Abstract
A number of elegant studies exploring the consequences of expression of various mutant forms of p53 in mice have been published over the last years. The results and conclusions drawn from these studies often contradict results previously obtained in biochemical assays and cell biology studies, questioning their relevance for p53 function in vivo. Owing to the multitude of post-translational modifications imposed on p53, however, the in vivo validation of their relevance for proper protein function and tumour suppression is constantly lagging behind new biochemical discoveries. Nevertheless, mouse genetics presents again its enormous power. Despite being relatively slow and tedious, it has become indispensable for researchers to sort out the wheat from the chaff in an endless sea of publications on p53.
Similar content being viewed by others
Main
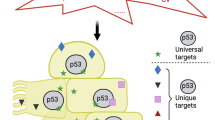
To this very moment, more than 42 500 papers have been published dealing with p53, in one or the other way, and still we are not quite sure what all of its biological functions are and how exactly it activates and coordinates them. What we assume is that p53 acts predominantly as a transcription factor, regulating the expression of more than 100 target genes to initiate apoptosis, cell cycle arrest, DNA-repair, cellular senescence as well as differentiation.1 We cannot be entirely certain that transcriptional activation of target genes is the only way by which p53 exerts its biological functions, because it has also been reported to translocate to the outer mitochondrial membrane where it interacts with pro- and antiapoptotic members of the Bcl-2 protein family.2
This review focuses on post-translational modifications, which have been reported to modulate p53's transcriptional activity and their influence on target gene expression, in particular those involved in cell cycle arrest and apoptosis. We have also tried to summarize attempts to verify the most important in vitro findings on p53 modification using knock-in mouse models that express certain variants of the molecule that lack key-residues subjected to post-translational modifications, or, entire portions of the protein considered relevant for proper function.
Target Genes Relevant for p53-Mediated Growth Arrest and Cell Death
The tumour suppressor function of p53 is based on its ability to regulate a range of cellular functions, including cell growth, cell cycle progression, DNA-repair, cellular senescence and cell death. We can assume today that deregulation of all these processes contributes to neoplastic transformation of p53-deficient cells.
The ability of p53 to induce cell cycle arrest depends on three critical target genes: p21,3 14-3-3σ4 and GADD45 (growth arrest and DNA damage-inducible protein 45).5 The transactivation of p21 triggers G1 cell cycle arrest through inhibition of G1 cyclin-dependent kinases (cyclinA/CDK2, cyclinE/CDK2 and cyclinD/CDK4 complexes).6 Maintenance of the Rb-E2F complex and consequent inhibition of S phase entry then stops cells with a damaged genome from undergoing faulty DNA replication, thereby preventing propagation of mutations.6 In response to DNA damage, cells lacking p21 fail to undergo G1 cell cycle arrest but they can still undergo cell death.7 Through induction of 14-3-3σ, a cytoplasmatic scaffold protein, and GADD45 p53 can also induce G2/M arrest in cells that have sustained DNA damage.4, 8 14-3-3σ has been shown to prevent nuclear import of cyclin B1 and CDC2, through sequestration in the cytoplasm,9 whereas GADD45 destabilizes CDC2/cyclinB complexes.10, 11 It appears that these two processes cooperate to prevent initiation of mitosis. Although the most prominent function of p21 is the mediation of G1 arrest, evidence has been presented that it also participates in the G2/M arrest after DNA damage,12 presumably by blocking PCNA function at replication forks.13
P53 target genes that are directly involved in the execution of apoptosis can be divided in two classes. First, components of the ‘extrinsic’ cell death signalling pathway, triggered by so-called death receptors (DR), members of the tumour necrosis factor (TNF) receptor (R) superfamily, after receptor-ligand (L) interaction.14 The TNF-R family members CD95/Fas/Apo-1, DR4, DR5 (also known as TRAIL-R1 and R2) were all reported to be transcriptionally induced by p53 and have been postulated to contribute to p53-induced apoptosis in certain model systems or in response to certain anticancer drugs (for details see Michalak et al.15). Analysis of mouse mutants lacking individual proteins of this family, such as CD95 or CD95L, however, demonstrated that these molecules are dispensable for p53-induced apoptosis in response to DNA damage (see Michalak et al.15 and citations therein). Lymphocytes from DR5-deficient animals were recently reported to resist radiation-induced apoptosis to a certain degree,16 although cells lacking the downstream signalling component of all DR able to induce apoptosis, that is, pro-caspase-8 were reported to respond normally to DNA damage.17 One might therefore speculate that the observed resistance of DR5-deficient cells in vivo may not be caused by defects in p53-induced apoptosis, as also evidenced by the normal sensitivity of E1A-transduced dr5−/− MEF to adriamycin,16 but may be due to enhanced clearance of apoptotic corpses in vivo or impaired JNK activation in response to DR5/TRAIL interaction. Maybe induction of DR by p53 may render cells with damaged genomes or under oncogenic stress more susceptible to immune surveillance by CTLs or NK cells.
The second class of p53 target genes relevant for apoptosis induction are central components of the ‘cell-intrinsic’ mitochondrial cell death pathway. APAF-1, the key component of the apoptosome, necessary for pro-caspase-9 recruitment and activation has been reported to be a primary p53 response gene.18, 19 APAF-1-deficiency confers significant resistance to p53-induced apoptosis in multiple cell types, albeit to various degrees.20, 21 Lymphocytes lacking APAF-1 or caspase-9, on the other hand, undergo DNA damage-induced apoptosis normally.22 Therefore, the relevance of the induction of APAF-1 by p53 remains unclear but may be required to enable a sustained DNA damage-induced apoptotic response.
More relevant for the proapoptotic p53 response appears to be its ability to regulate the abundance of a number of proapoptotic Bcl-2 family members, such as Bcl-2-associated protein X (Bax), and the BH3-only proteins p53-upregulated modulator of apoptosis (Puma) Noxa and Bcl-2-interacting domain death agonist (Bid). BH3-only proteins most likely trigger apoptosis by engaging multiple Bcl-2 pro-survival proteins in a stimulus- and possibly also cell-type-dependent manner, leading to Bax and/or Bcl-2 antagonist/killer (Bak) oligomerizaton, mitochondrial outer membrane permeabilization and subsequent cell death.23 Bax was the first Bcl-2-family member reported to be a p53 target,24 although this has been questioned later.25 Bax, in a manner largely redundant with Bak,26 is essential for apoptosis induction downstream of BH3-only proteins.27 Accordingly, Bax-deficient thymocytes are normally sensitive to DNA damage-induced apoptosis and MEFs from these knockout mice show only minor resistance to DNA-damaging agents.28, 29 However, in the context of malignant cells in which Bak function is impaired, for example, owing to mutation as reported for certain gastric and colon cancers,30 Bax will become rate limiting for p53-mediated (but also p53 independent) apoptosis. Whether induction of bax transcription by p53 in such cells will determine overall apoptosis sensitivity remains to be investigated. As for APAF-1, we speculate that p53-mediated induction of bax is not critical for apoptosis initiation but may rather serve to amplify apoptosis signalling.
There is now solid evidence that induction of BH3-only proteins is most essential for p53-induced apoptosis. The BH3-only protein Bid, which is considered to amplify apoptosis signalling, particularly by linking the DR pathway to the intrinsic pathway was reported to be induced transcriptionally by p53 in response to γ-irradiation.31 Although this finding is intriguing, it is unlikely that Bid is rate limiting for p53-induced killing. Bid activation requires caspase-8 or caspase-2-mediated cleavage, converting it into the truncated, active form, called tBid, to trigger apoptosis.32, 33 Caspase-8 or caspase-2-deficient lymphocytes and MEF, however, were all found to be normally sensitive to p53-dependent apoptotic stimuli.17, 34, 35 In general, the data reported on Bid in DNA damage-induced apoptosis are highly contradictory. Bid-deficient primary and hTERT immortalized MEF were shown to be refractory to cell death triggered by etoposide, UV- or γ-irradiation.36 In contrast, immortalized myeloid progenitor cells that lack Bid expression responded normally to apoptosis induced by the very same stimuli.37 Scrutinizing these findings in the originally published mouse strain as well as in an independently established bid−/− mouse line, generated on an inbred C57BL/6 background, did not reveal any evidence for Bid in the cellular response to DNA damage.38
Puma mRNA and protein were both reported to be induced in normal as well as malignant cells following DNA damage or in response to oncogenic stress.39, 40, 41 Its induction in response to DNA damage strictly depends on p53, as colon carcinoma cells expressing a human papillomavirus E6 protein or engineered to lack wt p53 (H1299 and HCT116 cells, respectively) are unable to express Puma.39, 41
The noxa promoter region contains a functional p53-binding site, as confirmed by promoter luciferase reportes assays.42 Human noxa has already previously been cloned from leukaemia cells treated with PMA and, therefore, was called ATL-derived PMA-responsive gene or APR, but no function was assigned to this gene.43 Noxa mRNA is also rapidly induced after adenovirus-mediated re-introduction of p53 into MEFs derived from p53−/− or wt mice and in wt thymocytes subjected to γ-irradiation but not in their p53−/− counterparts.42
In line with their role as p53 target genes, cells derived from Puma or Noxa-deficient mice show increased resistance to a range of p53-dependent apoptotic stimuli.44, 45, 46 Thymocytes and myeloid progenitors from Puma-deficient animals are abnormally resistant to DNA damage caused by treatment with etoposide or γ-irradiation. Although upregulation of noxa mRNA was also observed in these cells in response to these death stimuli, Noxa-deficient cells were normally sensitive. Similar results were obtained in an analysis of Puma-deficient bone marrow-derived pre-B cells, mature peripheral B and T cells as well as bone marrow-derived myeloid progenitors.44, 46 Again, loss of Noxa provided no protection against these death stimuli in these cell types.44, 45 In E1A-transformed MEFs, Noxa and Puma appear to play an overlapping role in mediating cell death in response to DNA damage caused by cytotoxic drugs, such as etoposide or γ-irradiation.44 Strikingly, in MEF and keratinocytes Noxa is the rate-limiting BH3-only protein for UV-irradiation-mediated apoptosis.47 In contrast, Noxa can act together with Bim (and not Puma) in triggering apoptosis in oncogene-transformed MEF after UV-induced DNA damage, whereas both, Noxa and Puma contribute to UV-induced death of primary MEF.47 This indicates that, depending on the cellular context, upstream signals induced by distinct forms of DNA damage, that trigger different post-translational modifications on p53 (e.g. by ATM (ataxia telangiectasia mutated) versus ATR (ataxia telangiectasia and Rad3-related) activation), induce either Puma and/or Noxa to promote apoptosis. Puma and Noxa often appear to act in concert with BH3-only proteins that are not directly regulated by p53 (e.g. Bim) but may be induced by parallel pathways triggered by DNA damage.48, 49
Besides the ability to transactivate proapoptotic target genes, evidence has been presented that p53 can trigger apoptosis directly by post-translational activation of proapoptotic Bax or Bak. As a possible mechanism, it has been proposed that cytoplasmic p53 is neutralized by Bcl-xL (or Bcl-2) in healthy cells and in response to DNA damage (e.g. caused by UV-radiation), nuclear p53 triggers expression of Puma that subsequently disrupts this cytoplasmic p53/Bcl-xL complex, allowing p53 to activate Bax and/or Bak.50, 51, 52, 53 It is, however, interesting to note that overexpression of Puma potently triggers cytochrome c release in p53−/− MEF54 and that deletion of the p53-response elements in the Puma promoter completely abrogates DNA damage-induced apoptosis in HCT-116 and DLD-1 carcinoma cells.55 This indicates that the transcription-independent proapoptotic potential of p53 may only be a minor one. Hence, we believe that under physiological conditions, similar to the effects of p53 on cell cycle, apoptosis induction by p53 mainly depends on the transcription of a distinct set of target genes that are central components of the cell death machinery. Which modifications in p53 promote the preferential expression of cell cycle arrest plus DNA repair genes versus induction of proapoptotic genes is poorly understood and under intense investigation (see below).
Structure–Function Relationships in p53
The p53 gene contains eleven exons with two alternative translation start sites in exon 2 and 4, respectively. Half a dozen domains are known to contribute to p53 protein function (Figure 1). At the N-terminus, two transactivation domains (TAD), required for transcriptional activation of target genes are encoded. The second TAD partially overlaps with the so-called prolin-rich domain (PRD), which is required for multiple protein–protein interactions. The central region of the protein contains the DNA-binding domain (DBD) that is subjected to heavy mutagenesis during tumour formation (>90% of all p53 mutations found in human cancer reside in this region). The DBD is followed by a domain that promotes oligomerization of p53 (tetramerization domain (4D)). Functional p53 acts as a transcription factor in tetrameric conformation, made up by two p53 homo-dimers. Importantly, the oligomerization domain also contains a nuclear export signal (NES). This region is followed by the C-terminal regulatory domain (CTD), harbouring three nuclear localization signals (NLS). The CTD is also able to bind nonspecifically to damaged DNA (see also http://p53.free.fr for further details). Post-translational modifications have been described to occur in all these domains with different effects on protein stability and function (Figure 1), but abundance, not so surprisingly, is also regulated at the mRNA level, a fact long ignored in p53 biology.
Domain-structure of human p53. The p53 protein consists of six major domains: TAD1 and 2; amino-acid residues 1–43 and 44–60; the PRD, residues 40–92; the DBD, residues 100–300, the 4D, residues 307–355, and the CTD, residues 356–393). NLS, nuclear localization signal; NES, nuclear export signal. Post-translational modifications are depicted in the individual domains of p53: P (phosphorylation), Ac (acetylation), Ub (ubiquitination), M (methylation), N (neddylation) and SU (sumoylation)
Transcriptional and Translational Control of p53 Function
In response to a broad range of cellular stresses, such as DNA damage, telomere erosion, aberrant oncogene activation or lack of oxygen (hypoxia), p53 accumulates in the cell and thereby becomes activated, indicating that protein abundance dictates function.1 Generally, p53 protein accumulation is not considered to be due to an enhanced transcriptional response of the cell but the consequence of p53 protein stabilization as a consequence of a range of post-translational modifications.56 However, this dogmatic view has been challenged recently by an elegant study, which showed for the first time that the translational regulation of p53 is modified in response to DNA damage by the ribosomal protein L26 and nucleolin, two interacting proteins found in the nucleolin-containing ribonucleocomplex, that bind to the 5′UTR of p53 mRNA and co-regulate its abundance with opposing functions.57 Furthermore, in germinal centre B cells, p53 mRNA and protein expression is repressed by the proto-oncogene BCL6, which appears to block p53 transcriptional activation in response to physiological levels of DNA damage, required for immunoglobulin class-switch recombination and somatic hypermutation.58
As p53 was cloned at a time when PCR technology was not yet available the possible existence of splice variants was ignored for a long time. This issue has been extensively investigated in the later discovered p53 relatives, p63 and p73. These proteins exist in multiple shapes, colours, flavours and sizes, which often display distinct biological properties. For example, the N-terminally truncated versions ΔNp63 and ΔNp73 appear to interfere in a dominant negative manner with the action of their corresponding full-length counterparts.59 This may have sparked the necessary interest to ‘re-clone’ p53.60 Until recently, only one alternative isoform has been described for mouse p53, generated by alternative splicing of intron 1061 and four isoforms have been reported in humans, p53, p53i9, Δ40p53 (also called ΔN-p53) and Δp53. No evidence was found that p53i9, lacking the last 60 aa of p53 would be functional or expressed endogenously in cells.62 The third isoform, ΔNp53, which lacks part of the first TAD (Figure 1), can still activate gene transcription when overexpressed, but acts in a dominant-negative manner against wt p53, interfering with p53-induced apoptosis as well as MDM2-mediated degradation.63 Δp53 is generated by alternative splicing lacking 66 aa residues (257–322) encoded by exons 7–9 and appears to be expressed only in primates.64 The deletion mutant, Δp53, reportedly can only form oligomers with itself, still binds to p53 response sites, but activates only a distinct subset of p53-target genes such as p21 and 14-3-3σ efficiently, thereby mediating ATR-dependent inhibition of S-phase progression in damaged cells.64
Re-evaluation of the p53 gene structure and RACE-PCR analysis revealed that at least six different mRNAs are expressed in normal human tissue, due to alternative splicing, as well as internal promoter usage. One of the novel isoforms, denoted p53β was investigated in greater detail and was shown to hetero-dimerize with wt p53 and to preferentially bind to the promoters of the bax and p21 genes but not to mdm2.60 Although p53β on its own was not able to activate p21 or bax expression in promoter reporter studies, it appeared to act in concert with wt p53 on the bax-gene promoter, particularly under conditions of stress. This finding did, however, not correlate with apoptosis induction, as co-expression of wt p53 together with p53β did not lead to increased apoptosis, when compared with cells overexpressing wt p53 alone.60 In this assay system, a different isoform of p53, Δ133p53, which is transcribed from the internal promoter found in exon 4 clearly acted as a potent dominant negative for wt p53 in apoptosis induction.60
At the moment, it is unclear what the biological function of the individual p53 isoforms might be. However, we can speculate on the possible role of Δ133p53, as a mouse model expressing a truncated version of p53 lacking the N-terminal 6 exons, generated from an ES cell clone that carried an aberrant gene-targeting event,65 called p53+/m, has been investigated. This truncated product, called p53m, lacking the TAD, PRD and a significant part of the DBD appeared to stabilize wt p53 in MEF from p53+/m mice and increase its transcriptional potential, at least on p21 expression, the only target gene analysed in this study. Although about 45% of wt and more than 80% of p53+/− mice developed malignant disease during their life, less than 10% of all p53+/m mice showed signs of malignancy. However, although the tumour resistance was significantly increased (subsequently shown to depend on the presence of wt p53), p53+/m mice died earlier than wt mice, displaying features of premature ageing such as strong weight loss and lordokyphosis but no other signs of overt disease.65 This implicates that tumour suppression comes at the price of premature ageing, a finding that has been regarded as controversial.66 Expression of extra copies of wild-type p53 in a BAC-transgenic mouse model did not cause such a premature ageing phenotype but showed clearly decreased tumour susceptibility in response to different carcinogenesis protocols.66 However, a possible explanation for these conflicting results could be that the truncated p53 protein may induce initiators of apoptosis, such as Puma or Noxa, more efficiently than wild-type p53 (even if present in excess), thereby causing a generalized organ atrophy leading to the premature ageing phenotype reported in the p53+/m mutant mouse strain. Unfortunately, the analysis of the impact of the p53m mutant on target-gene transcription was limited to p21 promoter–reporter assays. This observation, however, indicates that the p53m mutant, although similar in composition at the mRNA level, may be fundamentally different from Δ133p53, which appears to act as a dominant negative on p53-induced apoptosis (and presumably also target-gene expression), at least when overexpressed in vitro. Unfortunately, no common target genes were evaluated between these studies that would prove or disprove our theory.
The previously reported ΔNp53 (Δ40p53), lacking a substantial part of the first TAD, arises from an alternative translation initiation site encoded in exon 4, which is used when wt p53 inhibits its own translation by binding to a stem-loop structure in the 5′UTR of its mRNA.67 This amounts to approximately 5% of the level of full-length p53 transcripts in healthy cells and can be readily detected in various cell types.63 The auto-repression of p53 translation can be relieved by MDM2-mediated degradation of p53, bound to its own 5′UTR, boosting expression of the full-length transcript. Owing to the lack of the N-terminus, ΔNp53 is not subjected to MDM2-dependent proteolytic degradation and therefore has a much longer half-lives than full-length p53.63 A shorter p53 transcript encoding a mouse version of ΔNp53 was subsequently also detected using RNase protection assay and this was designated p44.68 Low-level expression of p44 appears to enhance p53 function, maybe due to increased overall stability of p53/p44 hetero-tetrameric complexes compared with p53-homo-tetramers. On the other hand, high-level expression of p44 was shown to have a negative effect on p53-mediated gene transcription, presumably by blocking p53-response elements.63 Interestingly, moderate levels of p44 expression in transgenic mice was reported to cause growth retardation, consistent with previously reported growth suppressive functions in cell culture, premature senescence and ageing.68 The latter feature was also reported in the p53+/m mouse model.65 In both studies, the effects were dependent on the presence of endogenous p53. Mice expressing the p44 transgene on the p53−/− background showed a tumour incidence similar to p53−/− mice. Whether p44 transgenic mice show reduced tumour formation, as reported for the p53+/m mice65 was not assessed due to the short lifespan of these animals of ∼60 weeks, a time frame where wt mice do not develop spontaneous tumours. The generation of p53+/− mice expressing the transgene or the analysis of radiation-induced lymphomas may have been useful to address this question. Overexpression of p44 appeared to enhance the transactivation of mdm2, IGF-BP3 and p21.68 Sustained levels of p21 in response to p53 activation were also reported in the p53+/m mice and we can therefore speculate that this may contribute to inefficient cell division in adult stem cells, negatively affecting tissue renewal and premature ageing. Unfortunately, in both studies, a possible relationship between increased spontaneous apoptosis and the observed organ atrophy was not investigated.
Taken together, these experiments indicate that p53 function is essential to prevent tumourigenesis, as appreciated for a long time, whereas the balance between the full length and N-terminally truncated p44 isoform of the molecule may determine its influence on ageing, cellular senescence and perhaps other processes.65, 68
Post-Translational Modifications of p53
The number of post-translational modifications on p53 that have been reported appears endless. Mono- and poly-ubiquitination, sumoylation and neddylation compete with methylation, acetylation and prolyl-isomerization as well as phosphorylation on multiple sites for physiological relevance (Figure 2). Below, we focus on these modifications, particularly those for which in vivo data based on the analyisis of p53 knock-in mouse models is available. We would also like to refer to a number of excellent reviews, which have summarized these findings lately.56, 69, 70 We try to give an update on this rapidly moving field and to add an additional angle by summing up information on how p53 target gene expression is affected in these mouse models and put this into the context of apoptosis induction and tumour suppression (Table 1).
Proteins known to modify p53 post-translationally. The p53-modifications of specific amino-acid residues are indicated in (a) with P (phosphorylation), and in (b) Ac (acetylation), Ub (ubiquitination), M (methylation), N (neddylation) and SU (sumoylation). Proteins responsible for the relevant modifications are depicted in boxes in matching shades of grey. Abbreviations: ATM, ataxia telangectasia mutated; ATR, ataxia telangectasia and Rad3-related protein; AURKA, aurora kinase A; CDK, cyclin-dependent kinase; Chk, checkpoint kinase; CK, casein kinase; CSN, cop-9 signalosome; DNA-PK, DNA-dependent protein kinase; DYRK2, dual-specificity tyrosine-phosphorylation-regulated kinase 2; E4F1, Ubiquitin E3 ligase; ERK, extracellular signal-regulated kinase; GSK3β, glycogen synthase kinase 3 β; HIPK2, homeodomain-interacting protein kinase2; hMOF, human ortholog of the Drosophila MOF gene (males absent on the first); JNK c-Jun NH2-terminal kinase; MAPKAPK2, mitogen-activated protein kinase-activated protein kinase2; p38, p38 kinase; MDM2, mouse double-minute 2 protein; PCAF, p300/CBP associated factor; PKC, protein kinase C; PKR, double stranded RNA-activated kinase; SET9, SET9 methyltransferase; SMYD2, SET/MYND domain-containing methyltransferase 2; SUMO, small ubiquitin-like modifier 1; TAF1, TATA-binding protein-associated factor1; Tip60, Tat-interactive protein. § constitutively phosphorylated and dephosphorylated in response to ionoizing radiation.* Modification caused by UV-light only. # Overexpression of AURKA causes phosphorylation of p53 at Ser315
Regulation of p53 by Ubiquitination and Proteasomal Degradation
In healthy (unstressed) cells, low concentrations of p53 protein are maintained by binding to E3 ubiquitin-ligases such as the Hdm2 (human homolog of Mdm2) protein (Mdm2 in mouse: mouse double minute 2),86 COP1 (constitutively photomorphogenic 1),87 Pirh2 (p53-induced, RING-H2 domain containing)88 and ARF-BP1 (ARF-binding protein 1),89 which target p53 for proteosomal degradation. Mdm2, COP1 and pirh2 are also transcriptional targets of p53, securing its self-destruction under physiological, stress-free conditions (for a recent review see also).90
P53 is subjected to both poly- and mono-ubiquitination. Poly-ubiqutination targets p53 for proteasomal degradation, whereas mono-ubiquitination promotes nuclear export of p53. Both processes can be catalysed by MDM2. Low levels of MDM2 activity induce mono-ubiquitination that allows for further modifications of p53, such as PIASy-mediated sumoylation, promoting nuclear export of p53, whereas high levels promote poly-ubiquitination and proteasomal degradation of p53.91, 92 It has therefore been proposed that MDM2 keeps p53 in check by mono-ubiqitination and nuclear export in unstressed cells, in which the E3-ligase is found at low levels, whereas polyubiquitination and degradation by MDM2 is considered to play a role during late phases of the DNA damage response when damage has been successfully repaired to allow re-entry into the cell cycle.92 Genetic evidence strongly supports the idea that MDM2 is the most critical regulator of p53 abundance due to the fact that embryonic lethality caused by loss of mdm2 can be completely rescued by concomitant loss of p53,93, 94 thus making us wonder how essential other E3 ligases such as COP1 may be for p53 regulation.
P53 can also be modified post-translationally by the ubiquitin-like molecules NEDD8 (neural precursor cell expressed developmentally downregulated 8) and SUMO-1 (small ubiquitin-like modifier-1). The functional consequences of these modifications are less well understood. Neddylation of human p53 at lysines in the CTD (Lys370, 372, 373) occurs in an MDM2-dependent manner and inhibits the transcriptional activity of p53, at least in overexpression studies.95 In contrast, neddylation of Lys320 or Lys321 in human p53 can be mediated by FBXO11, a member of the F-box protein family and a component of the Skp1.Cullin1.F-box (SCF) complex, which inhibits p53 function.96 SUMO-1 can be attached to human p53 at Lys386, which was report to enhance the transactivation activity of p53 in vitro.97, 98 Sumoylation is mediated by the SUMO-1 activating enzyme ubc9 and is promoted by MDM2 and ARF.99 Some studies found that a K386R sumoylation-resistant mutant of p53 had a weaker transcriptional activity and slightly impaired proapoptotic activity compared with wt p53,100 but others could not find any difference between the K386R mutant and wt p53 in transactivation and cell growth suppression assays.101
Attempts to Interfere with p53 Proteasomal Degradation in vivo
The PRD that follows TAD1 and overlaps with TAD2 in p53 (Figure 1) has been implicated in the modulation of p53 stability, mainly by prolyl-isomerase-mediated conformational changes of prolin residues (Figure 2). These are thought to reduce MDM2 binding and, hence, cause enhanced protein stability,102, 103 as well as transactivation potential via binding of the histone acetyl transferase (HAT) p300.104, 105, 106 Based on in vitro studies, the PRD was also considered essential for the proapoptotic activity of p53, as ΔPRD mutants of p53, lacking the entire PRD, were reported to induce cell cycle arrest but no longer apoptosis.107, 108 The PRD was also reported to be responsible for apoptosis induction in transactivation-deficient mutants of p53.51 Although it is unclear what consequences deletion of the PRD domains really has on overall structure of p53, mice expressing such a mutant were generated and heterozygous intercrosses gave rise to offspring at the expected Mendelian numbers, without gender bias.84 In contrast, a significant number (>50% on an inbred 129/Ola background) of female p53−/− mice die in utero due to defects in neuronal tube closure.109, 110 It has therefore been proposed that the ΔPRD version of p53 retains sufficient proapoptotic potential during embryogenesis to allow normal neural tube closure. The ΔPRD mutant of p53 was normally phophorylated in response to DNA damage but showed reduced transactivation potential on some target genes, most prominently noxa and p21.84 In contrast, induction of mdm2 or puma was not, or only mildly, impaired after doxorubicin treatment of MEF.84 In agreement with data from transfection studies, the overall half-life of the ΔPRD mutant was found to be significantly reduced compared with wt p53, due to increased sensitivity to MDM2-mediated degradation.84 Defects in target gene transcription, again, were most likely due to secondary effects on co-activator recruitment, as promoter binding appeared unaffected. Consistent with the most prominent effects on target gene transcription, cell cycle arrest functions were impaired in ΔPRD/ΔPRD homozygous cells. This was in contrast to in vitro studies, in which apoptotic responses in E1A-transduced MEF exposed to various forms of DNA damage inducing drugs were only significantly different to wt cells when low concentrations of these agents were used. This demonstrates that the ΔPRD mutant retains a reduced but still significant proapoptotic potential. This correlates well with the observation that expression of Noxa and Puma is impaired (albeit to varying degrees; noxa > puma), but not abrogated in response to DNA damage. Consistently, only very few ΔPRD/ΔPRD homozygous animals were reported to develop spontaneous tumours.84
Toledo et al. also addressed the question whether the internal deletion of the PRD may cause the observed phenotypes in p53ΔPRD/ΔPRD mice by interfering with the overall structure of p53 or by preventing the interaction with the co-activator p300 or the prolyl isomerase Pin1, respectively. In order to examine this, they generated two more p53 knock-in mouse strains: one lacking Pin1-binding sites (Thr81 in human p53), by mutating the relevant threonines in mouse p53, Thr76 and Thr86, to alanin (p53TTAA) and another strain, which retained these sites but lacked the four critical proline residues (79, 82, 84, 87) in this region, making up the tandem PXXP sites (p53AXXA). Animals carrying either of these mutations were born at the expected Mendelian ratio, indicating that the mutated p53 proteins maintain the majority of their proapoptotic functions during embryogenesis.85 Stabilization of p53 in response to DNA damage was comparable with wt p53 in the p53AXXA mutant but reduced in the p53TTAA mutant cells, consistent with the in vitro observations, showing that the interaction with Pin1 promotes p53 stabilization by interfering with MDM2-mediated degradation.102, 103 However, p53TTAA accumulated with delayed kinetics and Pin1 binding was observed to p53TTAA in response to UV-irradiation, indicating the existence of Pin1-binding sites outside the PRD. In contrast to the p53ΔPRD/ΔPRD mutant, activation of mdm2 and p21 in response to doxorubicin treatment was not affected, nor was replicative senescence, pointing to a functional cell cycle arrest in the p53AXXA and p53TTAA mutant animals. Most strikingly, apoptotic responses in MEF or primary thymocytes were unaffected by either of these two mutations. Consistently, oncogene-induced tumourigenesis in E1A/RAS xenograft assays was also efficiently suppressed.85
In summary, these data indicate that the deletion of the PRD has much more profound structural consequences for p53 than previously anticipated and that PXXP-dependent protein interactions are dispensable for transcriptional activation of p53 target genes and apoptosis induction. Pin1 interaction may fine-tune p53 abundance but does not interfere with its transcriptional potential. Although one would also have predicted that deletion of the four key-prolin residues in the PRD should substantially reduce the proposed transcription-independent proapoptotic action of p53, DNA damage-induced apoptosis occurred normally in these mutant cells.85
Overexpression experiments indicated that insertion of two point mutations in the TAD, changing leucin 25 to glutamin and tryptophan 26 to serine (residues 22 and 23 in human p53) disrupt MDM2-binding, leading to constitutive high-level expression of a transcription-deficient mutant, p53QS that was found to be able to bind constitutively to DNA.81 In agreement with previous data, implicating MDM2 in nuclear-cytoplasmic shuttling of p53, mutant p53QS was found to be abundant not only in the nucleus but also in the cytoplasm of MEF expressing this p53 mutant. The distorted distribution pattern did not change in response to DNA damage, highlighting the role of MDM2 in nuclear-cytoplasmic shuttling of p53. DNA damage-mediated induction of p21 was abrogated in these mutant cells, as was thymocyte apoptosis. Oncogenic transformation of MEF was as readily achieved in p53QS MEF as in p53−/− MEF and E1A/RasV12-transduced colonies formed tumours in nude mice with comparable efficiency as similarly transformed p53-deficient MEF. These results indicate that the p53QS mutant behaves like a null allele.81 Subsequent sequencing of the expressed mutant p53 revealed that the p53QS mutant carried an additional alanin to valine exchange at position 135. However, two additional laboratories reported the generation of cells expressing p53QS. Both studies observed essentially identical phenotypes as the first group, regarding apoptosis induction and cell cycle arrest in ES cells, MEF and thymocytes.82, 83
Collectively, these results demonstrate the importance of transcriptional transactivation of target genes by p53 for cell cycle arrest and apoptosis induction. Interestingly, despite retaining the intact PRD, which has been implicated in transcription-independent apoptosis, this p53 mutant was unable to induce apoptosis in response to DNA damage.83, 111 Surprisingly, bax mRNA induction in response to doxorubicin appeared largely unaffected, whereas induction of noxa or p21 was impaired in the p53QS-expressing cells. Overexposure of northern blots suggested, however, that this p53 mutant retains some transactivation capacity, at least on the p21 gene.83 Unfortunately, this was not assessed in direct comparison with p53−/− cells, which would have been informative given that p53-independent activation of p21 in response to DNA damage, for example by UV, has been described previously.112, 113 The observation that the p53QS mutant does not protect E1A-expressing MEF from hypoxia-induced cell death may simply be due to different target-gene modulation for cell death induction, rather than transcription-independent proapoptotic functions of p53QS. Consistent with this idea, p53 accumulation in response to hypoxia is not associated with the post-translational modifications seen after DNA damage and apoptosis under hypoxic conditions appears mediated mostly by trans-repression of target genes via recruitment of co-repressors, such as mSin3A.114 This function may still be fulfilled by p53QS especially, as it was found to bind constitutively to (at least certain) promoter sequences.
Regulation of p53 by Phosphorylation
Numerous serine and threonine residues, mainly located within the N-terminal TAD and the CTD of p53 are targets for phosphorylation (Figure 2). Various kinases, including ATM , ATR, Chk1 (checkpoint kinase 1), Chk2, CK1 (casein kinase 1), JNK, HIPK2 and DYRK2, have been shown to phosphorylate p53 after DNA damage (for a recent review see also).56 Most sites that are phosphorylated in response to a stress signal lead to p53 protein stabilization, and thereby enhance its function and/or affect the binding specificity of p53 to target sequences in the genome. In addition to stress-induced phosphorylation, there are also a few sites in p53, which are constitutively phosphorylated (e.g. Ser378) and certain sites are actually de-phosphorylated in response to stress signals, such as Ser376 and Thr55.115, 116
The phosphorylation of the amino-acid residues Ser15, Thr18 and Ser20 in the N-terminal TAD domain of human p53 has been studied most extensively. These residues are located in, or close to, the region in p53 that also binds to MDM2.56, 116 Ser15 is phosphorylated in an ATM-dependent manner early in response to γ-irradiation but is not phosphorylated upon exposure to UV-light.115, 117 Data from in vitro or overexpression studies indicate that phosphorylation at this position stimulates p53-dependent transactivation, growth arrest and apoptosis in response to DNA damage.118, 119, 120 However, there exist conflicting data on whether Ser15 phosphorylation affects MDM2 binding or not.118, 121 Phosphorylation of other residues, including Thr18 and Ser20, which occurs later in the response to DNA damage, was shown to depend on initial phosphorylation at Ser15.122, 123 Phosphorylation of Thr18 and Ser20 has been found in numerous studies to interfere with the interaction of p53 with MDM2, preventing the ubiquitination of p53 and thus promoting its stabilization.124, 125
In contrast to the proposed inhibition of MDM2/p53 interaction, phosphorylation of Ser46 by the kinases HIPK-2126 or DYRK-2127 was reported to mediate selectivity in promoter binding by p53 and to specifically promote the induction of apoptosis inducing genes, such as p53-regulated apoptosis-inducing protein 1 (p53AIP).128 Phosphorylation of Ser315, on the other hand, was reported to increase the transactivation potential of human p53 in response to irradiation damage,129 possibly by promoting nuclear retention, mediated via interaction of p53 with E2F1.130 Phosphorylation at the same site by aurora kinase A, however, was suggested to promote MDM2-dependent ubiquitination and proteolysis of p53,131 arguing for an inhibitory role of this modification.
Ser392 within the CTD of p53 (Ser389 in mouse p53) is phosphorylated by p38 MAPK or casein kinase 2 in response to UV-irradiation, but only very inefficiently after γ-irradiation.132, 133, 134, 135
In summary, the relevance of phosphorylation of p53 in its stabilization and activation remains controversial. Some studies investigating phosphorylation-deficient mutants of p53, generated by alanine-replacement of serine and threonine residues at single residues, or, combinations of up to twelve residues failed to show any effect on p53 stabilization, activation of target gene transcription or cellular responses.136, 137 However, the massive overexpression of these mutant proteins may have obscured effects that might be evident if p53 levels are limiting, that is in normal cells.
Investigations on the in vivo Relevance of p53 Phosphorylation
As mentioned above, in humans, a number of serine residues at the very N-terminus of p53, comprising the first TAD, become phosphorylated in response to DNA damage caused by γ- or UV-irradiation.56
Two groups established mouse models that express a mutant version of p53 that can no longer be phosphorylated at Ser18 by ATM in response to DNA damage (corresponding to Ser15 in human p53).71, 72 Both studies observed an abnormally reduced sensitivity of p53S18A homozygous mutant thymocytes to DNA damage induced apoptosis, and the extent of protection was similar to the one conferred by the loss of one p53 allele in mice. In cells from both mouse strains, stabilization of mutant p53S18A protein after DNA damage appeared normal. Gene expression profiling using RNA from γ-irradiated thymocytes revealed defects in the induction of the p53 targeted cell cycle regulators p21 and 14-3-3σ, but proapoptotic targets such as APAF-1, bax or the BH3-only protein bik/blk, previously reported to mediate E1A-induced cell death in a p53-dependent manner,138 were normally induced, suggesting a promoter-specific impact of the S18A mutation.72 In addition, qRT-PCR confirmed differential expression of most of the genes found differentially expressed on the micro-arrays and additionally revealed impaired induction of the mRNA coding for the BH3-only protein Noxa. The reduced induction of noxa is, however, irrelevant for the observed apoptosis resistance of thymocytes, as Noxa-deficient lymphoid cells are normally sensitive to γ-irradiation.44 Consistent with the abnormally increased resistance of p53S18A thymocytes to DNA damage, the other study reported reduced induction of Puma, the key p53-effector in lymphocyte apoptosis in response to γ-irradiation or DNA damage-inducing drugs.44 The differences observed in target gene induction between the two studies is most likely due to the fact that older generations of Affimetrix chips, as the ones used here, did not contain all relevant p53 target genes, in particular puma.
Contradicting results were observed regarding the effects of the p53S18A mutant on DNA damage-induced cell cycle arrest in MEF. This may, at least in part, be due to the fact that different stimuli were used to trigger cell cycle arrest. In response to UV-irradiation, p53S18A homozygous mutant MEF failed to undergo normal cell cycle arrest in one study, consistent with the reduced induction of p21 mRNA seen in the micro-array analysis. In contrast, in the other study, no defects in cell cycle arrest or differences in p21-induction in MEF subjected to γ-irradiation. This may point to differential requirements regarding Ser18 phosphorylation for p21-induction, depending on the form of DNA damage and the subsequently activated checkpoint kinases.
Interestingly, the p53S18A mutation did not appear to affect binding to a number of target gene promoters, as assessed by ChIP. This indicates that impaired co-regulator recruitment or promoter-specific transactivation defects may be responsible for the observed phenotypes. Both studies failed to observe the increased rates of spontaneous tumourigenesis in p53S18A homozygous mutant mice, indicating that phosphorylation of Ser18 alone, is not relevant for the tumour suppressor function.71, 72 However, given the fact that the most prominent defects were observed in response to γ-irradiation in thymocytes, investigation of radiation-induced thymic lymphoma development may have helped to unravel minor differences in tumour suppression more easily.
A number of studies have indicated that phosphorylation of Ser20 by Chk1 and/or Chk2 in human p53 (Ser23 in the mouse p53) is critical for its stabilization in response to DNA damage, most likely by interfering with MDM2 binding and subsequent proteasomal degradation (see above). Consistent with this notion, chk2−/− cells are defective in p53 stabilization in response to γ-irradiation.139 Using different blastocyst complementation approaches, Xu and co-workers generated MEFs and thymocytes that express a single p53S23A/− mutant allele of p53, using a p53+/− ES cell line where exons 2–4 were replaced by a loxP element on one allele, for gene targeting.73 Loss of p53 phosphorylation at the Ser23 site was confirmed in ES cells using phospho-site-specific antibodies but stabilization of p53 occurred normally in response to UV- or γ-irradiation. Induction of p21 and mdm2 in response to UV-irradiation in cells expressing mutant p53 did not differ from MEF or ES cells expressing wt p53.73 Furthermore, the amount of MDM2 that could be co-immunoprecipitated with p53 did not differ between wt and mutant p53. This indicates that Ser23 phosphorylation on its own cannot disrupt p53/MDM2 interaction. Consistent with this hypothesis, UV-irradiation induced apoptosis in ES cells or γ-irradiation induced apoptosis in thymocytes was normal in cells expressing mutant p53.73 This indicates that transcriptional activation of the relevant p53-effectors mediating UV- and γ-irradiation-induced apoptosis, that is Noxa and Puma respectively,44, 47 probably occurs normally in the p53 mutant cells. A small number of p53S23A rag2−/− chimeric mice were followed for up to 8 months, a time frame during which most p53-deficient animals develop thymic lymphomas or fibrosarcomas, but none of the p53S23A animals presented with malignant disease.73 This, however, does not prove that Ser23 phosphorylation is dispensable for p53 tumour suppressor function in vivo. It is conceivable that an unknown number of downstream effectors may not be regulated normally in p53S23A mutant cells and a more comprehensive analysis of p53 target gene expression and a longer follow-up of chimeric animals may have revealed subtle impairments of p53 function, as observed in a similar study by Jacks and colleagues.74 Mice that express mutated p53S23A protein in all somatic cells presented with increased B-cell lymphoma incidence in their second half of life and, although p53 stabilization in MEF appeared largely normal, as in the previous study, thymocytes were reported to be partially refractory to γ-irradiation-induced apoptosis. This partial resistance correlated with decreased protein stabilization in p53S23A homozygous mutant thymocytes exposed to γ-irradiation in vitro or in vivo.74 TUNEL analysis of spleens and the developing cerebellum, collected after whole body γ-irradiation revealed strongly reduced numbers of apoptotic cells in tissues from p53S23A mice compared with control wt animals. This indicates that the p53S23A mutation may cause a defect in puma induction. p53 protein accumulation appeared strongly impaired in the brains of γ-irradiated p53S23A mutant mice. We therefore assume that the overall transcriptional response activated by p53 must have been strongly impaired (due to its low levels) rather than mutant p53S23A exhibiting a specific defect in engaging target gene promoters. The molecular basis for the difference in apoptosis susceptibility of thymocytes between the two studies remains unknown.
It is interesting that p53S23A/S23A homozygous animals developed mostly B-cell lymphomas and some sarcomas but not the classical thymic lymphomas, observed in p53−/− mice. A possible explanation for these findings may be a cell type-specific threshold of p53 activity that is necessary to delete highly sensitive thymocytes versus mature T and B cells in response to DNA damage or oncogenic stress. Alternatively, this may also reflect the fact that these mice were kept on a mixed genetic background well known to modulate the tumour spectrum caused by the loss of p53.
Recently, a combined mutant mouse model expressing a p53S18/23A mutant protein was described.75 Accumulation of this mutant p53 in response to DNA damage in MEF appeared normal, although the overall stability appeared increased. Expression of classical p53 target genes, such as p21, noxa, bax and dr5, was significantly reduced, but not more than in p53S18A single mutant cells. This indicates that Ser18 and Ser23 phosphorylation may exert distinct functions in vivo. Upon γ-irradiation, cell cycle arrest was partially impaired in the p53S18/23A MEF, but this defect was clearly less pronounced compared with p53−/− MEF. The propensity to undergo replicative senescence was only mildly impaired, suggesting that the reduced target gene expression is still sufficient to execute at least certain key effector functions of p53.75 In contrast, thymocytes expressing p53S18/23A were almost as resistant to DNA damage-induced apoptosis as p53−/− cells, indicating overlapping functions for both amino acid residues in the induction of proapoptotic target genes, at least in this cell type. In contrast to the findings in MEF, stabilization of p53S18/23A in thymocytes was drastically impaired and the mutated protein appeared to be degraded more rapidly, which is consistent with the hypothesis that mainly Ser23 and Ser18 phosphorylation promote p53-stabilization and by interfering with MDM2-binding in response to DNA damage. In line with decreased p53 stabilization, induction of the proapoptotic targets noxa, bax, dr5 or puma was abnormally low (∼10–20% compared with wt cells).75 Again, it must be assumed that loss of puma induction may be the most critical event contributing to the apoptosis resistance of these p53S18/23A mutant thymocytes. The extent of protection observed in p53S18/23A thymocytes after γ-irradiation is comparable with those afforded by loss of Puma, but still lower than the protection observed in p53−/− cells.75 Which p53 target(s) mediate Puma-independent thymocyte apoptosis in response to γ-irradiation is unclear at present, but Noxa and/or Bim are possible candidates.49, 140
Consistent with impaired cell cycle arrest and apoptosis function of the p53S18/23A mutant, mice expressing this protein were abnormally prone to tumourigenesis, although tumour incidence was significantly lower and latency longer compared with p53−/− mice. Tumour latency and the heterogeneous tumour spectrum in these animals appeared to be comparable with the one observed in p53S23A/S23A mice and this fits with the observation that p53S18A/S18A mutant mice were not abnormally tumour prone.72, 74, 75, 141
Collectively, these results support the idea that phosphorylation of Ser23 predominantly interferes with MDM2 binding and subsequent p53 degradation. In addition, phosphorylation of both serines affects the expression of a number of p53 target genes required for p53-mediated cell death and cell cycle arrest, maybe by differential recruitment of co-activators to some but not all target gene promoters. This indicates that loss of both p53-induced cell cycle arrest and apoptosis are required for tumourigenesis. Moreover, it is possible that oncogene-induced senescence is also critical for the tumour suppressive function of p53, as indicated by studies with mice expressing conditionally active forms of p53.142, 143 It is presently not known whether activation of the senescence pathway is defective in cells expressing p53S18/23A or p53S23A mutant protein.
Phosphorylation of Ser46 in human p53 was reported to play a relevant role in the p53-mediated DNA damage response by inducing the proapoptotic factor p53AIP.128 Ser46 can be phosphorylated by at least two kinases, HIPK2144 and DYRK2.127 Remarkably, this residue does not appear to be conserved between mouse and man, but Ser37 (localized within the PRD) in mice may carry out an equivalent function to human Ser46. Therefore, Xu and co-workers took advantage of the human p53 knock-in ‘HUPKI’ mouse model (here referred to as: hki), in which a substantial portion of mouse p53 sequences was replaced by human p53 sequences.145 Introducing the relevant mutation in the previously used targeting construct, they generated p53hkiS46A mice.76 They found that the human-derived Ser46 residue was phophorylated in MEF in response to γ- or UV-irradiation, but the time of p53 stabilization appeared shorter in p53hkiS46A cells compared with cells expressing p53hki. Induction of the proapoptotic target genes noxa, dr5, pidd (a death domain containing protein involved in caspase-2 activation), puma as well as p21, perp and mdm2 revealed that expression of noxa and perp were most affected, whereas the induction of all other targets was, if anything, only moderately reduced compared with ‘wt’ p53hki cells.76 Unfortunately, p53AIP, a target specifically induced in response to Ser46 phosphorylation in human cells127 was not evaluated in this study. However, as Puma and Noxa account for most, if not all the proapoptotic activity of p53, the role of p53AIP in cell death is uncertain. In E1A/Ras transformed MEF, loss of Ser46 phosphorylation in p53 resulted in partially reduced apoptosis in response to DNA damage, and this correlated well with impaired induction of noxa and puma, whereas pidd or dr5 were unaffected. Loss of p53, again, proved more potent in inhibiting death of transformed MEF, indicating that other modifications in p53 must also contribute to the induction of Puma and Noxa expression or that additional targets required for killing, but not assessed by qRT-PCR, are induced normally in the p53hkiS46A cells. Death of thymocytes after γ-irradiation was only modestly affected, if at all, further pointing to cell type specific post-translational requirements for p53 function.76
Analysis of mouse ES cells expressing p53hkiS315A generated in the same lab, revealed that Ser315 phosphorylation is critical for activating p53 transcriptional activities during differentiation as exemplified by impaired induction of p21 and repression of the homeodomain protein gene nanog.146 Nanog is required to maintain ES cells in an undifferentiated stage. It was proposed that p53-mediated downregulation of nanog, that is also observed in ES cells exposed to DNA damage, serves as an alternative mechanism to maintain genomic stability, by promting ES cell differentiation into cell types that undergo p53-dependent cell cycle arrest or apoptosis more efficiently.146 Unfortunately, the impact of this post-translational modification on either of these functions was not addressed in this study and it will be interesting to see how this modification influences embryonic development and/or tumour suppression by p53S315A in mice.
Last, but not least, mice carrying a Ser389 to alanin mutation (Ser392 in human p53) were found to have increased sensitivity to UV-irradiation induced skin carcinogenesis, consistent with the UV-specific induction of Ser389 phosphorylation.135, 147 P53 protein stability, however, was not impaired in p53S389A mutant cells, but the DNA-binding efficiency of p53S389A was reduced after UV-treatment compared with wt p53.77 Whether this is due to impaired tetramerization is unclear, but it would be interesting to test if this modification modulates interaction of p53 with other p53 isoforms, such as ΔNp53, which is known to act as a dominant negative on some p53-regulated promoters (see above). Consistent with this hypothesis, activation of some target genes, such as mdm2 or p21, was reduced, but induction of the BH3-only protein Noxa was only mildly delayed. Given the prominent role of Noxa in UV-irradiation induced apoptosis of MEF and keratinocytes,47 this observation might explain why only minor apoptosis defects were observed in such cells from these p53 mutant mice.
Spontaneous tumourigenesis was not increased in p53S389A homozygous mice when compared with wt mice and the consequences of p53S389A expression on an already abnormally tumour prone background, such as p53S389A/−, was not assessed. However, after crossing of the animals onto the hairless back-ground (SKH : HR7), chronic UV-B-irradiation induced skin tumours (papillomas and squamous cell carcinomas) occurred significantly earlier in p53S389A mutant mice compared with control wt mice but still later than in p53+/− mice.77 It appears unlikely that this abnormal tumour predisposition is only due to reduced Noxa expression in UV-exposed keratinocytes, but it may well contribute to disease establishment and/or progression. In line with the specific requirement of Ser389 phosphorylation for the nucleotide excision repair response, p53S389A mutant mice were normally susceptible to γ-irradiation-induced thymic lymphomas but showed an increased tumour incidence when treated with carcinogens that cause bulky adducts, such as 2-actylaminofluoren.148
Acetylation and Methylation – can p53 Work without it?
P53 can be acetylated at several lysines by different HAT. CBP/p300 heterodimers acetylate p53 at lysines within the C-terminal domain (Lys370, 372, 373, 381, 382).149 In contrast, Lys320 and Lys305 in the nuclear localization domain are acetylated by PCAF (p300/CBP-associated factor) and p300, respectively.105, 141, 150 Moreover, the MYST family acetyl transferases, hMOF and TIP60, were recently shown to acetylate p53 at Lys120 in the DBD.151, 152 Lys120-acetylated p53 was reported to accumulate preferentially on the promoters of proapoptotic target genes, including bax and puma. Mutants of p53 that can no longer be modified at this residue showed impaired proapoptotic activity in overexpression experiments, whereas cell cycle arrest functions and induction of mdm2 were not affected.151, 152
Acetylation of p53 by p300 and PCAF occurs in response to DNA damaging agents, such as UV- and γ-irradiation. CBP/p300 and PCAF act as co-activators for p53-mediated transcription. However, although some studies could show an enhancement of sequence-specific DNA-binding activity of acetylated p53,105, 106, 153 as well as more potent promotion of transcriptional activation of target genes,154 others suggested that the enhancing effect of CBP/p300 on p53 transactivation function was independent of p53 acetylation.155 As the lysine residues within the CTD of p53 are also targets for ubiquitination, it was proposed that acetylation of these residues may promote the stabilization of p53 by interfering with proteasomal degradation.56, 156
Nevertheless, a number of recent studies have indicated that acetylation of different lysine residues has differential effects on which target genes are activated by p53. Using acetylation-mimicking lysine to glutamine mutations, functional differences between acetylation of Lys320 versus Lys373 were reported.157 Acetylation of Lys320 in p53 was shown to favour interaction with high-affinity p53-binding sites in target genes, promoting cell survival and cell cycle arrest. In contrast, acetylation of Lys373 led to a stronger interaction of p53 with low-affinity binding sites, which are found in proapoptotic target genes and therefore promoted cell death.157
Acetylation levels of p53 can be regulated via deacetylation by an HDAC1 (histone deacetylase 1)-containing complex or by the NAD-dependent histone deaceytylase Sir2α (silent information regulator 2α). Deacetylation was shown to repress p53-dependent transcriptional activation, apoptosis and growth arrest158, 159, 160 MDM2 also seems to interfere with the acetylation of p53 and was shown to inhibit p53 acetylation by p300 and PCAF161, 162 and to promote the HDAC1-mediated deacetylation of p53.163
Finally, lysine residues that can be subjected to acetylation are also targeted by methyl transferases. Methylation of p53 can occur at least two different sites, reported to lead to opposing effects on p53 function. Methylation at Lys372, by the methyl-transferase Set9, increases the stability of p53, restricts it to the nucleus and enhances p53 dependent transcription,164 whereas methylation of Lys370, mediated by another methyl transferase, Smyd2, leads to repression of transcriptional activity.165
Lessons from Mice Expressing Lysine-Deficient Mutants of p53
As mentioned above, a number of biochemical studies suggest that p53 protein stability is regulated by the competition of E3 ligase-mediated ubiquitination and HAT-mediated acetylation. Acetylation of lysine residues prevents ubiquitination (but not binding) by MDM2, thereby increasing p53 stability. A number of studies proposed that the acetylation status might influence co-regulator recruitment as well as sequence-specific DNA binding (see above).
Replacement of six or seven C-terminal lysine residues in p53 by gene targeting in mice (p53K6R and p537KR mice), including the five residues known to be targeted by p300, however, revealed that this caused only surprisingly mild abnormalities. On first sight, this indicates that p53 stabilization is not critically influenced by the proposed E3-ligase/HAT competition for lysine modification. Importantly, the overall ubiquitination of p53 appeared to be only slightly reduced by these mutations indicating that other lysine residues in p53 suffice to promote its MDM2-dependent degradation.79, 78
Consistent with the idea that lysine modifications are relevant for co-repressor recruitment and/or sequence-specific DNA binding, one study reported that expression of proapoptotic target genes (noxa, pidd, puma) was impaired after DNA damage in p53K6R ES cells and thymocytes, but surprisingly not in MEF.78 Along that line, in response to DNA damage, apoptosis was abnormally reduced in ES cells and thymocytes expressing mutant p53K6R but occurred normally in MEF.78 A minor resistance of thymocytes was reported in the K6R study and this correlated with reduced expression of dr5 and puma mRNA.78 In contrast, analysis of p537KR mutant mice showed that in thymocytes that have sustained DNA damage this p53 mutant was actually stabilized abnormally rapidly, and this correlated with accelerated transcription of the only common target analysed, puma.79 The difference regarding puma activation may be due to the fact that a non-conserved lysine in mouse, at position 394, was not mutated in the p536KR mutant mice and may still be a target for modification, impacting on Puma expression. In summary, these studies indicated that acetylation of C-terminal lysines may mediate cell-type-specific target gene expression but are not rate-limiting for modulation of p53 protein stability or apoptosis induction. As the same residues mutated in the aforementioned knock-in mice are targets for methyl-transferases, one might speculate that methylation of lysine residues is of minor importance for p53 regulation. However, as MDM2-mediated ubiquitination at the very same lysines is also not possible in the mutant mouse strains, such a conclusion might be premature. Inhibition of site-specific methyl-transferases or p53-acetylating enzymes in vivo will be necessary to clarify this issue.
Lysine modification by PCAF may affect nuclear localization of p53 and specificity of binding to target genes. However, expression of p21, noxa, dr5 and pidd and cell cycle arrest were found to be largely normal in MEF derived from mice expressing p53K317R (K320 in human p53).80 Interestingly, thymocytes and E1A transformed MEF from these mutant mice were found to have abnormally increased susceptibility to DNA damage-induced apoptosis. This hypersensitivity was also observed in vivo, as demonstrated by TUNEL staining of small intestine and retina derived from mice that were exposed to whole body γ-irradiation. In all cases analysed, proapoptotic target gene transcription was increased in p53K317R over wt cells, and noxa as well as puma were most prominently affected.80 A micro-array analysis performed on mRNA derived from wt and p53K317R mutant thymocytes revealed that about 88% of the p53 target genes affected by the K317R mutation were more strongly induced when compared with wt. PCAF-mediated modification of Lys317 appears to counterbalance high level expression of proapoptotic target genes, including noxa, pidd and puma, in a cell type-specific manner, whereas expression of other targets, including p21, mdm2 or gadd45, were not affected by this modification.80 In summary, these results support only in part published in vitro data, which proposed that this modification favours cell survival.157 Changes in cell cycle arrest efficiency, as also proposed, were not observed in vivo. Nevertheless, Lys317 modification by acetylation appears to play a crucial role in fine-tuning proapoptotic target gene expression in vivo. Consistently, neddylation of Lys320 in human p53 (Lys317 in mice) by the F-Box protein FBXO11 was also reported to exert inhibitory effects on p53's apoptotic function in vitro.96
A number of papers have demonstrated that mutations in the DBD of p53, which are frequently found in human tumours, impede its ability to act as a tumour suppressor. Mutant knock-in mice harbouring such mutations (e.g. p53R172H p53R172Pp53hkiR248W or p53R270H) proved to be excellent models of Li–Fraumeini syndrome and they will be useful tools to investigate the impact of these mutations on target gene transcription.166, 167, 168, 169 However, based on these observations, it will be difficult to speculate on the relevance of Lys120 modification by hMOF and TIP60, as it is located in the DBD. Although it was reported that the K120R mutant binds p53 target sites equally well as wt p53, this was only investigated for a limited number of target genes.151 The K120 residue in p53, however, interacts with the major groove of DNA170 and, therefore, possible phenotypes arising in putative K120R knock-in mice may not entirely be due to the effects on target gene expression caused by loss of p53 acetylation.
Conclusions
What can we extract from all these animal models regarding the relevance of post-translational modifications on p53? If we look at the relatively modest phenotypical changes in many of the knock-in mouse strains generated to validate the significance of in vitro observations from overexpression experiments (Table 1), one might conclude that biochemical studies describing various modifications are highly overrated. So far, ubiquitination of p53 by MDM2 is the only post-translational modification that has been confirmed to be also highly relevant in vivo.93, 94 This does not mean that the others are irrelevant or do not occur in vivo (meaning in a living organism, not cells in the culture dish), but nature appears to be able to compensate quite easily for the loss of one or even two or more of these modifications in p53. One might argue, that due to its importance for tumour suppression, there must be such a high degree of redundancy. Conversely, one might say that the in vivo studies only analysed a very restricted set of p53 target genes in a very limited number of cell types, that is ES cells, MEF and thymocytes only, and those that are genuinely influenced by post-translational modifications of p53, were overlooked.
What the in vivo studies show us, however, is that we are left within a dilemma. Biochemical analysis of post-translational protein modifications in cells is highly relevant and appreciated to help us to understand the multiple facets of cell biology and protein function but this technology is also confronted with a number of inherent problems. Overexpression of p53 or site-mutants, either alone or in combination with the potential modifying enzyme, for example, will never reflect the natural levels or balance of the interacting proteins. They are produced in vast excess and the natural spatial-temporal regulation of these molecules can no longer be maintained. In addition, the overall consequences of single or multiple amino acid changes or deletion in p53 on the overall structure and accessibility for modifying enzymes are still hard to predict. Cell lines used for these studies are often transformed or of malignant origin, sometimes carrying countless mutations that deregulate the control of their survival and proliferation in culture by unknown processes. Their response to enforced expression of modified p53 molecules will never reflect biological responses in primary cells or tissues. Maybe the comparative analysis of in vivo labelled proteomes of model organisms, such as mice, will allow us to pin down the relevant modifications of p53. However, even if technologies like this will guide the future, validation of biochemical observations in genetic model organism will always be of paramount importance to separate the wheat from the chaff.
Abbreviations
- TAD:
-
transactivation domains
- PRD:
-
proline-rich domain
- DBD:
-
DNA-binding domain
- 4D:
-
tetramerization domain
- CTD:
-
C-terminal regulatory domain
- NES:
-
nuclear export signal
- NLS:
-
nuclear localization signal
- p53AIP:
-
p53-regulated apoptosis-inducing protein 1
- Puma:
-
p53-upregulated modulator of apoptosis
- Bid:
-
Bcl-2-interacting domain death agonist
- Bak:
-
Bcl-2 antagonist/killer
- Bax:
-
Bcl-2-associated protein X
References
Vousden KH, Lane DP . p53 in health and disease. Nat Rev Mol Cell Biol 2007; 8: 275–283.
Chipuk JE, Green DR . p53's believe it or not: lessons on transcription-independent death. J Clin Immunol 2003; 23: 355–361.
El-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM et al. WAF1, a potential mediator of p53 tumor suppression. Cell 1993; 75: 817–825.
Hermeking H, Lengauer C, Polyak K, He TC, Zhang L, Thiagalingam S et al. 14-3-3 sigma is a p53-regulated inhibitor of G2/M progression. Mol Cell 1997; 1: 3–11.
Kastan MB, Zhan Q, El-Deiry WS, Carrier F, Jacks T, Walsh WV et al. A mammalian cell cycle checkpoint pathway utilizing p53 and lang2057GADD45 is defective in ataxia-telangiectasia. Cell 1992; 71: 587–597.
Harper JW, Adami GR, Wei N, Keyomarsi K, Elledge SJ . The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell 1993; 75: 805–816.
Brugarolas J, Chandrasekaran C, Gordon JI, Beach D, Jacks T, Hannon GJ . Radiation-induced cell cycle arrest compromised by p21 deficiency. Nature 1995; 377: 552–557.
Wang XW, Zhan Q, Coursen JD, Khan MA, Kontny HU, Yu L et al. GADD45 induction of a G2/M cell cycle checkpoint. Proc Natl Acad Sci USA 1999; 96: 3706–3711.
Chan TA, Hermeking H, Lengauer C, Kinzler KW, Vogelstein B . 14-3-3Sigma is required to prevent mitotic catastrophe after DNA damage. Nature 1999; 401: 616–620.
Jin S, Tong T, Fan W, Fan F, Antinore MJ, Zhu X et al. GADD45-induced cell cycle G2-M arrest associates with altered subcellular distribution of cyclin B1 and is independent of p38 kinase activity. Oncogene 2002; 21: 8696–8704.
Zhan Q, Antinore MJ, Wang XW, Carrier F, Smith ML, Harris CC et al. Association with Cdc2 and inhibition of Cdc2/Cyclin B1 kinase activity by the p53-regulated protein Gadd45. Oncogene 1999; 18: 2892–2900.
Bunz F, Dutriaux A, Lengauer C, Waldman T, Zhou S, Brown JP et al. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science 1998; 282: 1497–1501.
Ando T, Kawabe T, Ohara H, Ducommun B, Itoh M, Okamoto T . Involvement of the interaction between p21 and proliferating cell nuclear antigen for the maintenance of G2/M arrest after DNA damage. J Biol Chem 2001; 276: 42971–42977.
Danial NN, Korsmeyer SJ . Cell death: critical control points. Cell 2004; 116: 205–219.
Michalak E, Villunger A, Erlacher M, Strasser A . Death squads enlisted by the tumour suppressor p53. Biochem Biophys Res Commun 2005; 331: 786–798.
Finnberg N, Gruber JJ, Fei P, Rudolph D, Bric A, Kim SH et al. DR5 knockout mice are compromised in radiation-induced apoptosis. Mol Cell Biol 2005; 25: 2000–2013.
Varfolomeev EE, Schuchmann M, Luria V, Chiannilkulchai N, Beckmann JS, Mett IL et al. Targeted disruption of the mouse Caspase 8 gene ablates cell death induction by the TNF receptors, Fas/Apo1, and DR3 and is lethal prenatally. Immunity 1998; 9: 267–276.
Moroni MC, Hickman ES, Denchi EL, Caprara G, Colli E, Cecconi F et al. Apaf-1 fe1031 is a transcriptional target for E2F and p53. Nat Cell Biol 2001; 3: 552–558.
Fortin A, Cregan SP, MacLaurin JG, Kushwaha N, Hickman ES, Thompson CS et al. APAF1 is a key transcriptional target for p53 in the regulation of neuronal cell death. J Cell Biol 2001; 155: 207–216.
Cecconi F, Alvarez-Bolado G, Meyer BI, Roth KA, Gruss P . Apaf-1 (CED-4 homologue) regulates programmed cell death in mammalian development. Cell 1998; 94: 727–737.
Yoshida H, Kong Y-Y, Yoshida R, Elia AJ, Hakem A, Hakem R et al. Apaf1 is required for mitochondrial pathways of apoptosis and brain development. Cell 1998; 94: 739–750.
Marsden VS, O’Connor L, O’Reilly LA, Silke J, Metcalf D, Ekert PG et al. Apoptosis initiated by Bcl-2-regulated caspase activation independently of the cytochrome c/Apaf-1/caspase-9 apoptosome. Nature 2002; 419: 634–637.
Willis SN, Adams JM . Life in the balance: how BH3-only proteins induce apoptosis. Curr Opin Cell Biol 2005; 17: 617–625.
Miyashita T, Reed JC . Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell 1995; 80: 293–299.
Schmidt T, Korner K, Karsunky H, Korsmeyer S, Muller R, Moroy T . The activity of the murine bax promoter is regulated by Sp1/3 and E-box binding proteins but not by p53. Cell Death Differ 1999; 6: 873–882.
Lindsten T, Ross AJ, King A, Zong W, Rathmell JC, Shiels HA et al. The combined functions of proapoptotic Bcl-2 family members Bak and Bax are essential for normal development of multiple tissues. Mol Cell 2000; 6: 1389–1399.
Zong WX, Lindsten T, Ross AJ, MacGregor GR, Thompson CB . BH3-only proteins that bind pro-survival Bcl-2 family members fail to induce apoptosis in the absence of Bax and Bak. Genes Dev 2001; 15: 1481–1486.
Knudson CM, Tung KSK, Tourtellotte WG, Brown GAJ, Korsmeyer SJ . Bax-deficient mice with lymphoid hyperplasia and male germ cell death. Science 1995; 270: 96–99.
McCurrach ME, Connor TMF, Knudson CM, Korsmeyer SJ, Lowe SW . Bax-deficiency promotes drug resistance and oncogenic transformation by attenuating p53-dependent apoptosis. Proc Nat Acad Sci USA 1997; 94: 2345–2349.
Kondo S, Shinomura Y, Miyazaki Y, Kiyohara T, Tsutsui S, Kitamura S et al. Mutations of the bak gene in human gastri c and colorectal cancers. Cancer Res 2000; 60: 4328–4330.
Sax JK, Fei P, Murphy ME, Bernhard E, Korsmeyer SJ, El-Deiry WS . BID regulation by p53 contributes to chemosensitivity. Nat Cell Biol 2002; 4: 842–849.
Guo Y, Srinivasula SM, Druilhe A, Fernandes-Alnemri T, Alnemri ES . Caspase-2 induces apoptosis by releasing proapoptotic proteins from mitochondria. J Biol Chem 2002; 277: 13430–13437.
Gross A, Yin X-M, Wang K, Wei MC, Jockel J, Milliman C et al. Caspase cleaved BID targets mitochondria and is required for cytochrome c release, while BCL-XL prevents this release but not tumor necrosis factor-R1/Fas death. J Biol Chem 1999; 274: 1156–1163.
O’Reilly LA, Ekert P, Harvey N, Marsden V, Cullen L, Vaux DL et al. Caspase-2 is not required for thymocyte or neuronal apoptosis even though cleavage of caspase-2 is dependent on both Apaf-1 and caspase-9. Cell Death Differ 2002; 9: 832–841.
Marsden VS, Ekert PG, Van Delft M, Vaux DL, Adams JM, Strasser A . Bcl-2-regulated apoptosis and cytochrome c release can occur independently of both caspase-2 and caspase-9. J Cell Biol 2004; 165: 775–780.
Kamer I, Sarig R, Zaltsman Y, Niv H, Oberkovitz G, Regev L et al. Proapoptotic BID is an ATM effector in the DNA-damage response. Cell 2005; 122: 593–603.
Zinkel SS, Hurov KE, Ong C, Abtahi FM, Gross A, Korsmeyer SJ . A role for proapoptotic BID in the DNA-damage response. Cell 2005; 122: 579–591.
Kaufmann T, Tai L, Ekert PG, Huang DC, Norris F, Lindemann RK et al. The BH3-only protein bid is dispensable for DNA damage- and rep licative stress-induced apoptosis or cell-cycle arrest. Cell 2007; 129: 423–433.
Nakano K, Vousden KH . PUMA, a novel proapoptotic gene, is induced by p53. Mol Cell 2001; 7: 683–694.
Han J, Flemington C, Houghton AB, Gu Z, Zambetti GP, Lutz RJ et al. Expression of bbc3, a pro-apoptotic BH3-only gene, is regulated by diverse cell death and survival signals. Proc Nat Acad Sci USA 2001; 98: 11318–11323.
Yu J, Zhang L, Hwang PM, Kinzler KW, Vogelstein B . PUMA induces the rapid apoptosis of colorectal cancer cells. Mol Cell 2001; 7: 673–682.
Oda E, Ohki R, Murasawa H, Nemoto J, Shibue T, Yamashita T et al. Noxa, a BH3-only member of the bcl-2 fami ly and candidate mediator of p53-induced apoptosis. Science 2000; 288: 1053–1058.
Hijikata M, Kato N, Sato T, Kagami Y, Shimotohno K . Molecular cloning and characterization of a cDNA for a novel phorbol-12-myristate-13-acetate-responsive gene that is highly expressed in an adult T-cell leukemia cell line. J Virol 1990; 64: 4632–4639.
Villunger A, Michalak EM, Coultas L, Mullauer F, Bock G, Ausserlechner MJ et al. p53- and drug-induced apoptotic responses mediated by BH3-only proteins puma and noxa. Science 2003; 302: 1036–1038.
Shibue T, Takeda K, Oda E, Tanaka H, Murasawa H, Takaoka A et al. Integral role of Noxa in p53-mediated apoptotic response. Genes Dev 2003; 17: 2233–2238.
Jeffers JR, Parganas E, Lee Y, Yang C, Wang J, Brennan J et al. Puma is an essential mediator of p53-dependent and -independent apoptotic pathways. Cancer Cell 2003; 4: 321–328.
Naik E, Michalak EM, Villunger A, Adams JM, Strasser A . Ultraviolet radiation triggers apoptosis of fibroblasts and skin keratinocytes mainly via the BH3-only protein Noxa. J Cell Biol 2007; 176: 415–424.
Erlacher M, Michalak EM, Kelly PN, Labi V, Niederegger H, Coultas L et al. BH3-only proteins Puma and Bim are rate-limiting for {gamma} -radiation and glucocorticoid-induced apoptosis of lymphoid cells in vivo. Blood 2005; 131: 4131–4138.
Erlacher M, Labi V, Manzl C, Bock G, Tzankov A, Hacker G et al. Puma cooperates with Bim, the rate-limiting BH3-only protein in cell death during lymphocyte development, in apoptosis induction. J Exp Med 2006; 203: 2939–2951.
Chipuk JE, Maurer U, Green DR, Schuler M . Pharmacologic activation of p53 elicits Bax-dependent apoptosis in the absence of transcription. Cancer Cell 2003; 4: 371–381.
Chipuk JE, Kuwana T, Bouchier-Hayes L, Droin NM, Newmeyer DD, Schuler M et al. Direct activation of Bax by p53 mediates mitochondrial membrane permeabilization and apoptosis. Science 2004; 303: 1010–1014.
Mihara M, Erster S, Zaika A, Petrenko O, Chittenden T, Pancoska P et al. p53 has a direct apoptogenic role at the mitochondria. Mol Cell 2003; 11: 577–590.
Leu JI, Dumont P, Hafey M, Murphy ME, George DL . Mitochondrial p53 activates Bak and causes disruption of a Bak-Mcl1 complex. Nat Cell Biol 2004; 6: 443–450.
Kim H, Rafiuddin-Shah M, Tu HC, Jeffers JR, Zambetti GP, Hsieh JJ et al. Hierarchical regulation of mitochondrion-dependent apoptosis by BCL-2 subfamilies. Nat Cell Biol 2006; 8: 1348–1358.
Wang P, Yu J, Zhang L . The nuclear function of p53 is required for PUMA-mediated apoptosis induced by DNA damage. Proc Natl Acad Sci USA 2007; 104: 4054–4059.
Bode AM, Dong Z . Post-translational modification of p53 in tumorigenesis. Nat Rev Cancer 2004; 4: 793–805.
Takagi M, Absalon MJ, McLure KG, Kastan MB . Regulation of p53 translation and induction after DNA damage by ribosomal protein L26 and nucleolin. Cell 2005; 123: 49–63.
Phan RT, Dalla-Favera R . The BCL6 proto-oncogene suppresses p53 expression in germinal-centre B cells. Nature 2004; 432: 635–639.
Melino G, Lu X, Gasco M, Crook T, Knight RA . Functional regulation of p73 and p63: development and cancer. Trends Biochem Sci 2003; 28: 663–670.
Bourdon JC, Fernandes K, Murray-Zmijewski F, Liu G, Diot A, Xirodimas DP et al. p53 isoforms can regulate p53 transcriptional activity. Genes Dev 2005; 19: 2122–2137.
Wolf D, Harris N, Goldfinger N, Rotter V . Isolation of a full-length mouse cDNA clone coding for an immunologically distinct p53 molecule. Mol Cell Biol 1985; 5: 127–132.
Flaman JM, Waridel F, Estreicher A, Vannier A, Limacher JM, Gilbert D et al. The human tumour suppressor gene p53 is alternatively spliced in normal cells. Oncogene 1996; 12: 813–818.
Scrable H, Sasaki T, Maier B . DeltaNp53 or p44: prim ing the p53 pump. Int J Biochem Cell Biol 2005; 37: 913–919.
Rohaly G, Chemnitz J, Dehde S, Nunez AM, Heukeshoven J, Deppert W et al. A novel human p53 isoform is an essential element of the ATR-intra-S phase checkpoint. Cell 2005; 122: 21–32.
Tyner SD, Venkatachalam S, Choi J, Jones S, Ghebranious N, Igelmann H et al. p53 mutant mice that display early ageing-associated phenotypes. Nature 2002; 415: 45–53.
Garcia-Cao I, Garcia-Cao M, Martin-Caballero J, Criado LM, Klatt P, Flores JM et al. Super p53′ mice exhibit enhanced DNA damage response, are tumor resistant and age normally. EMBO J 2002; 21: 6225–6235.
Mosner J, Mummenbrauer T, Bauer C, Sczakiel G, Grosse F, Deppert W . Negative feedback regulation of wild-type p53 biosynthesis. EMBO J 1995; 14: 4442–4449.
Maier B, Gluba W, Bernier B, Turner T, Mohammad K, Guise T et al. Modulation of mammalian life span by the short isoform of p53. Genes Dev 2004; 18: 306–319.
Attardi LD, Donehower LA . Probing p53 biological functions through the use of genetically engineered mouse models. Mutat Res 2005; 576: 4–21.
Toledo F, Wahl GM . Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nat Rev Cancer 2006; 6: 909–923.
Sluss HK, Armata H, Gallant J, Jones SN . Phosphorylation of serine 18 regulates distinct p53 functions in mice. Mol Cell Biol 2004; 24: 976–984.
Sluss Chao C, Hergenhahn M, Kaeser MD, Wu Z, Saito S, Iggo R et al. Cell type- and promoter-specific roles of Ser18 phosphorylation in regulating p53 responses. J Biol Chem 2003; 278: 41028–41033.
Wu Z, Earle J, Saito S, Anderson CW, Appella E, Xu Y . Mutation of mouse p53 Ser23 and the response to DNA damage. Mol Cell Biol 2002; 22: 2441–2449.
MacPherson D, Kim J, Kim T, Rhee BK, Van Oostrom CT, DiTullio RA et al. Defective apoptosis and B-cell lymphomas in mice with p53 point mutation at Ser 23. EMBO J 2004; 23: 3689–3699.
Chao C, Herr D, Chun J, Xu Y . Ser18 and 23 phosphorylation is required for p53-dependent apoptosis and tumor suppression. EMBO J 2006; 25: 2615–2622.
Feng L, Hollstein M, Xu Y . Ser46 phosphorylation regulates p53-dependent apoptosis and replicative senescence. Cell Cycle 2006; 5: 2812–2819.
Bruins W, Zwart E, Attardi LD, Iwakuma T, Hoogervorst EM, Beems RB et al. Increased sensitivity to UV radiation in mice with a p53 point mutation at Ser389. Mol Cell Biol 2004; 24: 8884–8894.
Feng L, Lin T, Uranishi H, Gu W, Xu Y . Functional analysis of the roles of posttranslational modifications at the p53 C terminus in regulating p53 stability and activity. Mol Cell Biol 2005; 25: 5389–5395.
Krummel KA, Lee CJ, Toledo F, Wahl GM . The C-terminal lysines fine-tune P53 stress responses in a mouse model but are not required for stability control or transactivation. Proc Natl Acad Sci USA 2005; 102: 10188–10193.
Chao C, Wu Z, Mazur SJ, Borges H, Rossi M, Lin T et al. Acetylation of mouse p53 at lysine 317 negatively regulates p53 apoptotic activities after DNA damage. Mol Cell Biol 2006; 26: 6859–6869.
Jimenez GS, Nister M, Stommel JM, Beeche M, Barcarse EA, Zhang XQ et al. A transactivation-deficient mouse model provides insights into Trp53 regulation and function. Nat Genet 2000; 26: 37–43.
Chao C, Saito S, Kang J, Anderson CW, Appella E, Xu Y . p53 transcriptional activity is essential for p53-dependent apoptosis following DNA damage. EMBO J 2000; 19: 4967–4975.
Johnson TM, Hammond EM, Giaccia A, Attardi LD . The p53QS transactivation-deficient mutant shows stress-specific apoptotic activity and induces embryonic lethality. Nat Genet 2005; 37: 145–152.
Toledo F, Krummel KA, Lee CJ, Liu CW, Rodewald LW, Tang M et al. A mouse p53 mutant lacking the proline-rich domain rescues Mdm4 deficiency and provides insight into the Mdm2-Mdm4-p53 regulatory network. Cancer Cell 2006; 9: 273–285.
Toledo F, Lee CJ, Krummel KA, Rodewald LW, Liu CW, Wahl GM . Mouse mutants reveal that putative protein interaction sites in the p53 proline-rich domain are dispensable for tumor suppression. Mol Cell Biol 2007; 27: 1425–1432.
Scheffner M, Huibregtse JM, Vierstra RD, Howley PM . The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell 1993; 75: 495–505.
Dornan D, Wertz I, Shimizu H, Arnott D, Frantz GD, Dowd P et al. The ubiquitin ligase COP1 is a critical negative regulator of p53. Nature 2004; 429: 86–92.
Leng RP, Lin Y, Ma W, Wu H, Lemmers B, Chung S et al. Pirh2, a p53-induced ubiquitin-protein ligase, promotes p53 degradation. Cell 2003; 112: 779–791.
Chen D, Kon N, Li M, Zhang W, Qin J, Gu W . ARF-BP1/Mule is a critical mediator of the ARF tumor suppressor. Cell 2005; 121: 1071–1083.
Brooks CL, Gu W . p53 ubiquitination: Mdm2 and beyond. Mol Cell 2006; 21: 307–315.
Carter S, Bischof O, Dejean A, Vousden KH . C-terminal modifications regulate MDM2 dissociation and nuclear export of p53. Nat Cell Biol 2007; 9: 428–435.
Li M, Brooks CL, Wu-Baer F, Chen D, Baer R, Gu W . Mono- versus polyubiquitination: differential control of p53 fate by Mdm2. Science 2003; 302: 1972–1975.
Jones SN, Roe AE, Donehower LA, Bradley A . Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature 1995; 378: 206–208.
Montes de Oca Luna R, Wagner DS, Lozano G . Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature 1995; 378: 203–206.
Xirodimas DP, Saville MK, Bourdon JC, Hay RT, Lane DP . Mdm2-mediated NEDD8 conjugation of p53 inhibits its transcriptional activity. Cell 2004; 118: 83–97.
Abida WM, Nikolaev A, Zhao W, Zhang W, Gu W . FBXO11 promotes the Neddylation of p53 and inhibits its transcriptional activity. J Biol Chem 2007; 282: 1797–1804.
Gostissa M, Hengstermann A, Fogal V, Sandy P, Schwarz SE, Scheffner M et al. Activation of p53 by conjugation to the ubiquitin-like protein SUMO-1. EMBO J 1999; 18: 6462–6471.
Rodriguez MS, Desterro JM, Lain S, Midgley CA, Lane DP, Hay RT . SUMO-1 modification activates the transcriptional response of p53. EMBO J 1999; 18: 6455–6461.
Chen L, Chen J . MDM2-ARF complex regulates p53 sumoylation. Oncogene 2003; 22: 5348–5357.
Schmidt D, Muller S . Members of the PIAS family act as SUMO ligases for c-Jun and p53 and repress p53 activity. Proc Natl Acad Sci USA 2002; 99: 2872–2877.
Kwek SS, Derry J, Tyner AL, Shen Z, Gudkov AV . Functional analysis and intracellular localization of p53 modified by SUMO-1. Oncogene 2001; 20: 2587–2599.
Zheng H, You H, Zhou XZ, Murray SA, Uchida T, Wulf G et al. The prolyl is omerase Pin1 is a regulator of p53 in genotoxic response. Nature 2002; 419: 849–853.
Zacchi P, Gostissa M, Uchida T, Salvagno C, Avolio F, Volinia S et al. The prolyl isomerase Pin1 reveals a mechanism to control p53 functions after genotoxic insults. Nature 2002; 419: 853–857.
Dornan D, Shimizu H, Burch L, Smith AJ, Hupp TR . The proline repeat domain of p53 binds directly to the transcriptional coactivator p300 and allosterically controls DNA-dependent acetylation of p53. Mol Cell Biol 2003; 23: 8846–8861.
Sakaguchi K, Herrera JE, Saito S, Miki T, Bustin M, Vassilev A et al. DNA damage activates p53 through a phosphorylation-acetylation cascade. Genes Dev 1998; 12: 2831–2841.
Lill NL, Grossman SR, Ginsberg D, DeCaprio J, Livingston DM . Binding and modulation of p53 by p300/CBP coactivators. Nature 1997; 387: 823–827.
Roth J, Koch P, Contente A, Dobbelstein M . Tu mor-derived mutations within the DNA-binding domain of p53 that phenotypically resemble the deletion of the proline-rich domain. Oncogene 2000; 19: 1834–1842.
Baptiste N, Friedlander P, Chen X, Prives C . The proline-rich domain of p53 is required for cooperation with anti-neoplastic agents to promote apoptosis of tumor cells. Oncogene 2002; 21: 9–21.
Armstrong JF, Kaufman MH, Harrison DJ, Clarke AR . High-frequency developmental abnormalities in p53-deficient mice. Curr Biol 1995; 5: 931–936.
Sah VP, Attardi LD, Mulligan GJ, Williams BO, Bronson RT, Jacks T . A subset of p53-deficient embryos exhibit exencephaly. Nat Genet 1995; 10: 175–180.
Chao C, Saito S, Anderson CW, Appella E, Xu Y . Phosphorylation of murine p53 at ser-18 regulates the p53 responses to DNA damage. Proc Natl Acad Sci USA 2000; 97: 11936–11941.
Somasundaram K, Zhang H, Zeng YX, Houvras Y, Peng Y, Zhang H et al. Arrest of the cell cycle by the tumour-suppressor BRCA1 requires the CDK-inhibitor p21WAF1/CiP1. Nature 1997; 389: 187–190.
Haapajarvi T, Kivinen L, Heiskanen A, des Bordes C, Datto MB, Wang XF et al. UV radiation is a transcriptional inducer of p21(Cip1/Waf1) cyclin-kinase inhibitor in a p53-independent manner. Exp Cell Res 1999; 248: 272–279.
Koumenis C, Alarcon R, Hammond E, Sutphin P, Hoffman W, Murphy M et al. Regulation of p53 by hypoxia: dissociation of transcriptional repression and apoptosis from p53-dependent transactivation. Mol Cell Biol 2001; 21: 1297–1310.
Waterman MJ, Stavridi ES, Waterman JL, Halazonetis TD . ATM-dependent activation of p53 involves dephosphorylation and association with 14-3-3 proteins. Nat Genet 1998; 19: 175–178.
Appella E, Anderson CW . Post-translational modifications and activation of p53 by genotoxic stresses. Eur J Biochem 2001; 26: 2764–2772.
Banin S, Moyal L, Shieh S-Y, Taya Y, Anderson CW, Chessa L et al. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science 1998; 281: 1674–1677.
Shieh S-Y, Ikeda M, Taya Y, Prives C . DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell 1997; 91: 325–334.
Fiscella M, Ullrich SJ, Zambrano N, Shields MT, Lin D, Lees-Miller SP et al. Mutation of the serine 15 phosphorylation site of human p53 reduces the ability of p53 to inhibit cell cycle progression. Oncogene 1993; 8: 1519–1528.
Khanna KK, Keating KE, Kozlov S, Scott S, Gatei M, Hobson K et al. ATM associates with and phosphorylates p53: mapping the region of interaction. Nat Genet 1998; 20: 398–400.
Dumaz N, Meek DW . Serine15 phosphorylation stimulates p53 transactivation but does not directly influence interaction with HDM2. EMBO J 1999; 18: 7002–7010.
Saito S, Yamaguchi H, Higashimoto Y, Chao C, Xu Y, Fornace Jr AJ et al. Phosphorylation site interdependence of human p53 post-translational modifications in response to stress. J Biol Chem 2003; 278: 37536–37544.
Dumaz N, Milne DM, Meek DW . Protein kinase CK1 is a p53-threonine 18 kinase which requires prior phosphorylation of serine 15. FEBS Lett 1999; 463: 312–316.
Unger T, Sionov RV, Moallem E, Yee CL, Howley PM, Oren M et al. Mutations in serines 15 and 20 of human p53 impair its apoptotic activity. Oncogene 1999; 18: 3205–3212.
Chehab NH, Malikzay A, Stavridi ES, Halazonetis TD . Phosphorylation of Ser-20 mediates stabilization of human p53 in response to DNA damage. Proc Natl Acad Sci USA 1999; 96: 13777–13782.
D’Orazi G, Cecchinelli B, Bruno T, Manni I, Higashimoto Y, Saito S et al. Homeodomain-interacting protein kinase-2 phosphorylates p53 at Ser 46 and mediates apoptosis. Nat Cell Biol 2002; 4: 11–19.
Taira N, Nihira K, Yamaguchi T, Miki Y, Yoshida K . DYRK2 is targeted to the nucleus and controls p53 via Ser46 phosphorylation in the apoptotic response to DNA damage. Mol Cell 2007; 25: 725–738.
Oda K, Arakawa H, Tanaka T, Matsuda K, Tanikawa C, Mori T et al. p53AIP1, a potential mediator of p53-dependent apoptosis, and its regulation by Ser-46-phosphorylated p53. Cell 2000; 102: 849–862.
Blaydes JP, Luciani MG, Pospisilova S, Ball HM, Vojtesek B, Hupp TR . Stoichiometric phosphorylation of human p53 at Ser315 stimulates p53-dependent transcription. J Biol Chem 2001; 276: 4699–4708.
Fogal V, Hsieh JK, Royer C, Zhong S, Lu X . Cell cycle-dependent nuclear retention of p53 by E2F1 requires phosphorylation of p53 at Ser315. EMBO J 2005; 24: 2768–2782.
Katayama H, Sasai K, Kawai H, Yuan ZM, Bondaruk J, Suzuki F et al. Phosphorylation by aurora kinase A induces Mdm2-mediated destabilization and inhibition of p53. Nat Genet 2004; 36: 55–62.
Keller D, Zeng X, Li X, Kapoor M, Iordanov MS, Taya Y et al. The p38MAPK inhibitor SB203580 alleviates ultraviolet-induced phosphorylation at serine 389 but not serine 15 and activation of p53. Biochem Biophys Res Commun 1999; 261: 464–471.
Kapoor M, Hamm R, Yan W, Taya Y, Lozano G . Cooperative phosphorylation at multiple sites is required to activate p53 in response to UV radiation. Oncogene 2000; 19: 358–364.
Keller DM, Zeng X, Wang Y, Zhang QH, Kapoor M, Shu H et al. A DNA damage-induced p53 serine 392 kinase complex contains CK2, hSpt16, and SSRP1. Mol Cell 2001; 7: 283–292.
Lu H, Taya Y, Ikeda M, Levine AJ . Ultraviolet radiation, but not gamma radiation or etoposide-induced DNA damage, results in the phosphorylation of the murine p53 protein at serine-389. Proc Natl Acad Sci USA 1998; 95: 6399–6402.
Yamauchi M, Suzuki K, Kodama S, Watanabe M . Stabilization of alanine substituted p53 protein at Ser15, Thr18, and Ser20 in response to ionizing radiation. Biochem Biophys Res Commun 2004; 323: 906–911.
Fuchs B, O’Connor D, Fallis L, Scheidtmann KH, Lu X . p53 phosphorylation mutants retain transcription activity. Oncogene 1995; 10: 789–793.
Mathai JP, Germain M, Marcellus RC, Shore GC . Induction and endoplasmic reticulum location of BIK/NBK in response to apoptotic signaling by E1A and p53. Oncogene 2002; 21: 2534–2544.
Hirao A, Kong YY, Matsuoka S, Wakeham A, Ruland J, Yoshida H et al. DNA damage-induced activation of p53 by the checkpoint kinase Chk2. Science 2000; 287: 1824–1827.
Michalak E, Coultas L, Villunger A, Adams JM, Strasser A . The apoptosis induced by the tumour suppressor p53 relies largely on Puma but Noxa contributes 2007, submitted.
Liu L, Scolnick DM, Trievel RC, Zhang HB, Marmorstein R, Halazonetis TD et al. p53 sites acetylated in vitro by PCAF and p300 are acetylated in vivo in response to DNA damage. Mol Cell Biol 1999; 19: 1202–1209.
Ventura A, Kirsch DG, McLaughlin ME, Tuveson DA, Grimm J, Lintault L et al. Restoration of p53 function leads to tumour regression in vivo. Nature 2007; 445: 661–665.
Xue W, Zender L, Miething C, Dickins RA, Hernando E, Krizhanovsky V et al. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature 2007; 445: 656–660.
Rinaldo C, Prodosmo A, Mancini F, Iacovelli S, Sacchi A, Moretti F et al. MDM2-regulated degradation of HIPK2 prevents p53Ser46 phosphorylation and DNA damage-induced apoptosis. Mol Cell 2007; 25: 739–750.
Luo JL, Yang Q, Tong WM, Hergenhahn M, Wang ZQ, Hollstein M . Knock-in mice with a chimeric human/murine p53 gene develop normally and show wild-type p53 responses to DNA damaging agents: a new biomedical research tool. Oncogene 2001; 20: 320–328.
Lin T, Chao C, Saito S, Mazur SJ, Murphy ME, Appella E et al. p53 induces differentiation of mouse embryonic stem cells by suppressing Nanog expression. Nat Cell Biol 2005; 7: 165–171.
Kapoor M, Lozano G . Functional activation of p53 via phosphorylation following DNA damage by UV but not gamma radiation. Proc Natl Acad Sci USA 1998; 95: 2834–2837.
Hoogervorst EM, Bruins W, Zwart E, van Oostrom CT, van den Aardweg GJ, Beems RB et al. Lack of p53 Ser389 phosphorylation predisposes mice to develop 2-acetylaminofluorene-induced bladder tumors but not ionizing radiation-induced lymphomas. Cancer Res 2005; 65: 3610–3616.
Gu W, Roeder RG . Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 1997; 90: 595–606.
Wang YH, Tsay YG, Tan BC, Lo WY, Lee SC . Identification and characterization of a novel p300-mediated p53 acetylation site, lysine 305. J Biol Chem 2003; 278: 25568–25576.
Tang Y, Luo J, Zhang W, Gu W . Tip60-dependent acetylation of p53 modulates the decision between cell-cycle arrest and apoptosis. Mol Cell 2006; 24: 827–839.
Sykes SM, Mellert HS, Holbert MA, Li K, Marmorstein R, Lane WS et al. Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol Cell 2006; 24: 841–851.
Gu W, Shi X-L, Roeder RG . Synergistic activation of transcription by CBP and p53. Nature 1997; 387: 819–823.
Barlev NA, Liu L, Chehab NH, Mansfield K, Harris KG, Halazonetis TD et al. Acetylation of p53 activates trans cription through recruitment of coactivators/histone acetyltransferases. Mol Cell 2001; 8: 1243–1254.
Espinosa JM, Emerson BM . Transcriptional regulation by p53 through intrinsic DNA/chromatin binding and site-directed cofactor recruitment. Mol Cell 2001; 8: 57–69.
Brooks CL, Gu W . Ubiquitination, phosphorylation and acetylation: the molecular basis for p53 regulation. Curr Opin Cell Biol 2003; 15: 164–171.
Knights CD, Catania J, Di Giovanni S, Muratoglu S, Perez R, Swartzbeck A et al. Distinct p53 acetylation cassettes differentially influence gene-expression patterns and cell fate. J Cell Biol 2006; 173: 533–544.
Luo J, Nikolaev AY, Imai S, Chen D, Su F, Shiloh A et al. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell 2001; 107: 137–148.
Luo J, Su F, Chen D, Shiloh A, Gu W . Deacetylation of p53 modulates its effect on cell growth and apoptosis. Nature 2000; 408: 377–381.
Vaziri H, Dessain SK, Ng Eaton E, Imai SI, Frye RA, Pandita TK et al. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell 2001; 107: 149–159.
Ito A, Lai CH, Zhao X, Saito S, Hamilton MH, Appella E et al. p300/CBP-mediated p53 acetylation is commonly induced by p53-activating agents and inhibited by MDM2. EMBO J 2001; 20: 1331–1340.
Jin Y, Zeng SX, Dai MS, Yang XJ, Lu H . MDM2 inhibits PCAF (p300/CREB-binding protein-associated factor)-mediated p53 acetylation. J Biol Chem 2002; 277: 30838–30843.
Ito A, Kawaguchi Y, Lai CH, Kovacs JJ, Higashimoto Y, Appella E et al. MDM2-HDAC1-mediated deacetylation of p53 is required for its degradation. EMBO J 2002; 21: 6236–6245.
Chuikov S, Kurash JK, Wilson JR, Xiao B, Justin N, Ivanov GS et al. Regulation of p53 activity through lysine methylation. Nature 2004; 432: 353–360.
Huang J, Perez-Burgos L, Placek BJ, Sengupta R, Richter M, Dorsey JA et al. Repression of p53 activity by Smyd2-mediated methylation. Nature 2006; 444: 629–632.
Liu G, McDonnell TJ, Montes de Oca Luna R, Kapoor M, Mims B, El-Naggar AK et al. High metastatic potential in mice inheriting a targeted p53 missense mutation. Proc Nat Acad Sci USA 2000; 97: 4174–4179.
Lang GA, Iwakuma T, Suh YA, Liu G, Rao VA, Parant JM et al. Gain of function of a p53 hot spot mutation in a mouse model of Li–Fraumeni syndrome. Cell 2004; 119: 861–872.
Song H, Hollstein M, Xu Y . p53 gain-of-function cancer mutants induce genetic instability by inactivating ATM. Nat Cell Biol 2007; 9: 573–580.
Olive KP, Tuveson DA, Ruhe ZC, Yin B, Willis NA, Bronson RT et al. Mutant p53 gain of function in two mouse models of Li–Fraumeni syndrome. Cell 2004; 119: 847–860.
Wright JD, Noskov SY, Lim C . Factors governing loss and rescue of DNA binding upon single and double mutations in the p53 core domain. Nucleic Acids Res 2002; 30: 1563–1574.
Acknowledgements
The work in our laboratory is supported by fellowships and grants from the Austrian Science Fund (FWF), the Tiroler Wissenschaftsfond (TWF) and the Association for International Cancer Research (AICR). We are grateful to our lab members for many interesting discussions as well as to all members of the SFB021 ‘Proliferation and Cell Death in Tumours’ for their input into our research. We apologise to the many scientists in this field whose excellent research was not cited but was only referred to indirectly through reviews.
Author information
Authors and Affiliations
Corresponding author
Additional information
Edited by RA Knight
Rights and permissions
About this article
Cite this article
Olsson, A., Manzl, C., Strasser, A. et al. How important are post-translational modifications in p53 for selectivity in target-gene transcription and tumour suppression?. Cell Death Differ 14, 1561–1575 (2007). https://doi.org/10.1038/sj.cdd.4402196
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.cdd.4402196
Keywords
This article is cited by
-
p53 dry gene powder enhances anti-cancer effects of chemotherapy against malignant pleural mesothelioma
Gene Therapy (2024)
-
Timing and cell specificity of senescence drives postnatal lung development and injury
Nature Communications (2023)
-
Epigenetically silenced apoptosis-associated tyrosine kinase (AATK) facilitates a decreased expression of Cyclin D1 and WEE1, phosphorylates TP53 and reduces cell proliferation in a kinase-dependent manner
Cancer Gene Therapy (2022)
-
Protective effects of exosomes derived from lyophilized porcine liver against acetaminophen damage on HepG2 cells
BMC Complementary Medicine and Therapies (2021)
-
p53-induced ARVCF modulates the splicing landscape and supports the tumor suppressive function of p53
Oncogene (2020)