Abstract
The Epstein–Barr and Kaposi's sarcoma γ-herpesviruses (KSHVs) are associated with certain cancers, and encode B-cell leukemia/lymphoma 2 (BCL-2) homologs, BHRF-1 and KSHV BCL-2, respectively. Little is known, however, about the molecular interactions allowing viral BCL-2 homologs to mediate their anti-apoptotic function. Cellular anti-apoptotic proteins, such as BCL-2 and MCL-1, prevent death via selective interactions with pro-death BH3-only proteins. To investigate whether BHRF-1 and KSHV BCL-2 function similarly, we made recombinant BHRF-1 and KSHV BCL-2 proteins. We identified the individual binding patterns for BHRF-1 and KSHV BCL-2 to BH3 domains. These studies surprisingly showed that KSHV BCL-2 is more closely related to MCL-1 than to BCL-2, a result confirmed by sequence analysis. GST-BHRF-1 and GST-KSHV BCL-2 bound BH3-only family proteins from human cells. BHRF-1 protected mammalian cells from growth factor withdrawal, etoposide and adriamycin. We found that both BCL-2 and BHRF-1 sequestered pro-death BH3-only proteins under growth factor-deficient conditions. Finally, we tested the ability of a panel of BH3 peptides to inhibit BHRF-1 and KSHV BCL-2 function in a mitochondrial model of apoptosis. We found that each could be inhibited by the select group of BH3 peptides identified in our binding assay. Our studies define the biochemical interactions underlying BHRF-1 and KSHV BCL-2 anti-apoptotic function, and identify peptides that are prototypic inhibitors of this function.
Similar content being viewed by others
Main
The intrinsic, or mitochondrial, pathway of apoptosis is controlled by the B-cell leukemia/lymphoma 2 (BCL-2) family of proteins. This family is comprised of both pro- and anti-apoptotic proteins, which share homology in conserved BCL-2 homology (BH) regions.1 The anti-apoptotic proteins, such as BCL-2, BCL-XL and MCL-1, share homology in BH regions 1–4. The pro-apoptotic family members on the other hand, can be subdivided into ‘multi-domain’ proteins, such as BAX and BAK, which share homology in BH regions 1–3, and ‘BH3-only’ proteins, such as BIM, BID, BAD and NOXA. BH3-only proteins share homology only in the amphipathic α-helical BH3 region, which is required by pro-apoptotic family members for their pro-death function. The BH3-only proteins can be further subdivided into ‘activators’ and ‘sensitizers’.2 Activators, such as BIM and BID, induce BAX and BAK oligomerization resulting in mitochondrial outer membrane permeabilization (MOMP), release of cytochrome c and commitment to programmed cell death. To counteract this, anti-apoptotic proteins inhibit BAX and BAK activation and MOMP by sequestering activator BH3 domains such as BIM and BID, and perhaps also by sequestering activated forms of BAX and BAK. Sensitizers such as BAD and NOXA do not interact directly with BAX or BAK, but rather occupy the inhibitory pocket of anti-apoptotic BCL-2 family members, such as BCL-2 and MCL-1, displacing activator BH3 domains.2 It has been determined that mammalian orthologs of BCL-2 (including MCL-1, BCL-XL, BCL-w and BFL-1) each have a distinct pattern of binding of BH3 domains from pro-apoptotic BH3-only family members.2, 3, 4, 5, 6 It has further been demonstrated that this pattern of binding relates directly to the ability of these BH3 domains to antagonize the anti-apoptotic protection afforded by BCL-2 and orthologs.
Many γ-herpesviruses and a few other herpesviruses encode at least one homolog of BCL-2.7 Epstein–Barr virus (EBV) was discovered first in association with Burkitt's lymphoma and more recently has been found to be associated with Hodgkin's disease, T-cell lymphoma, nasopharyngeal carcinoma, and post-transplant lymphoproliferative disease.8, 9 Patients receiving immuno-suppressive anti-cancer therapy and individuals with HIV have an increased susceptibility to develop EBV-associated lymphomas. EBV encodes a viral homolog of BCL-2, anti-apoptotic BHRF-1,10 which has conserved BH1 and BH2 domains homologous to BCL-2.11 BHRF-1 can inhibit apoptosis induced by a number of death insults including serum depletion,12 death induced by tumor necrosis factor α and anti-Fas antibody,13, 14 γ irradiation, chemotherapeutic drugs,15, 16 deregulated c-myc,17 granzyme B,18 Sindbis virus infection19 and the tumor suppressor protein p53.20 BHRF-1 is expressed during the productive/lytic replication cycle.21, 22 It has been widely hypothesized that BHRF-1 may function in EBV infections to promote viral replication by maintaining survival of the host cell.
Like EBV, human herpesvirus 8, commonly called Kaposi's sarcoma herpesvirus (KSHV), a γ-herpesvirus, is associated with the development of human cancers. KSHV was first identified in Kaposi's sarcoma (KS) skin lesions of AIDS patients.23 Infection with KSHV is a significant risk factor in developing KS, primary effusion lymphomas, HIV-related lymphomas and Castleman's disease.24 KSHV BCL-2 shares homology in BH1 and BH2 domains with BCL-2.25 Like the EBV counterpart BHRF-1, KSHV BCL-2 is expressed early in the lytic replication cycle and can inhibit apoptosis induced by several stimuli, including BAX overexpression, viral cyclin overexpression and Sindbis virus infection.26, 27, 28 However, it is not known whether KSHV BCL-2 expression is required for development and maintenance of KS, or if KSHV BCL-2 is required for maintaining cell viability during lytic cycle replication. Of critical clinical importance, but equally obscure, is whether the function of BHRF-1 and KSHV BCL-2 is needed for the maintenance of tumors in which they are expressed.
Homologs of BCL-2 are conserved in species ranging from Caenorhabditis elegans to humans. Viruses seem to have adapted the mechanism of inhibiting cell death in the host through viral BCL-2 homologs, promoting their own survival in the host. BCL-2 homologs encoded by α- and γ-herpesvirus and the African swine fever virus share 20–30% homology with one another and cellular BCL-2.7 All viral homologs contain a BH1 sequence motif. The BH2 motif is also conserved in the almost all viral BCL-2 homologs except for murine γ-herpesvirus 68 (γHV68).7 Both BH1 and BH2 sequence motifs are critical for BH3 domain binding and the death repressor effects of the anti-apoptotic proteins.29 The BH3 domain, thought to be important for killing cells, is poorly conserved in the viral homologs. However, there is a similar pattern of bulky hydrophobic and charged residues in the BH3 domain of the viral homologs as in the cellular proteins. There is very little homology shared among the viral BCL-2 homologs in the BH4 domain, which is also true of the cellular proteins.7
Certain cellular anti-apoptotic proteins like BCL-2 are converted to pro-apoptotic proteins via caspase cleavage, likely as a positive feedback loop to accelerate apoptosis.26, 30 Most viral BCL-2 homologs cannot be converted into pro-apoptotic proteins because they are not cleavable by proteases or their C termini lack pro-apoptotic activity.31 Thus, viral BCL-2 proteins escape cellular control mechanisms.
An understanding of how BCL-2 proteins function to inhibit apoptosis and promote the virus life cycle necessitates the examination of how host BCL-2 family proteins regulate cell death. We examined the BH3 domain-binding properties of BHRF-1 and KSHV BCL-2, and found that, like their cellular homologs, they exhibit a selective interaction pattern. BHRF-1 and KSHV BCL-2 bind pro-apoptotic BH3-only proteins in cellular models. Their function can be inhibited by select BH3 domain peptides.
Results
BHRF-1 and KSHV show distinct patterns of interaction with BH3 domain peptides
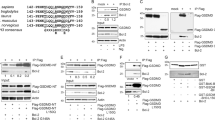
To investigate interactions of viral BCL-2 homologs, GST-BHRF-1 and GST-KSHV BCL-2 proteins were expressed and purified. The C terminus of both BHRF-1 and KSHV BCL-2 was truncated by removing the last 27 amino acids of BHRF-1 and 33 amino acids from KSHV BCL-2 (Figure 1a). These GST fusion proteins were characterized by size; BHRF-1, 46.5 kDa, and KSHV BCL-2, 45.2 kDa on a Coomassie-stained gel, and identified using anti-GST (Z-5; Santa Cruz) antibodies (Figure 1b). GST fusions with full-length KSHV BCL-2 or BHRF-1 were insoluble in aqueous buffers.
BHRF-1 and KSHV BCL-2 proteins were expressed and purified. GST-BHRF-1 and KSHV BCL-2 were truncated by 27 and 33 aa, respectively, at the C terminus to increase solubility (a). The protein sequence highlighted in parenthesis was removed, leaving the sequence shown in black in parenthesis in its place (b). To check purity and characterization, each protein was run on an SDS-PAGE gel, and stained with Coomassie blue, or transferred onto PVDF membrane and blotted for GST
Viral BCL-2 homolog interactions with BH3-only peptides were determined with the aid of fluorescence polarization assays (FPAs).2, 32 It has previously been determined that mammalian BCL-2 and orthologs (including MCL-1, BCL-XL, BCL-w and BFL-1) demonstrate distinct profiles of binding BH3 domains of BH3-only proteins.3, 4, 5, 6 This pattern of binding relates directly to the ability of these BH3 domains to antagonize the anti-apoptotic protection afforded by BCL-2 and orthologs. FPA showed that BHRF-1 binds to BH3-only peptides BIM, BID and PUMA but not BID BH3 mutant (BIDmut). This pattern of interaction most closely resembles that previously determined for the cellular anti-apoptotic protein BFL-1. In contrast, KSHV BCL-2 interacts with BH3 domains from BIM, BID, NOXA, BIK, PUMA and BMF, but not with BIDmut, or BAD BH3 (Table 1). This interaction pattern closely resembles that of the cellular anti-apoptotic proteins MCL-1, particularly the ability to bind NOXA BH3 peptides.3, 6 The results suggest that, at least functionally, KSHV BCL-2 more closely resembles MCL-1 than BCL-2. Of note, the binding affinities to interacting BH3 peptides is significantly lower than that found in prior studies with cellular anti-apoptotic proteins. While this may well reflect intrinsic properties of the different proteins, it is also possible that alterations in our bacterial expression, truncation or absence of post-translational modification have an effect as well.
As another way of investigating the relatedness of BHRF-1 and KSHV BCL-2 to the cellular anti-apoptotic proteins, BHRF-1 and KSHV BCL-2 full-length amino-acid sequences were aligned to the anti-apoptotic BCL-2 family proteins using ClustalW and TCOFFEE (EMBL-EBI research tools, http://www.ebi.ac.uk).33, 34 Interestingly, the cellular anti-apoptotic protein to which KSHV BCL-2 is most closely related is MCL-1. Alignment scores also show that KSHV BCL-2 shares most similarity with MCL-1 (Table 2a and b). In addition, when you compare the sequence for KSHV BCL-2 against all the proteins deposited in GenBank (http://ncbi.nih.gov), using the ‘BLAST’ tool, the most similar human sequence to KSHV BCL-2 was found to be MCL-1 (e value, 4e−05). The congruence of the functional as well as sequence similarities between KSHV BCL-2 and MCL-1 leads us to suggest that the KSHV anti-apoptotic protein more closely resembles MCL-1 than BCL-2.
As KSHV and EBV are involved in human disease, it may be of therapeutic benefit to be able to antagonize the function of KSHV BCL-2 or BHRF-1. ABT-737 is a small molecule antagonist of BCL-2, BCL-XL and BCL-w, binding, like the BAD BH3 peptide, with high affinity to the hydrophobic pocket that binds BH3 domains in these molecules.32, 35 We tested the ability of ABT-737 35 to displace BH3 peptides from BHRF-1 and KSHV BCL-2. It was found not to interact with BHRF-1 or KSHV BCL-2 (Figure 2), as would be predicted, given the lack of a BAD BH3 interaction with BHRF-1 and KSHV BCL-2 (Table 1).
BHRF-1 and KSHV BCL-2 interact with BH3-only proteins in human cells
Next, we investigated binding of BHRF-1 and KSHV BCL-2 to intact BH3-only proteins from human cells. To test whether viral BCL-2 homologs interact with BH3-only proteins, we treated lysates from human B lymphocyte (Ly-1) or Toledo or 293T cells with either GST-BHRF-1, GST-KSHV BCL-2, GST-BCL-XL, GST MCL-1 or GST alone. Lysates from Ly-1, Toledo and 293T cells were chosen based on the high endogenous levels of the proteins of interest present, namely BID, BIM, BAD and NOXA, respectively. Following immobilization on glutathione–agarose beads, protein complexes were identified by western blot (Figure 3). Purified GST was used as a control in this experiment, and does not show binding to any of the BCL-2 family proteins, as expected. Equal loading of GST proteins was determined by probing for GST (Figure 3c, e and h).
KSHV BCL-2 and BHRF-1 interact with sensitizer BH3-only proteins, consistent with their binding codes. GST pull-down assay. GST-KSHV BCL-2, GST-BHRF-1, GST alone and GST-BCL-XL or GST-MCL-1 were individually immobilized on glutathione–agarose beads and combined with either Ly-1 B lymphoblast cell lysate (a–c), Toledo cell lysate (d, e) (100 μg) or 293 T cell lysate (g, h) (1.5 mg). Proteins interacting with GST-KSHV BCL-2 and GST-BHRF-1 were analyzed by immunoblot. Densitometry was used to quantify levels of BAD (f) and NOXA (i)
BHRF-1 interacts strongly with activator BIM and BID proteins (Figure 3a and b, respectively). BHRF-1 and KSHV BCL-2 show nearly background binding to BAD, compared with a stronger signal for BCL-XL (Figure 3d). KSHV BCL-2 interacts relatively weakly with BIM and BID (Figure 3a and b). KSHV BCL-2 shows interaction with human NOXA, though weaker than that of MCL-1 with NOXA, consistent with the binding affinities in Table 1 (Figure 3g). Densitometry results are shown (Figure 3f and i). Binding of KSHV BCL-2 to NOXA is consistent with the binding code established in Table 1.
BHRF-1 protects against cell death caused by growth factor withdrawal and DNA damage
To establish a model in which BHRF-1 can protect cells from death in a cellular setting, we established cell lines stably overexpressing full-length FLAG-BHRF-1 and FLAG-BCL-2. We were unable to isolate cells either stably or transiently expressing full-length FLAG-KSHV BCL-2. The pro-lymphocytic murine FL5.12 cell line requires IL-3 to maintain survival. Consistent with prior results,32 apoptosis induced by IL-3 withdrawal is inhibited by overexpression of BCL-2 (Figure 4a). Cells stably expressing FLAG-BHRF-1 were examined after IL-3 withdrawal using FACS analysis with Annexin V staining. BHRF-1, like BCL-2 protects against cell death in an IL-3-depleted environment. BHRF-1- and BCL-2-transfected cells were also found to be protected from death induced by etoposide (50 μM) (Figure 4b) and adriamycin (300 nM) (Figure 4c) after 24 h. The increased level of protection afforded by BCL-2 is consistent with the observation that expression of BCL-2 exceeds that of BHRF-1, as indicated by the anti-FLAG immunoblot in Figure 5g. We have validated, in concurrence with prior results, that the presence of BHRF-1 can protect cells from death, following serum starvation, etoposide treatment and adriamycin treatment.
BHRF-1, like BCL-2, protects mammalian cells from diverse death stimuli. Survival was assigned to cells not staining with Annexin V by FACS analysis. Shown is average and standard deviation of three independent experiments. FLAG-BHRF-1- and FLAG-BCL-2-transfected FL5.12 single cell clones (BHRF-1 and BCL-2, respectively) were tested for survival following (a) IL-3 withdrawal (30 h), (b) etoposide treatment (24 h, 50 μM) and (c) adriamycin treatment (24 h, 300 nM)
BHRF-1 protects FL5.12-transfected cells from death by interaction with BIM. Cell lysates (1 mg) were prepared from FLAG-BHRF-1, FLAG-BCL-2 and vector-only transfected FL5.12 cells before and after IL-3 withdrawal. Lysates were combined with protein A beads and incubated with FLAG antibody (7 μg). Proteins interacting with BHRF-1, BCL-2 and vector-only, namely BIM (a), PUMA (b), BAK (c), BAX (d), BID (e), Actin (f) and FLAG (g) were analyzed by immunoblot as indicated
BHRF-1 protects against cell death by binding BIM
We have established that BHRF-1 protects against cell death following a variety of death inducers, and also have investigated potential binding partners of BHRF-1. It is unknown which BCL-2 family proteins BHRF-1 interacts with following a specific death stimulus. Here, we removed IL-3 from FL5.12 cells stably overexpressing FLAG-BHRF-1 or FLAG-BCL-2, prepared lysates from these cells and examined which BCL-2 family proteins could be found in complex with the two anti-apoptotic proteins. Following IL-3 withdrawal, BHRF-1 interacts strongly with activator BIM (Figure 5a), in the same way that BCL-2 does. PUMA can also be found in complex with BHRF-1 (Figure 5b), but BHRF-1 appears to interact weakly or not at all with BAK, BAX or BID (Figure 5c–e, respectively). It is important to note that BIM and PUMA have previously been shown to play important roles in death signaling downstream of IL-3 withdrawal.32, 36, 37 Equal loading in cell lysates was examined by probing for β-actin (Figure 5f). Notably, while similar amounts of FLAG-BCL-2 and FLAG-BHRF-1 are immunoprecipitated, the FLAG-BHRF-1 is expressed at levels significantly lower than FLAG-BCL-2 (Figure 5g). FLAG-BHRF-1, therefore, may well represent only the minority of net anti-apoptotic proteins expressed. Therefore, when pro-death BH3-only proteins are distributed across the total anti-apoptotic proteins, fewer pro-death proteins are available to be bound by the FLAG-BHRF-1. FLAG-BCL-2, expressed in higher quantities, can bind a larger proportion of the total pro-death protein pool. This is likely an explanation for the greater quantity of pro-death proteins in complex with the exogenous FLAG-BCL-2. Nonetheless, it is clear that removal of IL-3 results in an increase in the amounts of BIM and PUMA bound to FLAG-BHRF-1.
Anti-apoptotic function of BHRF-1 and KSHV BCL-2 can be opposed selectively by BH3 domain peptides
tBID, an activator BH3-only protein, can induce release of cytochrome c from mitochondria, and thereby cause cell death.38 The presence of anti-apoptotic proteins like BCL-2, MCL-1, BCL-XL, BFL-1 and BCL-w can protect the mitochondria from cell death induced by tBID.32 Sensitizer BH3 domain peptides, including BAD, NOXA, PUMA, BIK, BMF and HRK BH3 peptides, alone are unable to induce cytochrome c release from mitochondria.2, 32 However, we have shown that sensitizers, nonetheless, exhibit a pro-death function by displacing activators from anti-apoptotic proteins. Sensitizers therefore cause apoptosis by abrogating the function of anti-apoptotic cellular proteins like BCL-2 or MCL-1. We wanted to test whether function of viral homologs BHRF-1 and KSHV BCL-2 could be opposed in a similar fashion.
In our experiment, tBID, as before, induced cytochrome c release from mouse liver mitochondria (Figure 6a and b). Addition of either KSHV BCL-2 or BHRF-1 inhibited this cytochrome c release, as did BCL-2, BCL-XL, BCL-w, MCL-1 and BFL-1 in a previous study.32 BH3-only sensitizer peptides inhibited this protection in a pattern that recapitulated the binding pattern found in Table 1. It is important to note that BH3-only sensitizer peptides, alone, do not induce cytochrome c release, as previously described.2, 32 Cytochrome c release induced by the activator BH3 peptide BIM BH3 was prevented by addition of KSHV BCL-2 or BHRF-1, but not by addition of GST alone (Figure 6c). When compared with our prior study, it can again be seen that KSHV BCL-2 functions similarly to MCL-1, and BHRF-1 to BFL-1. Therefore, the viral anti-apoptotic proteins KSHV BCL-2 and BHRF-1 function like the cellular anti-apoptotic proteins to oppose apoptosis, by binding pro-apoptotic BH3-only proteins like tBID. Furthermore, their anti-death functions can be abrogated selectively by BH3 domain peptides that function as prototypic BHRF-1 and KSHV BCL-2 inhibitors.
Sensitizer BH3 peptides displace tBID protein from BHRF-1 and KSHV BCL-2, consistent with their binding codes. tBID competition assay. Mitochondria were prepared from wild-type mouse liver. Mitochondria were treated with 50 nM tBID or 13 nM tBID alone or in combination with GST-BHRF-1 (0.2 μM) (a) or KSHV BCL-2 (1.5 μM) (b), respectively, followed by the indicated sensitizer peptides (10 μM) at room temperature for 45 min. Release of cytochrome c was measured by ELISA. BIM-treated FL5.12 cells. Mitochondria were prepared from FL5.12 cells and treated with 500 nM BIM BH3 alone or in combination with GST-KSHV BCL-2 (2.4 μM), GST-BHRF-1 (1 μM) or GST alone (2 μM) (c). Release of cytochrome c was measured by ELISA
Discussion
While it has been understood for over a decade that KSHV and EBV express homologs of BCL-2, the details of the biological and biochemical functions of these proteins have remained somewhat obscure. It was clear that the overexpression of these proteins conferred resistance to apoptosis from numerous insults. However, interactions with pro-death BCL-2 family members seemed difficult to observe. KSHV BCL-2 was found not to interact with BAX or BAK.39 BHRF-1 was found not to interact with BAK, BAX, BAD or BIK,11 though another group found that it interacted with BAK, but not BAX.20 Our results demonstrate that both proteins do interact with pro-death BCL-2 family proteins, but the interaction pattern is quite selective. Both BHRF-1 and KSHV BCL-2 bind select pro-death BH3-only family members of the BCL-2 family to oppose apoptosis. In particular, using multiple assays using peptide binding, GST pull-down assays and co-immunoprecipitation, we show that BHRF-1 can interact with BIM, in contradiction to a previous finding.40 Moreover, like their cellular homologs, BHRF-1 and KSHV BCL-2 exhibit selective interaction with BH3-only proteins.
We use these interactions to show for the first time that KSHV, by both sequence and function, more closely resembles MCL-1 than the other cellular anti-apoptotic proteins. BHRF-1, on the other hand, more closely resembles BCL-2 by amino-acid sequence, though its binding pattern more closely resembles BFL-1 (Table 1). One way to make sense of this is to understand that while KSHV BCL-2 and BHRF-1 are functional homologs, they are not positionally homologous in their respective viral genomes. This suggests that the primordial anti-apoptotic genes were captured independently, perhaps from different cellular proteins, and then perhaps became more similar due to convergent evolutionary pressures.
The biological role of the viral BCL-2 homologs expressed by γ-herpesviruses is still undecided. It does seem clear that they are expressed at low levels during latent infection and are not necessary for maintaining latent infection. Some suggest that their expression is most important for the inhibition of apoptosis during initial cellular infection and transformation.41 These findings conflict with other results, however.42 Yet other work suggests that BCL-2 homologs are more important for reactivation or persistent infection.43 Their role in cancer is equally undecided, and understanding has been limited by the poor availability of good antibodies recognizing the proteins. However, transcripts of KSHV BCL-2 have been detected in cell lines and clinical samples from primary effusion lymphoma and KS.28 BHRF-1 transcripts have been detected in EBV-associated B-cell lymphomas.44 It has not been established whether the expression of the corresponding proteins is important in the tumor biology.
Given the possibility that antagonism of these BCL-2 homologs could be beneficial, we tested whether ABT-737, a high-affinity small molecule antagonist of BCL-2, had any effect on BHRF-1 or KSHV BCL-2 function. We found that ABT-737 was unable to displace a BH3 peptide from the binding site of either protein, consistent with a similar inability of BAD BH3 peptide to bind to either protein. We have previously shown that ability of a protein to bind the BAD BH3 peptide correlates with ability to bind the ABT-737 compound. However, the binding code in Table 1, combined with the evidence of BHRF-1 and KSHV BCL-2 inhibition in Figure 5, demonstrates that small molecule mimetics of interacting BH3 peptides could, nonetheless, be designed to selectively inhibit the two homologs. There is still much to learn about the role of viral BCL-2 homologs in tumor development, but our findings provide a stepping stone for identifying possible avenues that may be exploited to promote therapeutic programmed cell death.
Materials and Methods
Recombinant proteins
Constructs expressing BHRF-1 and KSHV BCL-2 were kind gifts of Professor Herbert Virgin's lab in Washington University School of Medicine, MO, USA. Using an IPTG-inducible GST fusion vector, PGEX-4T-1 (Amersham Biosciences), BHRF-1 and KSHV BCL-2 proteins were cloned and expressed in Escherichia coli BL-21 competent cells (Stratagene) and grown in Overnight ExpressTM Instant TB medium (EMD Biosciences) and affinity purified using glutathione–agarose (for GST-linked proteins) as previously described.2 The C-terminal transmembrane domain was truncated to maintain solubility in aqueous solution (Figure 1a). For binding assays, GST-linked proteins were used for BHRF-1 and KSHV BCL-2. Using a neomycin-resistant FLAG-tagged vector, pCMV3xFLAG (Sigma), BHRF-1 and KSHV BCL-2 were cloned and purified from DH5α bacteria (Invitrogen). Full-length FLAG-BHRF-1 was used in immunoprecipitation experiments. Recombinant tBID was made as previously described;6 it contained double cysteine to serine substitutions which maintains wild-type ability to induce cytochrome c release.
Peptides
Peptides were synthesized as previously described.6
Fluorescence polarization binding assays
Binding assays were performed using fluorescence polarization as previously described.2 Peptides at 5 nM were mixed with titrations of GST-BHRF-1 or GST-KSHV BCL-2 in binding buffer (140 mM NaCl, 10 mM Tris (pH 7.4)). An increase in fluorescence polarization measured on a Safire2 plate reader (Tecan) was quantitated to calculate binding. A nonlinear fit to a sigmoidal dose–response curve utilized the program Magellan version 6.1 to determine Kd. A minimum of three independent experiments was used to determine dissociation constants.
Cell culture
FL5.12 cells were cultured as described previously6 in Iscove's modified Dulbecco's medium, 10% fetal bovine serum, 1000 μg/ml G418 with or without IL-3 provided by 10% WEHI-3B supplement (supernatant of IL-3 secreting WEHI-3B cells). FL5.12 cells were stably transfected with a vector containing a neomycin-resistance construct and either FLAG-BHRF-1 cDNA (FL5.12-BHRF-1), human FLAG-BCL-2 cDNA (FL5.12-BCL-2) or no insert (wt).
Electroporation of cells
FL5.12 cells (1 ml of 8.0 × 106 cells/ml) were placed into 4 mm cuvette (Bio-Rad) and treated with either 50 μg of pCMV3x-FLAG-BHRF-1 and FLAG-BCL-2 plasmid and electroporated at 200 V, 975 μF. The cells were removed from cuvettes, and seeded onto 10 ml media in a T25 flask (Fisher) and incubated for 48 h. Media were then supplemented with 1 mg/ml G418 (Cellgro). To assess their apoptotic status, treated cells were stained with Annexin V and analyzed on a FACSCalibur machine (Becton-Dickinson).
Annexin V assays
Cells were stained with fluorescent conjugates of Annexin V (BioVision) and analyzed on a FACSCalibur machine (BD).
Immunoprecipitation
Cell lysates (1 mg) were incubated with anti-human FLAG antibody (10 μg) for at least 1 h at 4°C in 1% CHAPS buffer (5 mM sodium phosphate pH 7.4; 2.5 mM EDTA; 100 mM sodium chloride; 1% w/v CHAPS, in the presence of protease inhibitors (complete tablets; Roche). Protein A-sepharose beads (Sigma) were added to precipitate complexes containing BHRF-1. The beads were mixed with loading buffer prior to loading supernatant onto gel. Cell lysate (25 μg) in 1% CHAPS buffer was used per lane.
GST pull down
GST-KSHV BCL-2, GST-BHRF-1, GST-BCL-XL and GST-MCL-1 (10 μg) were incubated with glutathione–agarose beads for 1 h at 4°C in binding buffer (140 mM NaCl, 10 mM Tris pH 7.4). Beads were rinsed and incubated with approximately 100 μg of either Toledo cell lysate, Ly-1 cell lysate or 1.5 mg 293T cell lysate for 1 h at 4°C. Beads were washed again and loaded on a denaturing NuPAGE gel.
Immunoblots
Protein lysates were obtained by cell lysis in 1% CHAPS buffer. Protein samples were size fractionated on NuPAGE 10% Bis-Tris polyacrylamide gels (Invitrogen). Antibodies were used to detect the following proteins on membranes: BIM (Calbiochem; 22–40); PUMA (Prosci; NT); Bid (Santa Cruz; FL195); BAK (Upstate; NT); BAX (Santa Cruz; N-20); NOXA (Calbiochem; 114C307); GST (Santa Cruz; 425); BAD (Santa Cruz; 8044); FLAG (Sigma; M2) and actin (Chemicon; MAB1501).
Cytochrome c release
Mitochondria were purified from FL5.12 cells as previously described.2 Mitochondria were incubated with treatments for 45 min. Mitochondria from Fl5.12 cells were treated with 500 nM BIM BH3, followed by addition of 2.4 μM GST-KSHV BCL-2, 1 μM GST-BHRF-1 or 2 μM GST alone. Release of cytochrome c was determined by a comparison of cytochrome c in the pellet and supernatant following treatment, quantitated by ELISA (R&D Systems). When results of multiple experiments were averaged, results from solvent-only (DMSO) treatment values were subtracted from each, so that 0% release reflects that observed in solvent-only treatments.
tBID competition assay
Mitochondria prepared from mouse liver were treated with either GST-BHRF-1 (0.2 μM) or GST-KSHV BCL-2 (1.5 μM) and 50 nM cleaved Bid (tBID) and BH3-only sensitizer peptides (10 μM) at room temperature for 45 min. Cytochrome c release from triplicate experiments was assayed using ELISA.
Mice
Mitochondria were prepared from livers taken from wild-type mice as previously described.2
Abbreviations
- BCL-2:
-
B-cell leukemia/lymphoma 2
- EBV:
-
Epstein–Barr virus
- KSHV:
-
Kaposi's sarcoma herpesvirus
References
Adams JM, Cory S . The Bcl-2 protein family: arbiters of cell survival. Science 1998; 281: 1322–1326.
Letai A, Bassik MC, Walensky LD, Sorcinelli MD, Weiler S, Korsmeyer SJ . Distinct BH3 domains either sensitize or activate mitochondrial apoptosis, serving as prototype cancer therapeutics. Cancer Cell 2002; 2: 183–192.
Chen L, Willis SN, Wei A, Smith BJ, Fletcher JI, Hinds MG et al. Differential targeting of prosurvival Bcl-2 proteins by their BH3-only ligands allows complementary apoptotic function. Mol Cell 2005; 17: 393–403.
Kuwana T, Bouchier-Hayes L, Chipuk JE, Bonzon C, Sullivan BA, Green DR et al. BH3 domains of BH3-only proteins differentially regulate Bax-mediated mitochondrial membrane permeabilization both directly and indirectly. Mol Cell 2005; 17: 525–535.
Opferman JT, Letai A, Beard C, Sorcinelli MD, Ong CC, Korsmeyer SJ . Development and maintenance of B and T lymphocytes requires antiapoptotic MCL-1. Nature 2003; 426: 671–676.
Certo M, Del Gaizo Moore V, Nishino M, Wei G, Korsmeyer S, Armstrong SA et al. Mitochondria primed by death signals determine cellular addiction to antiapoptotic BCL-2 family members. Cancer Cell 2006; 9: 351–365.
Hardwick JM, Bellows DS . Viral versus cellular BCL-2 proteins. Cell Death Differ 2003; 10 (Suppl 1): S68–S76.
Rickinson AB, Kieff E . Epstein–Barr Virus. Lippincott Williams and Williams: Philadelphia, 2001.
Crawford DH . Biology and disease associations of Epstein–Barr virus. Philos Trans R Soc Lond B Biol Sci 2001; 356: 461–473.
Cleary ML, Smith SD, Sklar J . Cloning and structural analysis of cDNAs for bcl-2 and a hybrid bcl-2/immunoglobulin transcript resulting from the t(14;18) translocation. Cell 1986; 47: 19–28.
Huang Q, Petros AM, Virgin HW, Fesik SW, Olejniczak ET . Solution structure of the BHRF1 protein from Epstein–Barr virus, a homolog of human Bcl-2. J Mol Biol 2003; 332: 1123–1130.
Henderson S, Huen D, Rowe M, Dawson C, Johnson G, Rickinson A . Epstein–Barr virus-coded BHRF1 protein, a viral homologue of Bcl-2, protects human B cells from programmed cell death. Proc Natl Acad Sci USA 1993; 90: 8479–8483.
Kawanishi M . Epstein–Barr virus BHRF1 protein protects intestine 407 epithelial cells from apoptosis induced by tumor necrosis factor alpha and anti-Fas antibody. J Virol 1997; 71: 3319–3322.
Foghsgaard L, Jaattela M . The ability of BHRF1 to inhibit apoptosis is dependent on stimulus and cell type. J Virol 1997; 71: 7509–7517.
Khanim F, Dawson C, Meseda CA, Dawson J, Mackett M, Young LS . BHRF1, a viral homologue of the Bcl-2 oncogene, is conserved at both the sequence and functional level in different Epstein–Barr virus isolates. J Gen Virol 1997; 78 (Pt 11): 2987–2999.
Tarodi B, Subramanian T, Chinnadurai G . Epstein–Barr virus BHRF1 protein protects against cell death induced by DNA-damaging agents and heterologous viral infection. Virology 1994; 201: 404–407.
Fanidi A, Hancock DC, Littlewood TD . Suppression of c-Myc-induced apoptosis by the Epstein–Barr virus gene product BHRF1. J Virol 1998; 72: 8392–8395.
Davis JE, Sutton VR, Smyth MJ, Trapani JA . Dependence of granzyme B-mediated cell death on a pathway regulated by Bcl-2 or its viral homolog, BHRF1. Cell Death Differ 2000; 7: 973–983.
Nava VE, Cheng EH, Veliuona M, Zou S, Clem RJ, Mayer ML et al. Herpesvirus saimiri encodes a functional homolog of the human bcl-2 oncogene. J Virol 1997; 71: 4118–4122.
Theodorakis P, D'Sa-Eipper C, Subramanian T, Chinnadurai G . Unmasking of a proliferation-restraining activity of the anti-apoptosis protein EBV BHRF1. Oncogene 1996; 12: 1707–1713.
Hardwick JM, Lieberman PM, Hayward SD . A new Epstein–Barr virus transactivator, R, induces expression of a cytoplasmic early antigen. J Virol 1988; 62: 2274–2284.
Cox MA, Leahy J, Hardwick JM . An enhancer within the divergent promoter of Epstein–Barr virus responds synergistically to the R and Z transactivators. J Virol 1990; 64: 313–321.
Chang Y, Cesarman E, Pessin MS, Lee F, Culpepper J, Knowles DM et al. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 1994; 266: 1865–1869.
Boshoff C, Weiss RA . Epidemiology and pathogenesis of Kaposi's sarcoma-associated herpesvirus. Philos Trans R Soc Lond B Biol Sci 2001; 356: 517–534.
Huang Q, Petros AM, Virgin HW, Fesik SW, Olejniczak ET . Solution structure of a Bcl-2 homolog from Kaposi sarcoma virus. Proc Natl Acad Sci USA 2002; 99: 3428–3433.
Cheng EH, Kirsch DG, Clem RJ, Ravi R, Kastan MB, Bedi A et al. Conversion of Bcl-2 to a Bax-like death effector by caspases. Science 1997; 278: 1966–1968.
Ojala PM, Tiainen M, Salven P, Veikkola T, Castanos-Velez E, Sarid R et al. Kaposi's sarcoma-associated herpesvirus-encoded v-cyclin triggers apoptosis in cells with high levels of cyclin-dependent kinase 6. Cancer Res 1999; 59: 4984–4989.
Sarid R, Sato T, Bohenzky RA, Russo JJ, Chang Y . Kaposi's sarcoma-associated herpesvirus encodes a functional bcl-2 homologue. Nat Med 1997; 3: 293–298.
Cheng EH, Levine B, Boise LH, Thompson CB, Hardwick JM . Bax-independent inhibition of apoptosis by Bcl-XL. Nature 1996; 379: 554–556.
Clem RJ, Cheng EH, Karp CL, Kirsch DG, Ueno K, Takahashi A et al. Modulation of cell death by Bcl-XL through caspase interaction. Proc Natl Acad Sci USA 1998; 95: 554–559.
Bellows DS, Chau BN, Lee P, Lazebnik Y, Burns WH, Hardwick JM . Antiapoptotic herpesvirus Bcl-2 homologs escape caspase-mediated conversion to proapoptotic proteins. J Virol 2000; 74: 5024–5031.
Certo M, Moore Vdel G, Nishino M, Wei G, Korsmeyer S, Armstrong SA et al. Mitochondria primed by death signals determine cellular addiction to antiapoptotic BCL-2 family members. Cancer Cell 2006; 9: 351–365.
Notredame C, Higgins DG, Heringa J . T-Coffee: a novel method for fast and accurate multiple sequence alignment. J Mol Biol 2000; 302: 205–217.
Thompson JD, Higgins DG, Gibson TJ . CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 1994; 22: 4673–4680.
Oltersdorf T, Elmore SW, Shoemaker AR, Armstrong RC, Augeri DJ, Belli BA et al. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature 2005; 435: 677–681.
Harada H, Quearry B, Ruiz-Vela A, Korsmeyer SJ . Survival factor-induced extracellular signal-regulated kinase phosphorylates BIM, inhibiting its association with BAX and proapoptotic activity. Proc Natl Acad Sci USA 2004; 101: 15313–15317.
Shinjyo T, Kuribara R, Inukai T, Hosoi H, Kinoshita T, Miyajima A et al. Downregulation of Bim, a proapoptotic relative of Bcl-2, is a pivotal step in cytokine-initiated survival signaling in murine hematopoietic progenitors. Mol Cell Biol 2001; 21: 854–864.
Wei MC, Lindsten T, Mootha VK, Weiler S, Gross A, Ashiya M et al. tBID, a membrane-targeted death ligand, oligomerizes BAK to release cytochrome c. Genes Dev 2000; 14: 2060–2071.
Cheng EH, Nicholas J, Bellows DS, Hayward GS, Guo HG, Reitz MS et al. A Bcl-2 homolog encoded by Kaposi sarcoma-associated virus, human herpesvirus 8, inhibits apoptosis but does not heterodimerize with Bax or Bak. Proc Natl Acad Sci USA 1997; 94: 690–694.
O'Connor L, Strasser A, O'Reilly LA, Hausmann G, Adams JM, Cory S et al. Bim: a novel member of the Bcl-2 family that promotes apoptosis. EMBO J 1998; 17: 384–395.
Altmann M, Hammerschmidt W . Epstein–Barr virus provides a new paradigm: a requirement for the immediate inhibition of apoptosis. PLoS Biol 2005; 3: e404.
Marchini A, Tomkinson B, Cohen JI, Kieff E . BHRF1, the Epstein–Barr virus gene with homology to Bc12, is dispensable for B-lymphocyte transformation and virus replication. J Virol 1991; 65: 5991–6000.
Gangappa S, van Dyk LF, Jewett TJ, Speck SH, Virgin HW . Identification of the in vivo role of a viral bcl-2. J Exp Med 2002; 195: 931–940.
Oudejans JJ, van den Brule AJ, Jiwa NM, de Bruin PC, Ossenkoppele GJ, van der Valk P et al. BHRF1, the Epstein–Barr virus (EBV) homologue of the BCL-2 protooncogene, is transcribed in EBV-associated B-cell lymphomas and in reactive lymphocytes. Blood 1995; 86: 1893–1902.
Acknowledgements
We are grateful to Professor Herbert Virgin's laboratory of Washington University for providing plasmids containing cDNA from EBV BHRF-1 and KSHV BCL-2. We are grateful to Eric Johannsen and Ellen Cahir McFarland for sharing their expertise with γ-herpesviruses. AMF and AL acknowledge generous support from the V Foundation. AL also acknowledges generous support from K08 CA10254.
Author information
Authors and Affiliations
Corresponding author
Additional information
Edited by C Borner
Rights and permissions
About this article
Cite this article
Flanagan, A., Letai, A. BH3 domains define selective inhibitory interactions with BHRF-1 and KSHV BCL-2. Cell Death Differ 15, 580–588 (2008). https://doi.org/10.1038/sj.cdd.4402292
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.cdd.4402292
Keywords
This article is cited by
-
Sequence analysis of Epstein-Barr virus (EBV) early genes BARF1 and BHRF1 in NK/T cell lymphoma from Northern China
Virology Journal (2015)
-
Disruption of Bcl-2 and Bcl-xL by viral proteins as a possible cause of cancer
Infectious Agents and Cancer (2014)
-
The modulation of apoptosis by oncogenic viruses
Virology Journal (2013)
-
Structural biology of the Bcl-2 family and its mimicry by viral proteins
Cell Death & Disease (2013)
-
The matrix (M) protein of newcastle disease virus binds to human bax through its BH3 domain
Virology Journal (2011)