl-2 protects FL5.12 cells from IL-3 withdrawal-induced apoptosis, likely through sequestration of mouse Bim. (a) Zebrafish Bcl-2 interacts with mouse BIM and BID. A GST pull-down assay was performed to analyze zebrafish Bcl-2 binding interactions. GST-zBcl-2 was immobilized on glutathione-agarose beads and combined with LY-1 cell lysate (100 μg). Proteins interacting with GST-zBcl-2 were analyzed by western blot with antibodies recognizing BIM and BID. (b) Overexpression of zebrafish Bcl-2 protects FL5.12 cells from IL-3 withdrawal-induced apoptosis. FL5.12 cells were stably transfected with vector alone or pCMV-3XFLAG-zBcl-2 in the presence of IL-3. Cells were then cultured in the presence or absence of IL-3 for 30 h and analyzed by Annexin V assay. The average and standard deviation of three independent experiments are shown. (c) Zebrafish Bcl-2 binds more BIM in FL5.12 cells upon withdrawal of IL-3. Lysates were prepared from cells treated as described in (b). Lysates were combined with Protein A beads and incubated with FLAG antibody (7 μg). Proteins interacting with FLAG (vector) and FLAG-zBcl-2 were analyzed and compared by western blot with antibodies to the indicated BCL-2 family proteins
Discussion
The genetically tractable zebrafish model system holds great promise to further our understanding of the vertebrate mitochondrial-mediated apoptosis pathway, which is important for normal embryologic development and disrupted during the molecular pathogenesis of cancer.33 Within the zebrafish Bcl-2 family of genes, there is a high degree of sequence conservation within specific regions, like the BH3 domain, of the BH3-only proteins whereas other regions tend to be relatively divergent (Coultas et al.17 and Kratz et al.20; Figure 1 and data not shown). Thus, conserved synteny within the genome has contributed an important component to the identification of structural and functional zebrafish orthologs of the corresponding mammalian BH3-only genes.
Prior to our work, a BIM ortholog had not been identified in zebrafish, and it had been suggested that zebrafish bim existed only as a pseudogene.20 This posed a significant problem for those who wished to interpret results from zebrafish as relevant to mammalian apoptotic signaling. Mammalian BIM has been shown to play a critical role in the mitochondrial-mediated apoptosis pathway for many important types of cell death.29, 34, 35 We felt that identification of a BIM ortholog was absolutely essential to credential the zebrafish as a model system for studies of the vertebrate cell-death pathway. We found that compared to human and mouse BIM, zebrafish Bim shows 100% conservation of the BH3 domain (Figure 1). Importantly, zebrafish Bim shows conservation of the (human) serine 69, which is required for ERK1/2-dependent phosphorylation-mediated proteasomal degradation,21, 22 and possibly (human) serine 87 (depending on how zebrafish serine 68 is aligned), which has been shown to regulate activity of BIM through phosphorylation by AKT.23 Conservation of these serines, despite surrounding areas of divergent sequence, implicates their potential functional importance in the BIM EL domain. Notably, zebrafish Bim lacks an obvious dynein light chain-binding site (Figure 1). It will be interesting to determine whether localization of BIM to microtubules24 is conserved in zebrafish.
When overexpressed in early embryos, we found that zebrafish bim, noxa, and puma potently induced apoptosis before the advent of zygotic transcription. Death is likely induced through engagement of the intrinsic mitochondrial-mediated apoptotic pathway, as functional BH3 domains are required (Figure 2c) and Caspase-3 is activated (Figure 2a). It is intriguing to speculate why maternal mitochondria are already equipped with proteins to execute death signals during the first few hours of development. It suggests that either developmental modeling via apoptosis or the destruction of defective cells or even whole embryos is important even at very early stages of development.
As mammalian BH3-only proteins function in part by binding (via the BH3 domain) to antiapoptotic BCL-2 family members, we questioned whether zebrafish BH3-only protein-binding interactions were conserved. To assess the conservation of protein–protein interaction patterns, we analyzed the ability of each zebrafish BH3 domain peptide to bind to the full-length zebrafish Bcl-2 protein (Figure 5). Although zebrafish Bim and human BIM interacted strongly with zebrafish Bcl-2, we found that the other BH3 domain peptides diverged in their affinities for zebrafish Bcl-2. Remarkably, although the human BID peptide showed similar affinity for both human BCL-2 and zebrafish Bcl-2, the zebrafish Bid peptide failed to detectably bind to zebrafish Bcl-2, and zebrafish bid mRNA expression was only moderately pro-apoptotic in vivo (50% death by 24 h). Mammalian BID is normally activated by cleavage by caspase-8,36, 37 at a site that is conserved in zebrafish Bid (data not shown), and it is possible that zebrafish Bid is not sufficiently cleaved after its overexpression in embryos. However, the isolated zebrafish Bid BH3 domain peptide also failed to induce cytochrome c release in isolated mitochondria (Figure 4) suggesting that either its function has diverged from mammalian BID, or it is not in fact the true functional ortholog of BID. Notably, the identified zebrafish Bid ortholog is not syntenic with mammalian BID, and although it has been reported that the zebrafish Bid protein is 36% identical to human BID,20 we have been unable to show more than 23% identity between these two species using both the Jotun Hein and the ClustalW alignment methods (data not shown).
We have previously demonstrated in transgenic zebrafish engineered to express zebrafish Bcl-2 specifically in lymphoid cells that like mammalian BCL-2, zebrafish Bcl-2 is able to inhibit apoptosis induced by exposure to ionizing radiation and treatment with dexamethasone.38 We reasoned that if zebrafish Bcl-2 is highly functionally conserved, then it should function similar to human BCL-2 in a mammalian system. Indeed, we found that like human BCL-2, zebrafish Bcl-2 is able to bind to mammalian BID and BIM proteins and is capable of inhibiting IL-3 withdrawal-induced apoptosis in murine FL5.12 cells. Furthermore, zebrafish Bcl-2 bound more BIM in the absence of IL-3 suggesting that like human BCL-2, zebrafish Bcl-2 inhibits apoptosis in IL-3-deprived mammalian cells by sequestering the pro-apoptotic BIM protein.
Thus, we have demonstrated a high degree of functional conservation between zebrafish and mammalian BCL-2 family proteins, while simultaneously uncovering specific areas of divergence, such as in protein–protein interaction patterns. Ultimately, these findings will serve as a foundation for analysis of the BCL-2 family of proteins by way of future forward genetic and chemical modifier screens using the genetically tractable zebrafish model system.
Materials and Methods
Constructs
Zebrafish cDNAs were cloned into pCS2+ and digested with NotI. BH3 domain mutations were engineered into the coding sequences by site-directed mutagenesis (Quikchange II Site-directed Mutagenesis kit, Stratagene). The following primers were generated to perform the mutagenesis on pCS2+ constructs:
zbid-L96A (GCGAGAGAAATGGCGGCAGAGGCGATCAGAATAGCAGATC)
zbim-L128A (GGTGGTCGCTCGTGAAGCGCGACGCATAGGCGATG)
zbim-E132A, D133A (GCGACGCATAGCCGCTGAGTTCAATCGCC)
zbad-L99A (GCAGCTAAGAAATACGGCCAACAGGCGAGAAGAATGGCTGATGAG)
zbad-S84D (GGCCGAGATCCCGCGATGCTCCTCCTGCTTTGTGGGC)
zbad-S103D (CGGCCAACAGCTGAGAAGAATGGATGATGAGTTTGATAAAGGG)
zbad-S84A (GGCCGAGATCCCGCGCTGCTCCTCCTGCTTTGTGGGC)
zbad-S103A (CGGCCAACAGCTGAGAAGAATGGCTGATGAGTTTGATAAAGGG)
znoxa-L16A (GTGCGCGCAGCAGGCGCGCAACATTGG)
zpuma-L134A (GGAGAGGGTGGCCGTACAAGCGAGGACAATTGGGGACG)
zpuma-D139A (GCGAGGACAATCGGGGCCGCGATGAACGCTGTC)
zpuma-E140A (GGACAATCGGGGACGCGATGAACGCTGTCTTCC)
Messenger RNA was made using the SP6 Message Machine kit (Ambion) and purified for microinjection with NucAway Spin Columns (Ambion). Bacterial expression constructs were generated by subcloning the zebrafish bcl-2 coding sequence into either pGEX-4T-1 (to create GST-zBcl-2 protein) or pCMV-3XFLAG (to create FLAG-zBcl-2 protein). The C-terminal transmembrane domain of zebrafish Bcl-2 was truncated by 19 amino acids in length to maintain solubility in aqueous solution.
Cloning and alignments of full-length bim
A putative zebrafish bim ortholog was initially identified through synteny comparisons with human and mouse genomic sequences using the Ensembl genome database (Sanger, UK) as described in the Results section. The following primers were used to amplify full-length bim from 24 hpf zebrafish cDNA (ORF sequence in caps):
5′: ccgggaattcATGTCTGACACGTCCAGAGAGCAAA
3′: ccagctcgagTCATCTTCTTCGCAGGAAAAAG
The bim cDNA was initially TOPO-cloned (Invitrogen, USA) and subsequently transferred into pCS2+ using EcoRI and XhoI digestion (New England Biolabs). Alignment of zebrafish Bim and mouse and human BIM (EL) proteins (GenBank nos. AF032459 and AF032457, respectively) was performed using ClustalW and Boxshade from the EMBnet server. Individual pairwise alignments were used to determine identity and overall similarity between protein sequences.
Microinjections
Zebrafish AB strain one-cell stage embryos were injected with 100 ng/μl mRNA (approximately 300 pg per embryo). When dose responses were performed, egfp mRNA was supplemented to maintain the same total amount of mRNA injected. To control for natural variations in the percentage of surviving embryos following microinjection, an egfp control was included within each clutch of embryos injected, and experimental values for embryo survival at 2 h were calculated by dividing the percentage of embryos surviving in the experimental groups by the egfp control group. Unfertilized embryos were included in the calculations as the toxicity of BH3-only mRNAs occurred in both fertilized and unfertilized embryos. Where indicated, mRNA encoding mcherry (a monomeric variant of the fluorescent protein dsRED39) was injected as a negative control.
Whole-mount zebrafish immunofluorescence (Casp3 assay)
To perform the Casp3 assay, embryos were fixed in 4% paraformaldehyde overnight at 4°C and then transferred to 100% methanol overnight at −20°C. Embryos were then washed three times for 5 min with 1 × PBST (1 × PBS with 0.1% Tween) and transferred into 300 μl block (10% heat-inactivated FBS, 2% BSA in PBST) for 1 h at room temperature. A 1 : 500 dilution of antiactivated human Caspase-3 antibody (BD Biosciences, no. 559565) was added, and embryos were incubated at 4°C overnight. Embryos were then washed three times, 10 min each, with PDT buffer (1 × PBST, 0.3% Triton X, 1% DMSO) and then transferred back into block for 1 h at room temperature. A Molecular Probes goat anti-rabbit IGG Alexa Fluor 488 (green fluorescing, and used if embryos expressed mcherry) or 568 (red fluorescing, and used if embryos expressed egfp) was then added at a concentration of 1 : 200, and embryos were incubated at 4°C overnight. The next day, embryos were washed three times in PDT for 10 min each and visualized. All steps involved rocking the embryos except the methanol step. Embryos were stored in 80% glycerol and then results were documented by fluorescence microscopy using a Nikon Digital Sight DS-2MBWc black and white camera. All fluorescent pictures were taken at exactly the same exposure, gain, and magnification. Quantitation of fluorescence was performed with Volocity software.
Peptides and recombinant proteins
The pGEX-4T-1-zbcl-2 expression vector was expressed in BL-21 bacteria and affinity purified using glutathione-agarose (for GST-linked proteins) as previously described.9 The pCMV-3xFLAG-zbcl-2 expression vector was cloned into DH5α competent cells (Invitrogen). Peptides were designed and synthesized as shown in Figure 4a and Certo et al.14
Cytochrome c release
Mitochondria were purified from MEFs as previously described.9 Mitochondria were incubated with indicated peptides for 45 min. Release of cytochrome c was determined by a comparison of cytochrome c in the pellet and supernatant following treatment, quantified by ELISA (R&D systems). When results of multiple experiments were averaged, solvent-only (DMSO) treatment values were subtracted from each experimental value so that zero release reflects that observed in solvent-only treatments.
Fluorescence polarization binding assays
Binding assays were performed using fluorescence polarization as previously described.14 A minimum of three independent experiments was used to determine dissociation constants.
Cell culture
IL3-dependent murine pro-B lymphocytic (FL5.12) cells were transfected (by electroporation in a Gene Pulsar (BioRad), set at a conductance of 975 μF and a voltage of 0.25 kV) with either pCMV-3XFLAG-zBcl-2 or vector alone, and stable transfectants were selected for by growth in the presence of 1000 μg/ml G418. Stably transfected FL5.12 cells were cultured as described previously14 in Iscove's modified Dulbecco's medium, 10% fetal bovine serum, 1000 μg/ml G418 with or without IL-3 provided by 10% WEHI-3B supplement (supernatant of IL-3-secreting WEHI-3B cells). MEFs were cultured in Iscove's modified Dulbecco's medium and 10% fetal bovine serum.
GST pull-down
Ten micrograms of GST-zBcl-2 or GST alone were incubated with glutathione-agarose beads for 1 h at 4°C in binding buffer (140 mM NaCl, 10 mM Tris, pH 7.4). Beads were rinsed and incubated with approximately 100 μg human Ly-1 cell (human B-lymphocyte cells) lysate for 1 h at 4°C. Beads (1 mg) were washed again and loaded on a denaturing NuPAGE gel.
Immunoblots
Protein lysates were obtained by cell lysis in 1% CHAPS buffer. Protein samples (25 μg) were size-fractionated on NuPAGE 10% Bis-Tris polyacrylamide gels (Invitrogen) and transferred to immobilin-P PVDF membranes. The following antibodies were used for immunodetection of human and mouse proteins: BIM (Calbiochem, 22–40); PUMA (Prosci, NT); BID (Santa Cruz, FL195); BAK (Upstate, NT); BAX (Santa Cruz, N-20); and Actin (Chemicon, MAB1501).
Immunoprecipitation
FL5.12 cell lysates (1 mg) were incubated with anti-mouse FLAG antibody (7 μg, Sigma) for at least 1 h at 4 °C in 1% CHAPS buffer (5 mM sodium phosphate, pH 7.4; 2.5 mM EDTA; 100 mM sodium chloride; 1% w/v CHAPS, in the presence of protease inhibitors (Complete tablets; Roche)). Protein A-sepharose beads (Sigma) were added to precipitate complexes containing FLAG-zBcl-2. The beads (2 mg) were mixed with loading buffer prior to loading supernatant onto the gel. Twenty-five micrograms of cell lysate in 1% CHAPS buffer was used as a control.
Annexin V assay
Cells were stained with fluorescent conjugates of Annexin V (BioVision) and analyzed on a FACSCalibur machine (BD Biosciences).
Abbreviations
- BH:
-
Bcl-2 homology
- IRR:
-
irradiation
- hpf:
-
hours post-fertilization
References
Cory S, Adams JM . The Bcl2 family: regulators of the cellular life-or-death switch. Nat Rev Cancer 2002; 2: 647–656.
Danial NN, Korsmeyer SJ . Cell death: critical control points. Cell 2004; 116: 205–219.
Kelekar A, Thompson CB . Bcl-2-family proteins: the role of the BH3 domain in apoptosis. Trends Cell Biol 1998; 8: 324–330.
Puthalakath H, Strasser A . Keeping killers on a tight leash: transcriptional and post-translational control of the pro-apoptotic activity of BH3-only proteins. Cell Death Differ 2002; 9: 505–512.
Boyd JM, Gallo GJ, Elangovan B, Houghton AB, Malstrom S, Avery BJ et al. Bik, a novel death-inducing protein shares a distinct sequence motif with Bcl-2 family proteins and interacts with viral and cellular survival-promoting proteins. Oncogene 1995; 11: 1921–1928.
Wei MC, Zong WX, Cheng EH, Lindsten T, Panoutsakopoulou V, Ross AJ et al. Proapoptotic BAX and BAK: a requisite gateway to mitochondrial dysfunction and death. Science 2001; 292: 727–730.
Zong WX, Lindsten T, Ross AJ, MacGregor GR, Thompson CB . BH3-only proteins that bind pro-survival Bcl-2 family members fail to induce apoptosis in the absence of Bax and Bak. Genes Dev 2001; 15: 1481–1486.
Kuwana T, Bouchier-Hayes L, Chipuk JE, Bonzon C, Sullivan BA, Green DR et al. BH3 domains of BH3-only proteins differentially regulate Bax-mediated mitochondrial membrane permeabilization both directly and indirectly. Mol Cell 2005; 17: 525–535.
Letai A, Bassik MC, Walensky LD, Sorcinelli MD, Weiler S, Korsmeyer SJ . Distinct BH3 domains either sensitize or activate mitochondrial apoptosis, serving as prototype cancer therapeutics. Cancer Cell 2002; 2: 183–192.
Green DR, Kroemer G . The pathophysiology of mitochondrial cell death. Science 2004; 305: 626–629.
Wang X . The expanding role of mitochondria in apoptosis. Genes Dev 2001; 15: 2922–2933.
Cheng EH, Levine B, Boise LH, Thompson CB, Hardwick JM . Bax-independent inhibition of apoptosis by Bcl-XL. Nature 1996; 379: 554–556.
Cheng EH, Wei MC, Weiler S, Flavell RA, Mak TW, Lindsten T et al. BCL-2, BCL-X(L) sequester BH3 domain-only molecules preventing BAX- and BAK-mediated mitochondrial apoptosis. Mol Cell 2001; 8: 705–711.
Certo M, Del Gaizo Moore V, Nishino M, Wei G, Korsmeyer S, Armstrong SA et al. Mitochondria primed by death signals determine cellular addiction to antiapoptotic BCL-2 family members. Cancer Cell 2006; 9: 351–365.
Chen L, Willis SN, Wei A, Smith BJ, Fletcher JI, Hinds MG et al. Differential targeting of prosurvival Bcl-2 proteins by their BH3-only ligands allows complementary apoptotic function. Mol Cell 2005; 17: 393–403.
Opferman JT, Korsmeyer SJ . Apoptosis in the development and maintenance of the immune system. Nat Immunol 2003; 4: 410–415.
Coultas L, Huang DC, Adams JM, Strasser A . Pro-apoptotic BH3-only Bcl-2 family members in vertebrate model organisms suitable for genetic experimentation. Cell Death Differ 2002; 9: 1163–1166.
Inohara N, Nunez G . Genes with homology to mammalian apoptosis regulators identified in zebrafish. Cell Death Differ 2000; 7: 509–510.
Pyati UJ, Look AT, Hammerschmidt M . Zebrafish as a powerful vertebrate model system for in vivo studies of cell death. Semin cancer biol 2007; 17: 154–165.
Kratz E, Eimon PM, Mukhyala K, Stern H, Zha J, Strasser A et al. Functional characterization of the Bcl-2 gene family in the zebrafish. Cell Death Differ 2006; 13: 1631–1640.
Ley R, Balmanno K, Hadfield K, Weston C, Cook SJ . Activation of the ERK1/2 signaling pathway promotes phosphorylation and proteasome-dependent degradation of the BH3-only protein, Bim. J Biol Chem 2003; 278: 18811–18816.
Luciano F, Jacquel A, Colosetti P, Herrant M, Cagnol S, Pages G et al. Phosphorylation of Bim-EL by Erk1/2 on serine 69 promotes its degradation via the proteasome pathway and regulates its proapoptotic function. Oncogene 2003; 22: 6785–6793.
Qi XJ, Wildey GM, Howe PH . Evidence that Ser87 of BimEL is phosphorylated by Akt and regulates BimEL apoptotic function. J Biol Chem 2006; 281: 813–823.
Puthalakath H, Huang DC, O′Reilly LA, King SM, Strasser A . The proapoptotic activity of the Bcl-2 family member Bim is regulated by interaction with the dynein motor complex. Mol Cell 1999; 3: 287–296.
Detrich III HW, Westerfield M, Zon LI . Cell cycles and development in the embryonic zebrafish. Methods Cell Biol 1999; 59: 11–26.
Mok CL, Gil-Gomez G, Williams O, Coles M, Taga S, Tolaini M et al. Bad can act as a key regulator of T cell apoptosis and T cell development. J Exp Med 1999; 189: 575–586.
Seo SY, Chen YB, Ivanovska I, Ranger AM, Hong SJ, Dawson VL et al. BAD is a pro-survival factor prior to activation of its pro-apoptotic function. J Biol Chem 2004; 279: 42240–42249.
Datta SR, Katsov A, Hu L, Petros A, Fesik SW, Yaffe MB et al. 14-3-3 proteins and survival kinases cooperate to inactivate BAD by BH3 domain phosphorylation. Mol Cell 2000; 6: 41–51.
Certo M, Moore Vdel G, Nishino M, Wei G, Korsmeyer S, Armstrong SA et al. Mitochondria primed by death signals determine cellular addiction to antiapoptotic BCL-2 family members. Cancer Cell 2006; 9: 351–365.
Cheng EH, Levine B, Boise LH, Thompson CB, Hardwick JM . Bax-independent inhibition of apoptosis by Bcl-XL. Nature 1996; 379: 554–556.
Cheng EH, Wei MC, Weiler S, Flavell RA, Mak TW, Lindsten T et al. BCL-2, BCL-X(L) sequester BH3 domain-only molecules preventing BAX- and BAK-mediated mitochondrial apoptosis. Mol Cell 2001; 8: 705–711.
Letai A, Bassik MC, Walensky LD, Sorcinelli MD, Weiler S, Korsmeyer SJ . Distinct BH3 domains either sensitize or activate mitochondrial apoptosis, serving as prototype cancer therapeutics. Cancer Cell 2002; 2: 183–192.
Green DR, Evan GI . A matter of life and death. Cancer Cell 2002; 1: 19–30.
Bouillet P, Metcalf D, Huang DC, Tarlinton DM, Kay TW, Kontgen F et al. Proapoptotic Bcl-2 relative Bim required for certain apoptotic responses, leukocyte homeostasis, and to preclude autoimmunity. Science 1999; 286: 1735–1738.
O′Connor L, Strasser A, O′Reilly LA, Hausmann G, Adams JM, Cory S et al. Bim: a novel member of the Bcl-2 family that promotes apoptosis. EMBO J 1998; 17: 384–395.
Gross A, Yin XM, Wang K, Wei MC, Jockel J, Milliman C et al. Caspase cleaved BID targets mitochondria and is required for cytochrome c release, while BCL-XL prevents this release but not tumor necrosis factor-R1/Fas death. J Biol Chem 1999; 274: 1156–1163.
Li H, Zhu H, Xu CJ, Yuan J . Cleavage of BID by caspase 8 mediates the mitochondrial damage in the Fas pathway of apoptosis. Cell 1998; 94: 491–501.
Langenau DM, Jette C, Berghmans S, Palomero T, Kanki JP, Kutok JL et al. Suppression of apoptosis by bcl-2 overexpression in lymphoid cells of transgenic zebrafish. Blood 2005; 105: 3278–3285.
Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY . Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol 2004; 22: 1567–1572.
Acknowledgements
Funding for this work was provided by V foundation (AMF), and NIH grants RO1CA119066 (ATL), KO1DK0745551 by the National Institute of Diabetes and Digestive and Kidney Diseases (CAJ), 5T32HL07623-20 (UJP), K08CA10254 (AL), P01CA068484 (AL and ATL), and K99NS058608 (RAS). DML is the Safra Foundation Fellow from the Irvington Institute for Immunology. AL is a founder and member of the Scientific Advisory Board for Eutropics Pharmaceuticals.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Edited by RA Knight
Rights and permissions
About this article
Cite this article
Jette, C., Flanagan, A., Ryan, J. et al. BIM and other BCL-2 family proteins exhibit cross-species conservation of function between zebrafish and mammals. Cell Death Differ 15, 1063–1072 (2008). https://doi.org/10.1038/cdd.2008.42
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2008.42
Keywords
This article is cited by
-
BCL-G: 20 years of research on a non-typical protein from the BCL-2 family
Cell Death & Differentiation (2023)
-
Comparative Transcriptome Analysis of Head Kidney of Aeromonas hydrophila-infected Hypoxia-tolerant and Normal Large Yellow Croaker
Marine Biotechnology (2022)
-
Disassembly of dying cells in diverse organisms
Cellular and Molecular Life Sciences (2019)
-
All in one theranostic nanoplatform enables efficient anti-tumor peptide delivery for triple-modal imaging guided cancer therapy
Nano Research (2019)
-
A structural investigation of NRZ mediated apoptosis regulation in zebrafish
Cell Death & Disease (2018)