Abstract
Background:
Several studies in colorectal cancer (CRC) indicate a relationship between tumour immune infiltrates and clinical outcome. We tested the utility of a digital pattern recognition-based image analysis (DPRIA) system to segregate tissue regions and facilitate automated quantification of immune infiltrates in CRC.
Methods:
Primary CRC with matched hepatic metastatic (n=7), primary CRC alone (n=18) and primary CRC with matched normal (n=40) tissue were analysed immunohistochemically. Genie pattern recognition software was used to segregate distinct tissue regions in combination with image analysis algorithms to quantify immune cells.
Results:
Immune infiltrates were observed predominately at the invasive margin. Quantitative image analysis revealed a significant increase in the prevalence of Foxp3 (P<0.0001), CD8 (P<0.0001), CD68 (<0.0001) and CD31 (<0.0001) positive cells in the stroma of primary and metastatic CRC, compared with tumour cell mass. A direct comparison between non-metastatic primary CRC (MET−) and primary CRC that resulted in metastasis (MET+) showed an immunosuppressive phenotype, with elevated Foxp3 (P<0.05) and reduced numbers of CD8 (P<0.05) cells in the stroma of MET+ compared with MET− samples.
Conclusion:
By combining immunohistochemistry with DPRIA, we demonstrate a potential metastatic phenotype in CRC. Our study accelerates wider acceptance and use of automated systems as an adjunct to traditional histopathological techniques.
Similar content being viewed by others
Main
Conventional risk prediction in colorectal cancer (CRC) is usually achieved by histopathological evaluation of surgically removed tissue. Tumour staging (AJCC/UICC-TNM classification) summarises data on tumour burden (T), presence of cancer cells in draining and regional lymph nodes (N) and evidence for metastases (M). Increasing evidence supports the observation that immune cell infiltration in the tumour microenvironment is a strong parameter influencing clinical outcome, regardless of tumour burden (Naito et al, 1998; Pages et al, 2005, 2008; Galon et al, 2006; Camus et al, 2009; Halama et al, 2009; Salama et al, 2009; Mlecnik et al, 2011; Tosolini et al, 2011). The extent and nature of the immune reaction at the tumour site correlates with clinical outcome in CRC stages I and II (Pages et al, 2009). Within CRC, immune cells are distributed in different areas; in the tumour centre (CT), at the invasive margin (IM) and in any lymphoid structures present at the tumour periphery. The purpose of this study was to investigate the nature and extent of immune infiltrates in different tumour-associated areas of primary and metastatic CRC lesions using digital pattern recognition-based image analysis (DPRIA). Accurate assessment of immune infiltrate is fundamental to facilitating the use of the ‘immunoscore’ as prognostic and immune-monitoring tools in CRC (Galon et al, 2012). Studies have demonstrated that the immunoscore has a prognostic value that may be superior to the AJCC/UICC TNM-classification (Galon et al, 2007; Mlecnik et al, 2011). Thus, it is imperative to perform routine and robust immune score quantification and to introduce this parameter as a marker to classify cancers. Traditionally, analysis of immune infiltrates has been performed by subjective or semiquantitative by-eye assessment by a pathologist or trained scientist on tissue sections using a microscope. This activity can be time consuming. Analysis of digital images has the potential to identify more subtle changes or trends in the nature and extent of tumour-associated immune cell infiltrate than the human observer. DPRIA is inherently suited to high-throughput analysis, removing adverse human elements such as fatigue and is also more precise and reproducible (Jara-Lazaro et al, 2010; Camparo et al, 2012). However, the inherent complexity of quantitative IHC poses difficulties for reliable translation of markers into the clinic and despite significant advances in the development and application of image analysis in recent years, a number of challenges remain to be addressed.
Advanced computer image analysis capabilities can be used for the automated measurement of histopathological material. Whole-slide imaging (WSI) is a fast developing area that is transforming histopathology into the digitalised world (Jara-Lazaro et al, 2010). In addition, developments in image analysis tools are accelerating the transition of traditional pathology techniques of analysing histological specimens, into a more quantitative science (Randell et al, 2013).
We describe a platform of immunohistochemistry (IHC)-based assays for the detection of differential immune infiltrates in FFPE human tumours, combined with DPRIA for the quantification and localisation of infiltrating immune cell populations. This study aimed to investigate the potential of a DPRIA approach, across a cohort of CRC patients and to ascertain whether the density and orientation of immune cell populations within distinct tumour territories (CT, IM and stroma) changed between primary CRC and metastatic liver deposits.
Materials and methods
Patient samples
Non-metastatic (MET–) primary CRC (n=18), primary CRC that resulted in metastasis (MET+) with matched hepatic metastatic (n=7) and primary CRC with matched distant normal non-neoplastic (DNN; n=40) FFPE tissue were analysed. All cases were colorectal adenocarcinomas and we confirmed this on H&E sections as part of our internal quality control processes for samples acquired for exploratory research purposes. All samples used were sourced from the AstraZeneca Alderley Park Biobank, Cheshire, UK (Human Tissue Authority number 12109), which has National Research Ethics Service ethical approval as a tissue bank (07/MRE08/29). All third party providers to the Alderley Park Biobank passed an approval process at AstraZeneca and those providers acquired all samples with written informed consent.
Immunohistochemistry
FFPE 4 μm sections were stained using an Autostainer 720 (Lab Vision, kalamazoo, MI, USA). Following antigen retrieval using pH6 citrate buffer (Dako, S1699, Cambridge, UK) at 110 °C, quenching of endogenous peroxidase activity was achieved using 3% H2O2. Sections were treated with serum-free protein block (Dako, X0909) before a 60-min incubation with antibodies against: Foxp3 (5 μg ml–1, clone 236A/E7, Abcam, Cambridge, UK), CD8 (0.5 μg ml–1, clone 4B11, Leica NCL-CD8-4B11, Milton Keynes, UK), CD68 (0.2 μg ml–1, clone PG-M1, Dako M0876) or CD31 (0.27 μg ml–1, CHG-CD31-PI, AstraZeneca in house). Envision HRP-linked polymer (Dako, K4001) and 3,3-diaminobenzidine (D/K3468) were applied. Tissue sections were counterstained with Carazzi’s Haematoxylin Gill III (Surgipath, Lecia, Peterborough, UK; 01540BBE) on a Leica Autostainer XL. Isotype-matched mouse monoclonal antibodies were used as negative controls.
Image analysis
Digital images of IHC-stained glass slides were acquired using a ScanScope digital scanner (Aperio ePathology Solutions, Vista, CA, USA). Images were viewed and organised using ImageScope viewer and Spectrum slide manager system, respectively (Aperio ePathology Solutions). Each scanned image was annotated manually for analysis (by HKA) to exclude areas of glass or poor sample quality. Genie pattern recognition software was used to generate Genie classifiers for the separation of areas of tumour, stroma, immune aggregates, non-neoplastic tissue and glass. First, the pattern recognition software was trained using a montage of example images that had been manually marked by the operator to distinguish the individual areas of interest. Based on manufacturer recommendations, a 95% or greater training accuracy was required for each classifier in order to proceed with image analysis of each tissue region. Classifiers were combined with appropriate image analysis algorithms for each marker for the automated quantitative assessment of immune infiltrates within the specific regions of tissue, across whole-slide sections. DPRIA provides an accurate output of average percent positive cells for a cell type within a region of interest. All images were quality controlled for accuracy of the tissue recognition software, in addition to accuracy of the image analysis algorithm. Pathologist, CW, further confirmed that the tumour and stroma were being identified correctly.
Statistical analysis
Data were collated and displayed as mean percent positive cells (±s.d.). Further analysis was then carried out to quantify any trends in the data, using Prism 5 software (GraphPad, San Diego, CA, USA). If the data assumed a normal distribution pattern, a paired or unpaired t-test was carried out. If the data did not assume a normal distribution pattern, a ranked method was selected. For paired rank testing, a Wilcoxon test was carried out. For unpaired rank testing, a Mann–Whitney analysis was used.
Results
Tissue pattern recognition platform
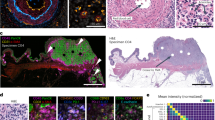
The combined workflow of the DPRIA platform is illustrated in Figure 1. Digital images were acquired using a ScanScope digital scanner (Figure 1A). Genie pattern recognition software was utilised to distinguish different areas of tissue. Specific tissue ‘classifiers’ were trained to identify distinct spatial-spectral features that discriminate image pixels for each region of tissue. This was achieved by the operator manually marking up example regions of segregated tissue to provide a training set of images for the pattern recognition software. Tissue regions included defined areas of tumour (red), stroma (yellow), lymphoid structures (green) and background glass (blue; Figure 1B). The assessment and validation of classifiers was achieved across example tissue images (Figure 1C). Trained Genie classifiers were then combined with appropriately tuned image analysis algorithms for the quantification of different markers within each specific area (Figure 1D). A nuclear algorithm (Aperio ePathology Solutions) was modified to quantify the number of immune cells and data expressed as a percentage of the total number of cells within the area of interest (percentage positive cells). A micro-vessel density (MVD) algorithm (Aperio ePathology Solutions) was tuned to assess the number of vessels per mm2 of each tissue region.
Genie analysis workflow. (A) Digitally acquired images were achieved for multiple slides using a ScanScope digital scanner (Aperio ePathology Solutions). (B) Genie pattern recognition software (Aperio ePathology Solutions) was used for automated assessment of specific tissue regions, where the software was trained to recognise patterns by manually marking up example areas. (C) Genie classifiers were generated for the separation of: tumour (red); stroma (yellow); lymphatic aggregates (green); non-neoplastic tissue (not shown) and glass (blue) and then tested for accuracy. (D) For each slide, the area of interest was manually marked (green line) to reduce the amount of background white space. Genie classifiers were then combined with a nuclear algorithm to quantify the percentage positive number of immune cells (brown) in each area. Images were viewed and organised using ImageScope viewer and Spectrum operating system.
Implementation of pattern recognition image analysis: the importance of the tumour IM
DPRIA distinguished areas of tumour (CT), tumour-associated stroma (TS), IM, DNN and DNN-associated stroma (DNNS). Primary CRC tumour with matched distant normal tissue (n=40) was evaluated for CD8+ cytotoxic T lymphocytes (CTLs), Foxp3+ Tregs, CD68+ macrophages and CD31+ vascular endothelium (Figure 2). CTLs, Tregs and macrophages were all observed in abundance at the IM, compared with the tumour (CT). The number of CTLs in the tumour (CT) was significantly lower than at the IM or DNN (P<0.001), however, there was no significant difference between the IM and DNN (Figure 2A). In contrast, there was an influx of Tregs at the IM significantly higher than both tumour (P<0.001) and DNN (P<0.001; Figure 2B). The same trend was also observed for macrophages (Figure 2C), however, the M1/M2 phenotype was not determined in this study. In addition, the MVD was calculated for the vascular endothelium infiltrating specific tissue regions (Figure 2D). Vessels resided predominately in the stroma, with reduced vascularisation seen in stroma throughout the tumour compared with DNN.
Location of immune cell infiltrates in primary CRC tissue. Primary CRC tissue was examined for (A) CD8, (B) Foxp3, (C) CD68 and (D) CD31, using IHC techniques. Digital images were acquired and Genie classifiers were used to distinguish areas of tumour (T), adjacent tumour IM and DNN tissue. Genie classifiers were combined with a modified nuclear algorithm to quantify the percentage positive cells or micro-vessel density (MVD). MVD was additionally compared in tumour-associated stroma (TS) and DNNS. Data were statistically analysed using a one-way ANOVA and Tukey’s multiple comparison post test (±s.d., *P<0.05, **P<0.01, ***P<0.001).
Elevated immune infiltrates in tumour-associated stroma
Successful segregation of tissue patterns enabled the quantification and comparison of immune infiltrates and vascular endothelium between areas of tumour and tumour-associated stroma (Figure 3). The importance of the stroma for infiltrating immune cells is highlighted by the significant prevalence of CTLs (P<0.001, Figure 3A), Tregs (P<0.001, Figure 3B) and macrophages (P<0.001, Figure 3C) in the stroma compared with those infiltrating the tumour. In addition, the density of vascular endothelium was significantly higher in the stroma, as alluded to previously (Figure 3D). Representative, corresponding images are illustrated (Figure 3E–H). A large variability in the density of immune infiltrates was observed, not only in different patients, but also in different regions of the same tumour. An additional four example images, for each marker, have been selected and composed in Supplementary Figure 1 to highlight the range of tumour/stroma staining.
Comparison of tumour and stroma signature in CRC (primary and metastatic tissue combined). Primary and metastatic CRC tissues were stained immunohistochemically, digital images were acquired and Genie classifiers were used to distinguish areas of tumour and stroma. An overall assessment of (A) CD8, (B) Foxp3, (C) CD68 and (D) CD31 was achieved for CRC (primary and metastatic data combined). A direct comparison was carried out between the percentage positive cells or micro-vessel density (MVD, vessels per mm2 of tissue). Corresponding example images are illustrated (E–H) at × 20 original magnification. (Paired t-test, normal distribution, ±s.d., ***P<0.001.)
Primary stromal immune content reveals a metastatic phenotype
The stroma of primary CRC tissue that had not metastasised at the time of surgery (MET–, n=18) was compared with primary CRC from patients with hepatic metastasis (MET+, n=7). Analysis of the tumour-associated stroma revealed an immunosuppressive phenotype in MET+ patients, with reduced numbers of CTLs (P<0.05, Figure 4A) and elevated Tregs (P<0.05, Figure 4B) in comparison with MET– patients. No significant difference was observed for CD68+ macrophages (Figure 4C). CD31+ MVD was significantly higher in the stroma of MET+ primary compared with MET– samples (Figure 4D). These data are in accordance with previous literature evaluating the impact of vascular endothelium, a key parameter for tumour progression and the metastasis of solid tumours (Yancopoulos et al, 2000; Lin et al, 2007).
Comparison of the stromal signature in MET+ and MET− primary CRC tissue. Primary CRC with no affiliated metastatic disease (MET−) and primary CRC that has since developed liver metastasis (MET+) were compared. A Genie classifier was used to distinguish areas of stroma and then combined with a nuclear algorithm to quantify the percentage positive cells or micro-vessel density (MVD). The stroma was examined for (A) CD8, (B) Foxp3, (C) CD68 and (D) CD31. (Paired t-test, normal distribution, ±s.d., *P<0.05.)
Discussion
Increasing understanding of the mechanisms underpinning the hallmarks of cancer have recognised the importance of the tumour microenvironment during tumourigenesis and in response to cancer therapy (Hanahan and Weinberg, 2011). Development of non-tumour cell-centric biomarkers is thus an emerging concept. Previous studies have alluded to the role of the innate and adaptive immune system providing protection against tumour development through mechanisms of immunosurveilance (Dunn et al, 2002). The development of stratified and personalised therapies for complex multifactorial diseases, such as cancer, requires a comprehensive understanding of the integration and diversity of contributing elements in human pathology. Advances in cancer immunotherapy aim to target tumours by activating immune effector mechanisms and/or neutralising immune-suppressive networks within the tumour microenvironment. However, translation of immunotherapeutic approaches into the clinic will require the development of reliable and robust immune-monitoring strategies for the identification of relevant immune markers; to identify suitable patients and/or monitor pharmacodynamic responses on treatment.
Here we have addressed specific problems relating to immune cell digital image analysis and quantification. Automated IHC staining procedures combined with increasingly sophisticated image analysis capability facilitated rigorous and systematic quantification of immune cell infiltrates. The platforms bring together state-of-the-art automated digitalisation, management and viewing of images, coupled with DPRIA. DPRIA contributes to an overall strategy to replace some of the more mundane and challenging microscope activities, to increase productivity, to improve quality and free up pathologist time. In application to oncology drug discovery, analysis of digital histopathology images with an automated platform was objective with an output comparable to traditional pathology techniques (AstraZeneca Oncology Molecular Pathology, unpublished observations). DPRIA is being used widely in a preclinical setting where it has been validated (data unpublished). The utilisation of an advanced image analysis platform described here reduces the limitations faced by traditional analysis of IHC-stained slides including inconsistent tissue region selection criteria, subjective assessment, combined with differences in qualitative and semiquantitative criteria applied by pathologists. Advantages include reduced timelines, reduced variability and improved assay robustness, reproducibility and reliability. Using digitalised WSI analysis facilitates a global assessment of the tissue section, in comparison with using representative or averaged fields of view under a microscope. The automation of immune cell quantification is fundamental to achieve the translation of IHC markers into the clinic (Galon et al, 2012). Thus, the development of robust immune-monitoring strategies to provide automated quantification in precise tissue regions is essential. DPRIA classifiers were able to separate the designated tissue regions with a high degree of accuracy. Within the selected patient cohort, the distinct and consistent morphology of CRC tumour necessitated the generation of only a few variations of tissue classifiers for different sets of tumour samples. Advancement in DPRIA has enabled a refined pathological approach to determine immune cell densities and their distribution accurately throughout tumour, stroma and non-neoplastic tissue, yielding an average immune cell infiltration score across whole sections for distinct regions of interest. The utilisation of pixel-based pattern recognition and segmentation facilitates an elegant assessment of WSIs and greatly reduces the labour-intensive workload for pathologists. The power of this platform resulted in immune cell quantification, in distinct morphological compartments, suggesting a metastatic phenotype (albeit in a small patient cohort). As to whether this technique could predict for metastasis is unknown at this stage and further studies are warranted to include a larger series of tissues with a wider range of matched primary and secondary samples. This would also enable the questions of reliable predictability, the confounding variability in immune infiltration and reference against pathologist qualitative or semiquantitative scoring to be addressed. Quantification of immune infiltrates into distinct morphological compartments within a tumour specimen has a distinct advantage over other multi-parameter cellular techniques, such as flow cytometry. Furthermore, the technique described is applicable to FFPE samples, which are routinely taken into a clinical oncology setting.
In accordance with previous literature, our results highlight the importance of the tumour IM (Galon et al, 2006). Studies suggest that tumour-related factors mediate Treg trafficking into the tumour microenvironment, which could explain the elevated Tregs observed at the IM. Chemotaxis experiments have demonstrated the ability of the macrophage-derived chemokine, CCL22, to induce Treg migration through its receptor CCR4 and impair antitumour immunity (Curiel et al, 2004). Further to this, the tumour microenvironment could provide a favourable setting for Treg expansion because of the complex mixture of chemokines (Yamaguchi et al, 2007).
In addition, we have demonstrated an inverse relationship between Tregs and CTLs within stroma-rich regions of MET+ tissue. An increase in the number of Tregs, combined with a decrease in the number of CTLs, was associated with a MET+ phenotype. The general consensus is that increased numbers of Tregs are associated with poor prognosis and tumour progression. Research has shown that patients with elevated Treg numbers showed reduced survival rates and the accumulation of Tregs was related to disease progression in ovarian carcinoma (Woo et al, 2001; Curiel et al, 2004), NSCLC (Woo et al, 2001), gastric (Mizukami et al, 2008) and hepatocellular carcinoma (Fu et al, 2007). The current data are in conflict with some previous literature showing no significant difference in the number of Tregs between MET+ and MET– when analysed using large-scale flow cytometry and defined by CD4+CD25hi expression (Camus et al, 2009). Analysis using large-scale flow cytometry loses the detail of spatial location of inflammatory cells demonstrated in this study and is a possible explanation for the lack of significance previously observed. Utilisation of IHC techniques allows the precise location of immune infiltrates to be scrutinised to a greater degree of accuracy.
An increase in Treg number may be responsible for a local imbalance in the immune system, which would favour an immune-suppressive tumour microenvironment. This situation may lead to tumour cells escaping immune cell recognition and killing (Colombo and Piconese, 2007). Tumour progression and metastasis could therefore result from Treg-mediated suppression of the antitumour immune response, such as CTLs (Bui et al, 2006). Previous evidence suggests that depleting Tregs leads to rejection of transplanted tumours in some mice models (Shimizu et al, 1999).
The analysis of CD8+ CTLs in this study is in concordance with previous studies demonstrating the CD8/CD3 T-cell density ratio in relation to clinical outcome, where high proportions of CD8 cytotoxic T cells within the primary tumour was associated with reduced risk of relapse (Mlecnik et al, 2011). The current data were converted into cells mm–2 where the numbers were comparable to those previously published in the literature (data not shown). Data presented here are in agreement, showing MET+ patients to have significantly lower levels of CD8+ cells compared with MET– patients (Camus et al, 2009).
The abundance of macrophages within the tumour microenvironment is well documented and has more recently been termed tumour-associated macrophages (TAM) (Erreni et al, 2011). Macrophages are able to elicit differing functional properties based on their exposure to various conditions, characterised as proinflammatory (M1) or immunomodulatory (M2). M2-polarised macrophages have been shown to influence tumour biology, by promoting tissue remodelling, angiogenesis and the secretion of growth hormones (Mantovani et al, 2002). Here we have analysed macrophages based on CD68, which does not distinguish the M1/M2 phenotype. There was no statistical significance between macrophages in MET+ and MET− samples. This is in agreement with previous data demonstrating that CD68+ cell density resulted in no correlation to survival or metastatic development (DeNardo et al, 2011). Differences in the prognostic significance of TAM could, in part, be explained by their ability to elicit either pro- or antitumour effects via their sub-populations (Ruffell et al, 2010).
There is an increasing need to improve prognostic classifiers in CRC. This could be achieved by utilising multiple relevant IHC biomarkers combined with using multivariate analysis methods to facilitate reliable stratification and exploit the broad dynamic range of IHC data. Here we present, in a small cohort of FFPE tissues, the potential of DPRIA to select and evaluate IHC staining to reveal biologically meaningful interdependencies of the CRC immunophenotype in relation to different components of the tumour microenvironment. Additional studies are warranted to further validate this technology, however, there is significant scope for the role of quantitative and automated image analysis and visualisation methods in automated digital pathology, diagnosis, prognosis and treatment planning.
Change history
17 September 2013
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Bui JD, Uppaluri R, Hsieh CS, Schreiber RD (2006) Comparative analysis of regulatory and effector T cells in progressively growing versus rejecting tumors of similar origins. Cancer Res 66 (14): 7301–7309.
Camparo P, Egevad L, Algaba F, Berney DM, Boccon-Gibod L, Comperat E, Evans AJ, Grobholz R, Kristiansen G, Langner C, Lopez-Beltran A, Montironi R, Oliveira P, Vainer B, Varma M (2012) Utility of whole slide imaging and virtual microscopy in prostate pathology. APMIS 120 (4): 298–304.
Camus M, Tosolini M, Mlecnik B, Pages F, Kirilovsky A, Berger A, Costes A, Bindea G, Charoentong P, Bruneval P, Trajanoski Z, Fridman WH, Galon J (2009) Coordination of intratumoral immune reaction and human colorectal cancer recurrence. Cancer Res 69 (6): 2685–2693.
Colombo MP, Piconese S (2007) Regulatory-T-cell inhibition versus depletion: the right choice in cancer immunotherapy. Nat Rev Cancer 7 (11): 880–887.
Curiel TJ, Coukos G, Zou L, Alvarez X, Cheng P, Mottram P, Evdemon-Hogan M, Conejo-Garcia JR, Zhang L, Burow M, Zhu Y, Wei S, Kryczek I, Daniel B, Gordon A, Myers L, Lackner A, Disis ML, Knutson KL, Chen L, Zou W (2004) Specific recruitment of regulatory T cells in ovarian carcinoma fosters immune privilege and predicts reduced survival. Nat Med 10 (9): 942–949.
DeNardo DG, Brennan DJ, Rexhepaj E, Ruffell B, Shiao SL, Madden SF, Gallagher WM, Wadhwani N, Keil SD, Junaid SA, Rugo HS, Hwang ES, Jirstrom K, West BL, Coussens LM (2011) Leukocyte complexity predicts breast cancer survival and functionally regulates response to chemotherapy. Cancer Disc 1 (1): 54–67.
Dunn GP, Bruce AT, Ikeda H, Old LJ, Schreiber RD (2002) Cancer immunoediting: from immunosurveillance to tumor escape. Nat Immunol 3 (11): 991–998.
Erreni M, Mantovani A, Allavena P (2011) Tumor-associated macrophages (TAM) and inflammation in colorectal cancer. Cancer Microenviron 4 (2): 141–154.
Fu J, Xu D, Liu Z, Shi M, Zhao P, Fu B, Zhang Z, Yang H, Zhang H, Zhou C, Yao J, Jin L, Wang H, Yang Y, Fu YX, Wang FS (2007) Increased regulatory T cells correlate with CD8 T-cell impairment and poor survival in hepatocellular carcinoma patients. Gastroenterology 132 (7): 2328–2339.
Galon J, Costes A, Sanchez-Cabo F, Kirilovsky A, Mlecnik B, Lagorce-Pages C, Tosolini M, Camus M, Berger A, Wind P, Zinzindohoue F, Bruneval P, Cugnenc PH, Trajanoski Z, Fridman WH, Pages F (2006) Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science 313 (5795): 1960–1964.
Galon J, Fridman WH, Pages F (2007) The adaptive immunologic microenvironment in colorectal cancer: a novel perspective. Cancer Res 67 (5): 1883–1886.
Galon J, Pages F, Marincola FM, Thurin M, Trinchieri G, Fox BA, Gajewski TF, Ascierto PA (2012) The immune score as a new possible approach for the classification of cancer. J Translational Med 10: 1.
Halama N, Michel S, Kloor M, Zoernig I, Pommerencke T, von Knebel Doeberitz M, Schirmacher P, Weitz J, Grabe N, Jager D (2009) The localization and density of immune cells in primary tumors of human metastatic colorectal cancer shows an association with response to chemotherapy. Cancer Immunity 9: 1.
Hanahan D, Weinberg RA (2011) Hallmarks of cancer: the next generation. Cell 144 (5): 646–674.
Jara-Lazaro AR, Thamboo TP, Teh M, Tan PH (2010) Digital pathology: exploring its applications in diagnostic surgical pathology practice. Pathology 42 (6): 512–518.
Lin EY, Li JF, Bricard G, Wang W, Deng Y, Sellers R, Porcelli SA, Pollard JW (2007) Vascular endothelial growth factor restores delayed tumor progression in tumors depleted of macrophages. Mol Oncol 1 (3): 288–302.
Mantovani A, Sozzani S, Locati M, Allavena P, Sica A (2002) Macrophage polarization: tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol 23 (11): 549–555.
Mizukami Y, Kono K, Kawaguchi Y, Akaike H, Kamimura K, Sugai H, Fujii H (2008) CCL17 and CCL22 chemokines within tumor microenvironment are related to accumulation of Foxp3+ regulatory T cells in gastric cancer. Int J Cancer 122 (10): 2286–2293.
Mlecnik B, Tosolini M, Kirilovsky A, Berger A, Bindea G, Meatchi T, Bruneval P, Trajanoski Z, Fridman WH, Pages F, Galon J (2011) Histopathologic-based prognostic factors of colorectal cancers are associated with the state of the local immune reaction. J Clin Oncol 29 (6): 610–618.
Naito Y, Saito K, Shiiba K, Ohuchi A, Saigenji K, Nagura H, Ohtani H (1998) CD8+ T cells infiltrated within cancer cell nests as a prognostic factor in human colorectal cancer. Cancer Res 58 (16): 3491–3494.
Pages F, Berger A, Camus M, Sanchez-Cabo F, Costes A, Molidor R, Mlecnik B, Kirilovsky A, Nilsson M, Damotte D, Meatchi T, Bruneval P, Cugnenc PH, Trajanoski Z, Fridman WH, Galon J (2005) Effector memory T cells, early metastasis, and survival in colorectal cancer. N Engl J Med 353 (25): 2654–2666.
Pages F, Galon J, Fridman WH (2008) The essential role of the in situ immune reaction in human colorectal cancer. J Leukoc Biol 84 (4): 981–987.
Pages F, Kirilovsky A, Mlecnik B, Asslaber M, Tosolini M, Bindea G, Lagorce C, Wind P, Marliot F, Bruneval P, Zatloukal K, Trajanoski Z, Berger A, Fridman WH, Galon J (2009) In situ cytotoxic and memory T cells predict outcome in patients with early-stage colorectal cancer. J Clin Oncol 27 (35): 5944–5951.
Randell R, Ruddle RA, Mello-Thoms C, Thomas RG, Quirke P, Treanor D (2013) Virtual reality microscope versus conventional microscope regarding time to diagnosis: an experimental study. Histopathology 62 (2): 351–358.
Ruffell B, DeNardo DG, Affara NI, Coussens LM (2010) Lymphocytes in cancer development: polarization towards pro-tumor immunity. Cytokine Growth Factor Rev 21 (1): 3–10.
Salama P, Phillips M, Grieu F, Morris M, Zeps N, Joseph D, Platell C, Iacopetta B (2009) Tumor-infiltrating FOXP3+ T regulatory cells show strong prognostic significance in colorectal cancer. J Clin Oncol 27 (2): 186–192.
Shimizu J, Yamazaki S, Sakaguchi S (1999) Induction of tumor immunity by removing CD25+CD4+ T cells: a common basis between tumor immunity and autoimmunity. J Immunol 163 (10): 5211–5218.
Tosolini M, Kirilovsky A, Mlecnik B, Fredriksen T, Mauger S, Bindea G, Berger A, Bruneval P, Fridman WH, Pages F, Galon J (2011) Clinical impact of different classes of infiltrating T cytotoxic and helper cells (Th1, th2, treg, th17) in patients with colorectal cancer. Cancer Res 71 (4): 1263–1271.
Woo EY, Chu CS, Goletz TJ, Schlienger K, Yeh H, Coukos G, Rubin SC, Kaiser LR, June CH (2001) Regulatory CD4(+)CD25(+) T cells in tumors from patients with early-stage non-small cell lung cancer and late-stage ovarian cancer. Cancer Res 61 (12): 4766–4772.
Yamaguchi T, Hirota K, Nagahama K, Ohkawa K, Takahashi T, Nomura T, Sakaguchi S (2007) Control of immune responses by antigen-specific regulatory T cells expressing the folate receptor. Immunity 27 (1): 145–159.
Yancopoulos GD, Davis S, Gale NW, Rudge JS, Wiegand SJ, Holash J (2000) Vascular-specific growth factors and blood vessel formation. Nature 407 (6801): 242–248.
Acknowledgements
This work was supported by the University of Nottingham, the EPSRC (EP/D501849/1) and AstraZeneca. We acknowledge the third party providers to the Alderley Park Biobank, Indivumed GmbH and the Manchester Cancer Research Centre (MCRC) Biobank. We acknowledge the scientific guidance of her PhD supervisors Professor David Pritchard and Dr Robert Wilkinson.
Author contributions
HKA performed all experiments and wrote the manuscript. CW, DIP, RWW and MC provided scientific guidance and participated in drafting and writing of the manuscript. CW provided pathological support. Neil Gray died on 31 July 2012. All other authors read and approved the final manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Information accompanies this paper on British Journal of Cancer website
Supplementary information
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Angell, H., Gray, N., Womack, C. et al. Digital pattern recognition-based image analysis quantifies immune infiltrates in distinct tissue regions of colorectal cancer and identifies a metastatic phenotype. Br J Cancer 109, 1618–1624 (2013). https://doi.org/10.1038/bjc.2013.487
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bjc.2013.487
Keywords
This article is cited by
-
A template to quantify the location and density of CD3 + and CD8 + tumor-infiltrating lymphocytes in colon cancer by digital pathology on whole slides for an objective, standardized immune score assessment
Cancer Immunology, Immunotherapy (2021)
-
Data integration from pathology slides for quantitative imaging of multiple cell types within the tumor immune cell infiltrate
Diagnostic Pathology (2017)
-
Quantitative assessment of pancreatic cancer precursor lesions in IHC-stained tissue with a tissue image analysis platform
Laboratory Investigation (2016)
-
Image analysis-derived metrics of histomorphological complexity predicts prognosis and treatment response in stage II-III colon cancer
Scientific Reports (2016)
-
Comparison of Monofractal, Multifractal and gray level Co-occurrence matrix algorithms in analysis of Breast tumor microscopic images for prognosis of distant metastasis risk
Biomedical Microdevices (2016)