Abstract
RNA interference (RNAi) has emerged as one of the most powerful tools for functionally characterizing large sets of genomic data. Capabilities of RNAi place it at the forefront of high-throughput screens, which are able to span the human genome in search of novel targets. Although RNAi screens have been used to elucidate pathway components and discover potential drug targets in lower organisms, including Caenorhabditis elegans and Drosophila, only recently has the technology been advanced to a state in which large-scale screens can be performed in mammalian cells. In this review, we will evaluate the major advancements in the field of mammalian RNAi, specifically in terms of high-throughput assays. Crucial points of experimental design will be highlighted, as well as suggestions as to how to interpret and follow-up on potential cell death targets. Finally, we assess the prospective applications of high-throughput screens, the data they are capable of generating, and the potential for this technique to further our understanding of human disease.
Similar content being viewed by others
Main
The sequencing of the human genome ushered a new era into the field of modern biology; it is now possible to elucidate molecular pathways relevant to development and disease with a breadth never before seen. The revelation of the genome has proved to be an invaluable tool for scientists, giving them the ability to functionalize genetic events and to study their relevance on a biochemical level. Large-scale RNA interference (RNAi) screens have the capability of producing genome-wide loss-of-function phenotypes that can place previously uncharacterized mutations into the context of a specific pathway within the cell, and ultimately within the context of human disease. As the majority of diseases arise from not one, but a multitude of mutations, the ability to monitor the global effects of such abnormalities provides an invaluable resource to modern biologists.
Aberrancies associated with the balance between cell death and cell survival account for a large number of known malignancies. Indeed, the ability to evade apoptosis has been labeled one of the hallmarks of cancer,1 and defects in the apoptotic machinery have also been linked to a multitude of illnesses, including neurodegenerative and autoimmune disorders as well as AIDS and other viral infections.2 Recent findings have also implicated ‘non-apoptotic cell death,’ most commonly agreed upon as autophagic or necrotic cell death, as critical in tumorigenesis,3, 4 atherosclerosis,5, 6 diabetes,7, 8 Alzheimer's,9, 10, 11 and Parkinson's diseases.12 It is evident that maintaining the balance between cell survival, cell death, and autophagy represents a crucial regulatory point in human health and that when this balance is tipped, a variety of maladies can arise in many different disease contexts.
Although the specific events encompassing cell death have been uncovered over the past two decades, much still remains unknown with regards to the specific molecular mechanisms determining cell death outcomes, as well as global effects associated with their differential regulation. RNAi has the potential to address these issues in a thorough and efficient manner, and, when applied skillfully, can generate comprehensive data sets that would have never been possible without a high-throughput approach. The technical challenges associated with RNAi itself have largely been addressed in the past few years, allowing this method to be exploited by almost every scientific subset of researchers, so long as they understand the limitations and capabilities associated with each specific approach. As the potential for genome-wide discoveries allows our knowledge to grow exponentially, tremendous effort is now made in utilizing RNAi screens to address prominent, unanswered questions in the field of cell death. Many of the necessary experimental considerations regarding RNAi screening, suggestions for the interpretation and validation of data sets, as well as the scope of the potential knowledge to be gained from such screens are discussed in more detail below.
The Advent of RNAi and Optimization in Model Organisms
RNAi initially took the stage as a potential tool for modulating gene expression in metazoans when Andrew Fire and Craig Mello showed that double-stranded RNA (dsRNA) conferred specific gene silencing in Caenorhabditis elegans.13 The striking phenotype resulting from RNAi was first seen when these organisms were fed long strands of dsRNA, which unexpectedly showed a two-fold higher potency than either sense or antisense single-stranded molecules alone.13 Although techniques for eliminating gene expression in yeast, worms, and invertebrates had been well established for a few years, researchers had few options as to how to easily knock down and target specific genes within mammalian genomes. Knockout mice, although extremely effective in creating whole-organism phenotypes as well as gene-deficient embryonic fibroblasts, took months to years to engineer and breed. Even in its earliest stages, RNAi proved to be more robust than the then current antisense technologies. RNAi-mediated knocked down decreased protein levels more often, more potently, and at concentrations several orders of magnitude lower than antisense.14, 15 The incredible significance of this discovery has not gone overlooked within the scientific community, as both Fire and Mello were awarded the 2006 Nobel Prize in Medicine for their revolutionary finding, less than a decade after the original research was published.
One of the most striking characteristics of dsRNA-mediated gene knockdown is its incredible potency. RNAi is surprisingly robust, as injection of dsRNA into the tail of the animal provided gene silencing throughout the entire organism, with knockdown persisting through the F1 progeny. It was later discovered that knockdown in C. elegans could be induced by a wide variety of mechanisms, including bathing the animals in a solution containing dsRNA or feeding them the RNA directly.16 The ease of delivering dsRNA to C. elegans proved similar to that of transfecting Drosophila embryos17 and invertebrate cultured cell lines,18 allowing both model organisms to be exploited for experimental analysis by multiple groups and subsequent genome-wide RNAi screens.
The success of RNAi-based screens in lower organisms directly correlates to the relative ease of designing appropriate experiments. In addition to the convenience of being able to effectively diffuse dsRNA into the cells of interest, both model organisms (C. elegans and Drosophila) are able to use large strands of dsRNA (up to 2 kb) to mediate gene knockdown. Although originally thought to be the agent responsible for RNAi, these long strands were soon found to be cut into small active species—known as short interfering RNAs (siRNAs)—by an RNase III class riboendonuclease called Dicer.19 The 21- to 23-nucleotide products of this cleavage were similar to those seen in plants undergoing post-transcriptional gene silencing,20 suggesting a conserved mechanism of gene-specific knockdown mediated by dsRNA. Indeed, RNAi has been shown to work in a wide variety of species, including Trypanosomes,21 nematodes,13 mouse oocytes,22 and various mammalian cell lines.15
Perhaps one of the most interesting twists to the advancement of RNAi has been the discovery of an endogenous class of siRNA-like molecules termed microRNAs (miRNAs). A handful of ‘small temporal RNAs’ had been fairly well characterized in the regulation of developmental processes in C. elegans,23, 24 when it was realized that a large class of these 21–25 base-pair (bp) noncoding RNA molecules exist.25 Renamed miRNAs, orthologs of these molecules have been found in numerous species, suggesting an important role within evolution. Indeed, miRNAs have been implicated in the regulation of gene expression, as a recent study suggests the presence of at least 340 sequences constituting unique, mature transcripts in the human genome.26 Each of these sequences is postulated to repress the expression of up to 200 mRNA transcripts, suggesting an enormous regulatory network of endogenous RNAi.
As miRNAs are endogenous and found throughout the genome, their transcription and processing differ from that of siRNA or short hairpin RNA (shRNA) sequences; shRNA molecules are produced ectopically and siRNAs are delivered exogenously to the cell. miRNAs are transcribed by the class II RNA polymerases into an approximately 100 bp structure termed primary miRNA (pri-miRNA), which has significant secondary structure. This secondary structure is recognized by the Drosha–DGCR8 complex, which cleaves the miRNA into its second immature form, a 70 nucleotide hairpin containing a 2 bp overhang on its 3′ end. This ‘pre-miRNA’ is then exported from the nucleus to the cytoplasm, where it is processed by Dicer into its fully active form.
The processing of siRNA duplexes, shRNA, and miRNA converge into a conserved RNAi-modulating pathway at the point of cleavage by Dicer, as siRNA and miRNA moieties utilize the same cellular machinery to target mRNA transcripts. After processing by Dicer, mature siRNAs and miRNAs associate with proteins to create an RNA-induced silencing complex (RISC). The proteins present in RISC vary between species, but a core of proteins including Dicer and the Argonaute protein family seems to be consistently found within the silencing complex.
Although both siRNAs and miRNAs utilize RISC to induce RNAi, the mechanisms in which they abrogate gene expression differ. siRNAs are phosphorylated on their 5′ end and subsequently assembled into RISC, where the guide strand is chosen by the cleavage of the passenger strand by the argonaute family member Ago2. The guide strand is then able to target its exact complementary mRNA sequence within the cell, allowing it to be cleaved by Ago2.27 miRNAs utilize RISC in a slightly different manner: once processed by Dicer, the miRNA duplex is assembled into the silencing complex; the selection of either the 5′ or 3′ arm of the hairpin to be processed and loaded into the active positions of RISC largely determines miRNA function. The antisense strand in siRNAs acts as the guide strand to allow mRNA degradation, whereas miRNA duplexes are almost always asymmetric and thus do not have an ‘antisense’ stretch of nucleotides. Thus, the selection of the guide strand within miRNAs does not recapitulate that of siRNAs because of the inherent base pair mismatches within the molecule. Although the mechanism for this selection is not yet well understood, it is thought that once a strand is selected and loaded into RISC, the other strand is destroyed. The remaining RNA then guides RISC to the 3′ untranslated regions (UTRs) of various mRNA transcripts, leading to the repression of protein expression. As ribosomes on repressed mRNA dissociate more rapidly than those on control transcripts, it has been postulated that the inhibition of gene expression is due to ribosomal drop off during translation.28
Although the mechanism of RNAi is widely conserved, the advancement of this method from lower organisms to mammalian cells had to overcome a major obstacle. In both Drosophila and C. elegans, dsRNA is cleaved by Dicer, resulting in siRNAs that act as a template for degradation of the corresponding mRNA sequence within the cell. When long strands of dsRNA were delivered to mammalian cells, however, the reduction in gene expression seen in model organisms could not be reproduced. It was soon realized that as the long dsRNA molecules entered mammalian cells, they triggered the interferon response, a mechanism thought to be the immune system's defense against viruses that carry their genomes on such dsRNA.29 Tuschl and co-workers15 demonstrated that the direct delivery of 21-bp dsRNA molecules mimicking the cleavage products of Dicer could avoid this response and mediate gene-specific knockdown, enabling the application of RNAi in mammalian cells and, in doing so, provided an easily exploitable tool for biochemical and cellular analysis.
Designing Potent siRNAs
Whether using an siRNA-, shRNA-, or miRNA-based approach to modulate RNAi, optimization of the target sequence is paramount in ensuring the strongest knockdown obtainable, simultaneously limiting the potential promiscuous activity that leads to nonspecific off-target effects. Designing potent siRNAs requires a set of rules that can be applied to efficient and specific siRNA design. A basic familiarity with these rules can simplify the efforts of researchers trying to design or optimize siRNA sequences to knockdown any number of targets.
The first consideration while designing an siRNA sequence should be the nature of the target sequence. Sequences should be chosen to incorporate or exclude splice variants and various isoforms as the assay dictates. Care should be taken to choose sequences within the coding regions of the target sequence, and not in any sections of its intronic genomic sequences, as gene silencing is an exclusively cytoplasmic process.30 Sequences corresponding to the 3′ UTR of mRNA have been demonstrated to have potent activity within certain cell types and can also be an option if targeting highly conserved proteins such as two homologous isoforms, as the 3′ UTR is likely to have less conservation than the coding sequence in these instances.
The most straightforward approach to determine the efficacy of knocking down is to directly test a handful of siRNAs in the cell type of interest or to determine through quantitative PCR or western blotting which molecules give the strongest knockdown. However, this is not practical in a high-throughput format in which thousands of siRNAs will be probed. Thus, the ability to predict the efficacy of the siRNA before actually testing the molecule proves to be an important, if not crucial, tool for high-throughput experiments. In light of this, many groups have devised algorithms to predict both the efficacy and the specificity of RNAi sequences.31, 32, 33 Properties such as thermodynamic values, sequence asymmetry, and polymorphisms that contribute to RNA duplex stability are taken into account within these databases.34
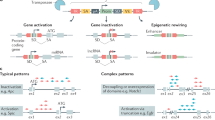
Artificial neural networks have been utilized to train algorithms based on the analysis of randomly selected siRNAs.32 These programs siphon significant trends from large sets of RNA sequences, whose efficacies are known and validated. As shown in Figure 1, certain base pair positions have a tendency to possess distinct nucleotides. In effective sequences, site 1 is preferentially an A or U, and many strands are enriched with these nucleotides along the first 6–7 bps of sequence.34 This is presumed to be due to weak hydrogen bonding that allows RISC to easily unravel the double-stranded duplex and load the guide strand. Site 10 tends to be an A, which is thought to be the conserved RISC cleavage site. Other nucleotides have been overrepresented at distinct sites throughout potent sequences, including (7)=U, (11)=U, (19)=C, and (21)=G. Although the rational for these sequence preferences is unknown, these bp positions probably confer thermodynamic or stability advantages for effective loading and targeting of the complementary mRNA. Table 1 lists a selection of websites containing currently available algorithms that incorporate many of these sequence biases and that can be used for quick and efficient siRNA design.
Conventional versus statistical siRNA design. Standard siRNA design rules originally suggested only 2 bp are conserved at each end of the siRNA, an AA at the 5′ end and TT at the 3′ end. Algorithmic and statistical analyses have shown that the most effective siRNAs have a bias for distinct nucleotides at certain points1, 4, 7, 10 throughout the 21-mer. Base-pair preferences at positions 1(A or U), 2(U), 7(U), 10(A), 11(U), 19(C), and 21(G) are illustrated above
Although the efficacy of various siRNA molecules has been associated with trends within its primary sequence, the physical and chemical properties of any given oligonucleotide also affect its potency within the cell. Statistical analysis of siRNA and miRNA molecules demonstrated a bias toward lower internal stabilities within the 5′ end of the antisense strand.35 As siRNA duplexes contain internal symmetry, these thermodynamic considerations play a large role in determining which siRNA strand is incorporated as the guide strand into RISC. Should the incorrect strand be consistently loaded as the template within RISC, gene silencing will fail to occur on account of the lack of complementarity found within the strands and the subsequent inability to target and form a helix with the target mRNA. It is crucial, therefore, to design siRNAs with the less stable duplex at the 5′ end of the molecule, as this is the end preferentially loaded into RISC. Other various properties of siRNA have been brought to light and should also be taken into consideration when designing oligonucleotides. It has been postulated that an excessive G/C content within a sequence will render it fairly ineffective based solely on its thermodynamics,36 and sequences rich in these base pairs should be avoided when possible. The various properties of standard oligonucleotide design should also be taken into account, such as the reduction of palindromes and repetitive sequences to alleviate potential hairpins and dimers, which could effectively block processing or loading of the siRNA at any step of the silencing process.
The efficacy of an siRNA sequence highly depends on where it hybridizes on its target, and, more importantly, where this sequence lies within the secondary structure of the mRNA targeted for degradation. Although the parameters outlined above can eliminate many of the sequence and thermodynamically unfavorable oligonucleotides, it has been difficult to elucidate a perfect algorithm on account of every mRNA having a unique secondary structure. Until such advanced algorithms are available, we recommend choosing three to four distinct sequences that span the length of the coding sequence of each gene, so as to increase the chances of targeting an mRNA sequence fragment that is easily accessible to the siRNA-loaded RISC. When screened in a high-throughput manner, this can be accomplished by using four distinct siRNA target sequences against a single gene into one well. Although proper controls and procedures must be carried out to appropriately interpret the data, this method will greatly cut back on materials and time while ensuring knockdown of most, if not all gene targets.
Reducing Off-target Effects Associated with Sequence Design
One of the largest concerns within the field of RNAi is that of unintended gene silencing caused by nonspecific mRNA targeting. Such promiscuous activity is usually referred to as ‘off-target effects,’ and although a very real concern, is not considered to be an unreasonable challenge to overcome.37 In 2004, independent investigation of 360 published siRNA sequences found that almost 75% of these oligonucleotides had the potential to have off-target effects.38 We concluded that the use of inappropriate programs such as basic local alignment and search tool (BLAST) to design effective oligos led to a potential abundance of unintended silencing. A precise homologous stretch of 6 or 7 bp is necessary for detection through BLAST,38 eliminating the detection of very closely related sequences of 5–10 bp that could have one or two mismatches, allowing moderate but substantial hybridization to off-target sequences. The use of BLAST in designing siRNAs should be limited to searching for stretches of exact homology between sequences, which are only likely within isoforms of genes or between homologues of genes within different species. Although the use of this database can detect extremely obvious sequence similarities, a portion of applicable alignments will not be detected, rendering BLAST too insensitive to detect less readily foreseen nonspecific targets.
Recent findings have demonstrated that many sequence-associated off-target effects, usually associated with RNAi, are caused not by homology to the coding sequences of other genes, but rather by short stretches of 6–7 bp within the 5′ end of the guide strand.39, 40 These so-called ‘seed regions’ perfectly align with regions in the 3′ UTRs of other mRNA molecules.41 Interestingly, seed regions were originally found within miRNAs. Several groups, therefore, have suggested that many siRNAs that impart off-target effects do so in a manner analogous to that of endogenous miRNAs within a cell. Indeed, analysis a thousand siRNA sequences known to nonspecifically degrade mRNA showed a bias for complementation to 3′ UTR regions within positions 2–7 of their guide strands.40 Such off-target effects were abrogated neither by decreasing the concentration of siRNA used nor by the time of transfection; these effects were seen in both siRNA- and shRNA-based systems within a variety of cell types.39 This implicates that sequence-specific off-target effects can strongly overcome the safeguards usually followed by researchers, and that these sequences should be identified and eliminated from experiments.
Through analysis of the human transcriptome, it has been estimated that approximately 83% of the possible ‘21-mers’ within the coding sequences of the genome are unique.37 This percentage suggests that one out of every five 21 bp siRNA sequences shows some homology compared with one-mRNA transcript. This potential for off-target effects presents itself as a significant barrier to overcome. In light of this, researchers must take care to recapitulate phenotypes observed within an RNAi experiment through repetition (i.e., more than two potent siRNA sequences gives the same phenotype) or through rescue (i.e., the phenotype can be restored by the re-expression of the gene of interest).
Experimental Design: siRNA, shRNA, and miRNA
One of the most significant parameters within an RNAi experiment, whether targeting an individual gene or an entire genome, is the reagent used to carry out the knockdown. Although each agent is capable of mediating knockdown, the decision between the use of siRNA-, shRNA-, or miRNA-based gene targeting is extremely important. To make an informed decision on which experimental approach to employ, researchers must consider the length of the proposed experiment, the cell type to be used, and any time-related constraints within each individual assay. After these parameters have been taken into account, the best available reagent for the question at hand should present itself as a natural choice for experimental use.
As siRNAs have been the most extensively characterized tools of RNAi, we recommend their use in a highly transfectable cell type within a transient setting whenever an experiment will allow. Chemically synthesized siRNAs are readily available from a large number of vendors, and many of these corporations have databases of sequences that have been selected for potency or pre-validated knockdown. The use of oligonucleotides eliminates the need for the time-consuming process of cloning the sequence into a plasmid. Although the delivery of the reagent must be highly optimized and the experiment carried out in a short amount of time, the advantages in utilizing such a system outweigh the potential setbacks, especially in a large screen-based setting.
The delivery method chosen for the RNAi agent in any particular experiment will heavily depend on the cell type being used and the nature of the assay. Groups have had success with lipid-based transfections,42, 43, 44, 45, 46 electroporation,47 and viral transduction48, 49, 50, 51, 52 of both siRNA and shRNAs. We believe that most non-primary and non-neuronal cell types can attain effective uptake of siRNA with a lipid-based or an electroporation-based approach. Lipid-based transfection reagents are the most used means of delivery and have had high success rates, although extensive optimization is needed for each individual cell line. A wide array of lipids designed specifically for the transfection of siRNAs into mammalian cells is now commercially available, and, from our experience, most transformed cell types are amenable to at least one of the lipids with high (>80%) efficiency (NW and JM, unpublished data).
When choosing a reagent for a lipid-based approach, the lipid must be able to efficiently knock down gene expression at relatively dilute concentrations to avoid lipid-mediated toxicity. Furthermore, the delivery of the siRNA must be achieved in a relatively short time, again to avoid toxicity due to the presence of the lipid. We have found that most cell types are transfectable with 1–4 μl of lipid per milliliter of total transfection volume for less than 4 h with one of the commercial lipids, although results vary greatly among cell types and lipids. We highly recommend titrating a handful of lipids at various time points and concentrations to optimize conditions for each cell type being used.
In addition to the temporal advantages associated with the use of siRNAs, the method itself has been very well characterized, which allows for efficient and optimal experimental design and follow-up. The specific amount of siRNA delivered to the cell is much easier to control than from a plasmid- or viral-based system. Assuming a uniform transfection efficiency, siRNAs can often be titrated down into the low nanomolar range and still allow substantial knockdown of the gene of interest, an especially important consideration so as to limit potential off-target effects. Determining the lowest effective dose of siRNA that gives a substantial phenotype minimizes the potential off-target effects caused by an overabundance of oligonucleotide present.
For experiments requiring the use of cultured cells that are especially difficult to transfect, viral plasmids containing shRNA motifs have been engineered with sequences spanning almost the entire genome. Retroviral-,48, 49, 51, 52 adenoviral-,53, 54 and lentiviral- based50, 55 systems have been utilized by numerous groups with a high success rate. As lenti- and adenovirus are capable of infecting both dividing and nondividing cells, transduction of shRNA via these viruses may be the only feasible way to achieve knockdown in extremely slow-growing and nondividing cell types.
Viral plasmids allow the generation of a stable knockdown, as their shRNA-containing genomes integrate into the host cell and thus replicate with each division of the cell. This is an important distinguishing characteristic of a viral-based approach, and is necessary for experiments that require longer than the 5–7 days window of knockdown that the transient siRNAs provide. Researchers must pay particular attention to validating experiments and eliminating the possibilities of off-target effects, which can afflict viral-based systems on account of the relative difficulty in determining how much virus is necessary for sufficient knockdown. The determination of the lowest effective dose using a viral shRNA transduction has yet to be perfected, deeming this method much riskier in terms of potential nonspecific phenotypes. The titration of viral-based shRNAs is much more difficult than that of a single population of oligonucleotides, as it has never precisely been established how many viral integration events are necessary for sufficient knockdown of a target gene. Until a greater understanding of the mechanics of virally transduced knockdown is achieved, researchers should determine the multiplicity of infection of each viral titer so as to normalize the amount of each virus to which cells are exposed. Although this cannot assuredly eliminate all off-target effects, it can get rid of the potential over-infection of any particular viral construct.
It is worth noting that certain groups have attempted to clone shRNA sequences into a plasmid that does not use the viral machinery for entry into cells. Although the plasmid DNA does represent a renewable resource that could be more cost-effective in the long run, the nonviral plasmid will be much more difficult to optimize for transfection and expression. Many cell types that are highly amenable to transfection with siRNAs are extremely difficult to transfect with plasmid-based DNA. For instance, we routinely see transfection efficiencies over 95% with alexaflour-conjugated siRNAs in U2OS osteosarcoma cells, whereas the transfection of a plasmid containing enhanced green fluorescent protein will yield only a quarter of the population fluorescent (NW and JM, unpublished data). In addition to transfectability issues, cloning siRNA sequences into vectors can be challenging, as a single base pair mismatch can render the sequence ineffective. Thus, unless using the viral-based transduction of shRNA plasmids, there is no observable advantage in cloning a sequence capable of modulating RNAi into a plasmid.
Although siRNA- and shRNA-mediated gene knockdown has been the most thoroughly characterized, groups have set out to optimize the sequences with miRNA-like properties, which has been shown to increase their potency and/or specificity. Using miRNA precursors as the backbone for the delivery of hairpins, ‘shRNA-mirs’ contain unique hairpin loops of complementary sense and antisense strands, just as shRNA molecules do. However, these hairpins are flanked by the stem sequences found within miRNAs, providing extensive secondary structure beyond the early-designed shRNA molecules.
shRNA-mirs have been shown to efficiently and specifically target and inhibit gene expression in both transient56 and stable57 settings. In both of these settings, shRNA-mirs produce a more potent gene silencing effect than traditionally designed shRNAs.58, 59 Notably, when shRNA-mir cassettes integrate into the genome as a low-number or even single copy, effective gene silencing is still detected.57 This efficiency is especially important for lowering the potential for off-target effects and mediating sufficient knockdown to observe phenotypes. As these vectors are available in viral vectors, shRNA-mirs could provide an excellent option for those who require either a long-term knockdown or delivery into a difficult to transfect cell type. As libraries have become available spanning the both the human and mouse genomes,59 we suggest one to consider the use of shRNA-mirs in addition to the traditionally designed shRNAs on account of their differences in efficiency and specificity. It is worth noting, however, that shRNA-mirs, like their shRNA-based precursors, failed to show effective knockdown for some genes probed within the context of a Pol II-driven viral-based vector.57 It is plausible that the constructs that failed to effectively silence gene expression were tied up within the long sequence of events necessary for virally mediated shRNA knockdown as mentioned above. Therefore, whenever possible, we recommend utilizing siRNA-based transient transfections whenever an experimental setting will allow.
Choosing a Library for RNAi-based Screens
The identification of the subset of genes to be examined is an important factor in the implementation of a successful screen. Although many genome-wide screens have been carried out in both C. elegans and Drosophila,60 this is made possible by the experimental parameters associated with these screens, such as the ease of delivery and administration of large strands of dsRNA. No group has yet published a screen spanning every single mammalian gene, presumably on account of the need for multiple RNA constructs per gene and all possible splice isoforms of each human gene. Thus, most groups have settled on a particular subset of genes that contains the most relevant target genes for their specific hypothesis.
Currently available libraries usually self-organize into subsets of gene families based on gene ontology. These categories span a wide variety of genes implicated in numerous processes including kinases,44, 45 phosphatases,44, 45, 50 tumor suppressors,50 DNA-modifying enzymes,50 cell cycle components,48 a variety of signaling molecules,48 and miscellaneous molecules implicated in important cellular processes such as biosynthesis, proteolysis, and metabolism.48 Within the past few years, most of these libraries have become available commercially, with companies often allowing a hand-picked selection of various genes of interest as well as offering large pre-organized gene clusters. The number and nature of the genes selected will reflect the individual goals of each screen and should be chosen with the intention to maximize potential results while minimizing irrelevant targets. One must remember, however, to factor in multiple sequences targeting a single gene, as replication of the phenotype with multiple siRNAs is one of the most crucial factors in experimental success.
Of course, when limiting gene families within any RNAi screen, researchers dismiss some level of potential novelty to their findings. When searching to find novel targets within a biochemical pathway or cellular process, we highly recommend keeping the range of siRNA sequences broad and experimentally manageable. However, when looking to answer questions within the context of a specific molecular mechanism, the paring down of a RNAi library to only specifically relevant genes is both practical and efficient. For instance, if a researcher wanted to identify a novel regulator of a RhoGTPase, probing a hand-picked set of human GTPase-activating proteins and guanine nucleotide exchange factors expressed within the relevant cell type would minimize the cost and effort put into a large experiment, yet still retains enough information to make the screen highly pertinent. Customized library design must be painstakingly thorough, preferably beyond the scope of a bioinformatics search, to guarantee a comprehensive gene list. Pre-validated as well as algorithmically derived sequences can be found for siRNAs, shRNAs, and shRNA-mirs, which span the majority of the human genome.
Recent efforts have been made to enzymatically prepare RNAi libraries from cDNA templates.61, 62 Endoribonuclease-prepared siRNAs (esiRNAs) are prepared from digestion of long dsRNA molecules by RNase III or recombinant Dicer, resulting in a pool of small RNAs capable of modulating the RNAi phenotype. Utilizing esiRNAs recapitulates the ability of researchers studying model organisms to administer a long dsRNA transcript that correlates with an entire gene of interest to modulate the potent and specific knockdown seen in both C. elegans and Drosophila. It has been suggested that the use of esiRNAs dramatically reduces the off-target effects seen with many siRNA sequences; one report demonstrates a 13-fold reduction in unintended gene silencing.63 As the software to predict effective esiRNA pools as well as many primer pairs generated by this software become publicly available, this could present an affordable method for acquiring RNAi-modulating sequences. It is worth noting, however, that utilizing a pool of various transcripts still presents the potential for off-target effects and that any given esiRNA will contain a handful of sequences that could modulate off-target effects. Once again, this stresses the importance of validation of all RNAi work through repetition and rescue.
Determining Hits, Follow-up, and Validation
After the selection of the specific techniques to be employed and a subset of genes have been chosen to screen, one must determine how to quantify potential hits associated with the phenotype of interest. The design of a reporter assay or molecular sensor to distinguish positive results can prove to be the most challenging of the steps in a successful RNAi screen; the method must be highly reproducible while giving the smallest amount of assay noise so as to minimize standard error and allow even minor hits to be revealed with a decent amount of sensitivity. The actual techniques chosen for this screen will depend largely on the question being probed; however, many considerations must be taken for all screens in order for them to be successful.
The most straightforward screens carried out thus far have utilized well-established experimental methods whose relative ease has allowed the rapid identification of a manageable number of hits from a large pool of genes. The plausibility of such methods will vary from screen to screen, but a number of groups that have explored large cellular processes have exploited experimental methods that are easily translated into a large-scale setting, such as fluorescence-based cell-cycle analysis, DNA-fragmentation ELISAs for apoptosis, and soft agar assays for transformation. Such experiments fall into a broad list of generalizations needed for successful assays. First, the output must be robust so as to diminish background and assay noise. Second, the output must be reproducible to identify potential hits accurately. Third, the assay must be sensitive enough to detect genuine changes in the phenotype caused by RNAi without being masked by assay noise. And finally, the experimentation must be relatively inexpensive and easily expandable to a large number of samples so as to be amenable to a large-scale screen.
Table 2 illustrates a large number of previously examined phenotypes and a handful of cell-based assays chosen to quantify changes associated with RNAi knockdown, as well as some ideas for assays that have not yet been used experimentally. All of these screening methods can be tailored to explore a large number of well-characterized processes in different cell types. Although these easily exploitable experimental methods have allowed a relatively straight-forward way to screen RNAi libraries, there exist phenotypes that require a more specialized and innovative method for detection. Certain groups have overcome this challenge by creating specialized assays to probe a specific phenotype and whose ideas can be used as a springboard for a countless number of potential screening assays that have yet to be imagined. The most promising methods thus far have been based on fluorescence, which has application in antibody-based detections, GFP-based reporter assays, and fluorescence-assisted cell sorting (FACS) techniques. In addition to the potential for high-throughput applications in FACS, which can quickly process hundreds of thousands of cells, automated fluorescence microscopy has shown great promise for the quantitation of both static and dynamic processes. Individual phenotypes can be fixed and numerically measured through software optimized to detect differences in fluorescence, or time-lapse microscopy can monitor the process as it occurs, demonstrating the activity of the process and thus allowing the observation of time as an additional parameter to be analyzed. As the construction of GFP-based reporter genes is very economical, many groups have turned to this methodology as a reliable and reproducible determination of RNAi-mediated phenotypes.
A large-scale RNAi screen will deliver a certain number of positive hits, but the nature and quality of these hits will largely depend on the cut-off stringency and statistical relevance. The numerical determination of the cut-off value has proven to be one of the more controversial aspects of screening, but it is usually accepted that a hit will fall more than three standard deviations above or below the mean value of the assay reporting the phenotype. In addition to the quantification of the primary assay, most groups employ secondary and even tertiary screens to validate the hits from the primary assay. Follow-up cell-based assays must be used as a method of validation and confirmation of knockdown, and the associated phenotype must be completed for a subset of genes pulled out in the screen.
When screening for novel modulators of a particular process, such as apoptosis, it is almost always necessary to validate the cell death assay by demonstrating that well-known components of the process behave as expected. For instance, when looking for novel negative modulators of the PI3K/AKT pathway, a crucial step in the validation of the assay would be the demonstration that PTEN behaves in the same manner as your expected RNAi hits. This should be carried out with a handful of established genes. It is worth noting, however, that although positive hits will, with enough stringency, provide a rather convincing set of data, the same cannot be said for the interpretation of negative results. There are many points within the experiment that expected results could be skewed, from mechanical error due to transfection or screening methods to insufficient knockdown to mediate the expected phenotype. All of these potential errors must be considered when interpreting negative results, and, unless each siRNA is followed up individually, each hit must sit as an unknown rather than a conclusive negative.
Overall, two methods should be utilized to follow up any hits within a RNAi screen so as to validate that the phenotype of interest is a product of specific gene knockdown and not simply an off-target effect. The first of these is the demonstration of the phenotype with multiple siRNA sequences to the target of interest. The demonstration that two, or preferably three, unique sequences that modulate sufficient knockdown in the cell type of interest give the same phenotype is almost indisputable evidence that the knockdown and phenotype correlate strongly. There is very little chance of off-target effects taking on the same phenotype in multiple samples, so this can be a very good way to follow up observations seen originally with one siRNA sequence.
The other unequivocal validation method is that of rescue. There are two main strategies for the rescue of a knockdown phenotype, the first of which involves siRNAs that are targeted to the 3′UTR of the gene of interest. The 3′UTR siRNA will only affect the endogenous expression of the gene of interest, allowing an ectopic plasmid to be translated without degradation, thus allowing a phenotype to be reversed. The challenge with this approach is finding siRNAs targeted to the 3′UTR that modulate sufficient knockdown. We have observed that most of these siRNAs give only a partial knockdown, averaging between 30 and 60% (NW and JM, unpublished data). Thus, if a 3′UTR cannot give a sufficient knockdown to recapitulate the phenotype of interest, researchers can make a silently mutated version of their gene for ectopic expression within cells. We have found that the mutation of five base pairs along the stretch of the 21-mer to which the siRNA corresponds is sufficient to allow expression of a plasmid containing a gene of interest in up to 100 nM of siRNA. The base pairs chosen to mutate were those at wobble positions of the codons within the sequence, and should, if possible, correlate with the conserved residues known to be important for silencing (as shown in Figure 1), as much as possible.
The validation of hits within a screen requires the demonstration that the phenotypes observed were neither false-positives, false-negatives, or off-target effects. Through validation by either repetition or rescue, researchers are able to show that the hits they observe are truly caused only by the knockdown of the specific gene of interest. As RNAi-based screens are a large investment both monetarily and temporally, solid follow-up of the hits found stands as a crucial step in the success of any genome-wide assay.
RNAi Screening and Human Disease
Many groups have exploited high-throughput RNAi to focus on issues regarding human disease, such as cancer. The strategies employed to carry out these screens, as well as the fundamental questions addressed, vary widely from study to study, as well as between organisms, yet many of the experiments already carried out have implicated genes in novel molecular pathways and cell death processes. These identifications have greatly increased our knowledge of the fundamental workings in the cell, which can be applied to better understanding and treating diseases with dysregulated cell death.
The advent of genome-wide RNAi screens in model organisms coincided almost simultaneously with the completion of the sequencing of their respective genomes (Figure 2). With the capability for simple design and delivery of siRNAs, large-scale screens were completed well before those in mammalian cell lines. Although the experimental techniques inherently differ from those used with the mammalian cell, the data sets generated from these organisms still shed light on fairly complex processes in evolutionarily conserved pathways in a surprising amount of detail. As well as identifying potential homologues, the screens provide a basis for the fundamentals of screen-based assays, and many of the techniques can be translated into useful methods in higher organisms.
The first large-scale screens in model organisms characterized the phenotypic knockdown of genes within an entire C. elegans chromosome, chromosome III64 and chromosome I.65 Less than three years later, a truly genome-wide loss-of-function screen had been carried out in which about 1800 mutant phenotypes were characterized, two-thirds of which were novel.60 These screens sought to identify interesting open reading frames within the nematode genome and place them into a cellular process with which they had not been previously associated. A small percentage of the genes identified in these screens have homologues in humans that have been implicated in human disease, demonstrating the power of such a method in identifying potentially conserved homologues involved in the etiology of disease. Homologues to both APC and TSC1 were identified as growth-defective mutant phenotypes in the nematode, and importantly, both genes are characterized as tumor suppressors in mammals.66, 67 This identification shows the exciting potential of RNAi screening in lower organisms.
While many RNAi screens have used model organisms to distinguish novel gene products and place them in the context of a broad cellular process, many groups have exploited the simplicity of model organisms to illuminate previously unidentified regulators of well-characterized cellular pathways and processes. Proteins implicated in the regulation of the DNA damage response,68 cell-cycle progression,69 and migration70 have been discovered, as well as novel constituents of the Hedgehog71 and Wnt17 pathways. Such identifications will surely be explored within the context of mammalian signaling, and could provide the identification of a homologue in a context that has not yet been discovered or fully appreciated.
While model organisms can be useful for the identification of genes associated with human disease, the complexities associated with genetic aberrancies found in mammals often cannot be recapitulated through the simplicity of a lower organism. In these instances, only screens in selected cell types can provide a greater insight to the intricacies of mammalian cell biology. The first screen carried out in a mammalian cell probed a library of siRNAs against selected targets in the human genome to identify regulators of TRAIL-induced apoptosis.42 Within two years, RNAi based screens had identified multiple tumor suppressors,49, 52 as well as kinases and phosphatases implicated in cell survival and chemoresistance.44 These studies show the vast potential granted by large-scale RNAi screens in target identification and develop of new therapeutic targets. Importantly, the identification of genes implicated in apoptosis, cell survival, and tumor suppression allows us to understand the molecular pathways in which they act. Not to be overlooked, the identification of genes implicated in chemoresistance provides the opportunity to understand the therapeutic hurdle and design novel methods to overcome such difficulties.
Conclusions
Although most screens carried out thus far have focused on the etiology of cancer, the door for increased understanding of other diseases has been opened wide. RNAi-based screens have already identified novel targets for a handful of diseases, including Alzheimer's,72 metabolic syndrome,73 and, most notably, cancer.74 This information will allow researchers to better understand the molecular basis of these diseases, which, in time, will help to identify new and improved therapeutic targets. As the regulation of cell death processes is crucial in many elements of human disease, we believe that RNAi has the potential to address questions associated with these processes in a high-throughput fashion. Although RNAi screens have been useful in identifying proteins that could be implicated in certain facets of cell death, a handful of tumor suppressors for instance, the field has not yet utilized this technology to its full capacity in the context of studying cell death processes. Very little is known about many proteins which play a role in caspase-independent cell death and even the fairly well characterized process of apoptosis. Although certain global approaches have been used to study these processes, the study of cell death processes on a large scale has lagged. siRNA-based screens allow researchers to address questions in a global context, and, in doing so, can elucidate much more about the function of a specific gene than in a more focused manner. We believe that RNAi has the potential to address almost any question within the field of apoptosis, with results that will be able to steer researchers in new and exciting directions. Hopefully, within a few years, many will have used this powerful new technique to unlock the secrets still hidden in the processes of cell death and use this knowledge to better understand and treat human disease.
Abbreviations
- BLAST:
-
basic local alignment and search tool
- bp:
-
base pair
- dsRNA:
-
double-stranded RNA
- esiRNA:
-
endoribonuclease-prepared siRNA
- FACS:
-
fluorescence assisted cell sorting
- GEF:
-
guanine nucleotide exchange factor
- miRNA:
-
microRNA
- RISC:
-
RNA-induced silencing complex
- RNAi:
-
RNA interference
- shRNA:
-
short hairpin RNA
- siRNA:
-
short interfering RNA
- UTR:
-
untranslated region
References
Hanahan D, Weinberg RA . The hallmarks of cancer. Cell 2000; 100: 57–70.
Thompson CB . Apoptosis in the pathogenesis and treatment of disease. Science 1995; 267: 1456–1462.
Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, Chen G et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell 2006; 10: 51–64.
Jin S . p53, autophagy and tumor suppression. Autophagy 2005; 1: 171–173.
Jia G, Cheng G, Agrawal DK . Autophagy of vascular smooth muscle cells in atherosclerotic lesions. Autophagy 2007; 3: 63–64.
Mistiaen WP, Somers P, Knaapen MW, Kockx MM . Autophagy as mechanism for cell death in degenerative aortic valve disease. Autophagy 2006; 2: 221–223.
Castino R, Isidoro C, Murphy D . Autophagy-dependent cell survival and cell death in an autosomal dominant familial neurohypophyseal diabetes insipidus in vitro model. FASEB J 2005; 19: 1024–1026.
Kaniuk NA, Kiraly M, Bates H, Vranic M, Volchuk A, Brumell JH . Ubiquitinated-protein aggregates form in pancreatic beta-cells during diabetes-induced oxidative stress and are regulated by autophagy. Diabetes 2007; 56: 930–939.
Zheng L, Roberg K, Jerhammar F, Marcusson J, Terman A . Oxidative stress induces intralysosomal accumulation of Alzheimer amyloid beta-protein in cultured neuroblastoma cells. Ann NY Acad Sci 2006; 1067: 248–251.
Zheng L, Marcusson J, Terman A . Oxidative stress and Alzheimer disease: the autophagy connection? Autophagy 2006; 2: 143–145.
Zhu X, Raina AK, Perry G, Smith MA . Apoptosis in Alzheimer disease: a mathematical improbability. Curr Alzheimer Res 2006; 3: 393–396.
Webb JL, Ravikumar B, Atkins J, Skepper JN, Rubinsztein DC . Alpha-Synuclein is degraded by both autophagy and the proteasome. J Biol Chem 2003; 278: 25009–25013.
Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC . Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998; 391: 806–811.
Bass BL . RNA interference. The short answer. Nature 2001; 411: 428–429.
Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T . Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 2001; 411: 494–498.
Timmons L, Fire A . Specific interference by ingested dsRNA. Nature 1998; 395: 854.
Kennerdell JR, Carthew RW . Use of dsRNA-mediated genetic interference to demonstrate that frizzled and frizzled 2 act in the wingless pathway. Cell 1998; 95: 1017–1026.
Clemens JC, Worby CA, Simonson-Leff N, Muda M, Maehama T, Hemmings BA et al. Use of double-stranded RNA interference in Drosophila cell lines to dissect signal transduction pathways. Proc Natl Acad Sci USA 2000; 97: 6499–6503.
Bernstein E, Caudy AA, Hammond SM, Hannon GJ . Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 2001; 409: 363–366.
Hamilton AJ, Baulcombe DC . A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 1999; 286: 950–952.
Djikeng A, Shi H, Tschudi C, Ullu E . RNA interference in Trypanosoma brucei: cloning of small interfering RNAs provides evidence for retroposon-derived 24–26-nucleotide RNAs. RNA 2001; 7: 1522–1530.
Wianny F, Zernicka-Goetz M . Specific interference with gene function by double-stranded RNA in early mouse development. Nat Cell Biol 2000; 2: 70–75.
Lee RC, Feinbaum RL, Ambros V . The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993; 75: 843–854.
Wightman B, Ha I, Ruvkun G . Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 1993; 75: 855–862.
Lau NC, Lim LP, Weinstein EG, Bartel DP . An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 2001; 294: 858–862.
Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 2007; 129: 1401–1414.
Rana TM . Illuminating the silence: understanding the structure and function of small RNAs. Nat Rev Mol Cell Biol 2007; 8: 23–36.
Petersen CP, Bordeleau ME, Pelletier J, Sharp PA . Short RNAs repress translation after initiation in mammalian cells. Mol Cell 2006; 21: 533–542.
Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD . How cells respond to interferons. Annu Rev Biochem 1998; 67: 227–264.
Martinez J, Patkaniowska A, Urlaub H, Luhrmann R, Tuschl T . Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell 2002; 110: 563–574.
Boese Q, Leake D, Reynolds A, Read S, Scaringe SA, Marshall WS et al. Mechanistic insights aid computational short interfering RNA design. Methods Enzymol 2005; 392: 73–96.
Huesken D, Lange J, Mickanin C, Weiler J, Asselbergs F, Warner J et al. Design of a genome-wide siRNA library using an artificial neural network. Nat Biotechnol 2005; 23: 995–1001.
Yuan B, Latek R, Hossbach M, Tuschl T, Lewitter F . siRNA Selection Server: an automated siRNA oligonucleotide prediction server. Nucleic Acids Res 2004; 32 (Web Server issue): W130–W134.
Pei Y, Tuschl T . On the art of identifying effective and specific siRNAs. Nat Methods 2006; 3: 670–676.
Khvorova A, Reynolds A, Jayasena SD . Functional siRNAs and miRNAs exhibit strand bias. Cell 2003; 115: 209–216.
Reynolds A, Leake D, Boese Q, Scaringe S, Marshall WS, Khvorova A . Rational siRNA design for RNA interference. Nat Biotechnol 2004; 22: 326–330.
Qiu S, Adema CM, Lane T . A computational study of off-target effects of RNA interference. Nucleic Acids Res 2005; 33: 1834–1847.
Snove Jr O, Holen T . Many commonly used siRNAs risk off-target activity. Biochem Biophys Res Commun 2004; 319: 256–263.
Jackson AL, Bartz SR, Schelter J, Kobayashi SV, Burchard J, Mao M et al. Expression profiling reveals off-target gene regulation by RNAi. Nat Biotechnol 2003; 21: 635–637.
Jackson AL, Burchard J, Schelter J, Chau BN, Cleary M, Lim L et al. Widespread siRNA ‘off-target’ transcript silencing mediated by seed region sequence complementarity. RNA 2006; 12: 1179–1187.
Birmingham A, Anderson EM, Reynolds A, Ilsley-Tyree D, Leake D, Fedorov Y et al. 3′ UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat Methods 2006; 3: 199–204.
Aza-Blanc P, Cooper CL, Wagner K, Batalov S, Deveraux QL, Cooke MP . Identification of modulators of TRAIL-induced apoptosis via RNAi-based phenotypic screening. Mol Cell 2003; 12: 627–637.
Li S, Wang L, Berman MA, Zhang Y, Dorf ME . RNAi screen in mouse astrocytes identifies phosphatases that regulate NF-kappaB signaling. Mol Cell 2006; 24: 497–509.
MacKeigan JP, Murphy LO, Blenis J . Sensitized RNAi screen of human kinases and phosphatases identifies new regulators of apoptosis and chemoresistance. Nat Cell Biol 2005; 7: 591–600.
Paddison PJ, Silva JM, Conklin DS, Schlabach M, Li M, Aruleba S et al. A resource for large-scale RNA-interference-based screens in mammals. Nature 2004; 428: 427–431.
Pelkmans L, Fava E, Grabner H, Hannus M, Habermann B, Krausz E et al. Genome-wide analysis of human kinases in clathrin- and caveolae/raft-mediated endocytosis. Nature 2005; 436: 78–86.
Brummelkamp TR, Nijman SM, Dirac AM, Bernards R . Loss of the cylindromatosis tumour suppressor inhibits apoptosis by activating NF-kappaB. Nature 2003; 424: 797–801.
Berns K, Hijmans EM, Mullenders J, Brummelkamp TR, Velds A, Heimerikx M et al. A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature 2004; 428: 431–437.
Kolfschoten IG, van Leeuwen B, Berns K, Mullenders J, Beijersbergen RL, Bernards R et al. A genetic screen identifies PITX1 as a suppressor of RAS activity and tumorigenicity. Cell 2005; 121: 849–858.
Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell 2006; 124: 1283–1298.
Ngo VN, Davis RE, Lamy L, Yu X, Zhao H, Lenz G et al. A loss-of-function RNA interference screen for molecular targets in cancer. Nature 2006; 441: 106–110.
Westbrook TF, Martin ES, Schlabach MR, Leng Y, Liang AC, Feng B et al. A genetic screen for candidate tumor suppressors identifies REST. Cell 2005; 121: 837–848.
Subramanian T, Chinnadurai G . Temperature-sensitive replication-competent adenovirus shRNA vectors to study cellular genes in virus-induced apoptosis. Methods Mol Med 2006; 130: 125–134.
Cao HB, Wang A, Martin B, Koehler DR, Zeitlin PL, Tanawell AK et al. Down-regulation of IL-8 expression in human airway epithelial cells through helper-dependent adenoviral-mediated RNA interference. Cell Res 2005; 15: 111–119.
Golding MC, Long CR, Carmell MA, Hannon GJ, Westhusin ME . Suppression of prion protein in livestock by RNA interference. Proc Natl Acad Sci USA 2006; 103: 5285–5290.
Zeng Y, Wagner EJ, Cullen BR . Both natural and designed micro RNAs can inhibit the expression of cognate mRNAs when expressed in human cells. Mol Cell 2002; 9: 1327–1333.
Dickins RA, Hemann MT, Zilfou JT, Simpson DR, Ibarra I, Hannon GJ et al. Probing tumor phenotypes using stable and regulated synthetic microRNA precursors. Nat Genet 2005; 37: 1289–1295.
Boden D, Pusch O, Silbermann R, Lee F, Tucker L, Ramratnam B . Enhanced gene silencing of HIV-1 specific siRNA using microRNA designed hairpins. Nucleic Acids Res 2004; 32: 1154–1158.
Silva JM, Li MZ, Chang K, Ge W, Golding MC, Rickles RJ et al. Second-generation shRNA libraries covering the mouse and human genomes. Nat Genet 2005; 37: 1281–1288.
Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 2003; 421: 231–237.
Buchholz F, Kittler R, Slabicki M, Theis M . Enzymatically prepared RNAi libraries. Nat Methods 2006; 3: 696–700.
Shirane D, Sugao K, Namiki S, Tanabe M, Iino M, Hirose K . Enzymatic production of RNAi libraries from cDNAs. Nat Genet 2004; 36: 190–196.
Kittler R, Surendranath V, Heninger AK, Slabicki M, Theis M, Putz G et al. Genome-wide resources of endoribonuclease-prepared short interfering RNAs for specific loss-of-function studies. Nat Methods 2007; 4: 337–344.
Gonczy P, Echeverri C, Oegema K, Coulson A, Jones SJ, Copley RR et al. Functional genomic analysis of cell division in C. elegans using RNAi of genes on chromosome III. Nature 2000; 408: 331–336.
Fraser AG, Kamath RS, Zipperlen P, Martinez-Campos M, Sohrmann M, Ahringer J . Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature 2000; 408: 325–330.
Yeung RS, Xiao GH, Everitt JI, Jin F, Walker CL . Allelic loss at the tuberous sclerosis 2 locus in spontaneous tumors in the Eker rat. Mol Carcinog 1995; 14: 28–36.
Powell SM, Zilz N, Beazer-Barclay Y, Bryan TM, Hamilton SR, Thibodeau SN et al. APC mutations occur early during colorectal tumorigenesis. Nature 1992; 359: 235–237.
van Haaften G, Romeijn R, Pothof J, Koole W, Mullenders LH, Pastink A et al. Identification of conserved pathways of DNA-damage response and radiation protection by genome-wide RNAi. Curr Biol 2006; 16: 1344–1350.
Bjorklund M, Taipale M, Varjosalo M, Saharinen J, Lahdenpera J, Taipale J . Identification of pathways regulating cell size and cell-cycle progression by RNAi. Nature 2006; 439: 1009–1013.
Cram EJ, Shang H, Schwarzbauer JE . A systematic RNA interference screen reveals a cell migration gene network in C. elegans. J Cell Sci 2006; 119 (Part 23): 4811–4818.
Lum L, Yao S, Mozer B, Rovescalli A, Von Kessler D, Nirenberg M et al. Identification of Hedgehog pathway components by RNAi in Drosophila cultured cells. Science 2003; 299: 2039–2045.
Yang JP, Fan W, Rogers C, Chatterton JE, Bliesath J, Liu G et al. A novel RNAi library based on partially randomized consensus sequences of nuclear receptors: identifying the receptors involved in amyloid beta degradation. Genomics 2006; 88: 282–292.
Tang X, Guilherme A, Chakladar A, Powelka AM, Konda S, Virbasius JV et al. An RNA interference-based screen identifies MAP4K4/NIK as a negative regulator of PPARgamma, adipogenesis, and insulin-responsive hexose transport. Proc Natl Acad Sci USA 2006; 103: 2087–2092.
Bartz S, Jackson AL . How will RNAi facilitate drug development? Sci STKE 2005: pe39.
Henschel A, Buchholz F, Habermann B . DEQOR: a web-based tool for the design and quality control of siRNAs. Nucleic Acids Res 2004; 32: W113–W120.
Jiang P, Wu H, Da Y, Sang F, Wei J, Sun X et al. RFRCDB-siRNA: improved design of siRNAs by random forest regression model coupled with database searching. Comput Methods Programs Biomed 2007; 87: 230–238.
Yamada T, Morishita S . Accelerated off-target search algorithm for siRNA. Bioinformatics 2005; 21: 1316–1324.
Naito Y, Yamada T, Ui-Tei K, Morishita S, Saigo K . siDirect: highly effective, target-specific siRNA design software for mammalian RNA interference. Nucleic Acids Res 2004; 32: W124–W129.
Ding Y, Chan CY, Lawrence CE . Sfold web server for statistical folding and rational design of nucleic acids. Nucleic Acids Res 2004; 32: W135–W141.
Friedman A, Perrimon N . Genome-wide high-throughput screens in functional genomics. Curr Opin Genet Dev 2004; 14: 470–476.
Boutros M, Kiger AA, Armknecht S, Kerr K, Hild M, Koch B et al. Genome-wide RNAi analysis of growth and viability in Drosophila cells. Science 2004; 303: 832–835.
Kiger AA, Baum B, Jones S, Jones MR, Coulson A, Echeverri C et al. A functional genomic analysis of cell morphology using RNA interference. J Biol 2003; 2: 27.
Nollen EA, Garcia SM, van Haaften G, Kim S, Chavez A, Morimoto RI et al. Genome-wide RNA interference screen identifies previously undescribed regulators of polyglutamine aggregation. Proc Natl Acad Sci USA 2004; 101: 6403–6408.
Author information
Authors and Affiliations
Corresponding author
Additional information
Edited by RA Knight
Rights and permissions
About this article
Cite this article
Wolters, N., MacKeigan, J. From sequence to function: using RNAi to elucidate mechanisms of human disease. Cell Death Differ 15, 809–819 (2008). https://doi.org/10.1038/sj.cdd.4402311
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.cdd.4402311