Abstract
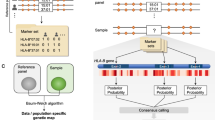
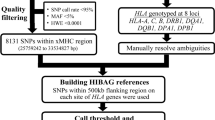
Four single nucleotide polymorphism (SNP)-based human leukocyte antigen (HLA) imputation methods (e-HLA, HIBAG, HLA*IMP:02 and MAGPrediction) were trained using 1000 Genomes SNP and HLA genotypes and assessed for their ability to accurately impute molecular HLA-A, -B, -C and –DRB1 genotypes in the Human Genome Diversity Project cell panel. Imputation concordance was high (>89%) across all methods for both HLA-A and HLA-C, but HLA-B and HLA-DRB1 proved generally difficult to impute. Overall, <27.8% of subjects were correctly imputed for all HLA loci by any method. Concordance across all loci was not enhanced via the application of confidence thresholds; reliance on confidence scores across methods only led to noticeable improvement (+3.2%) for HLA-DRB1. As the HLA complex is highly relevant to the study of human health and disease, a standardized assessment of SNP-based HLA imputation methods is crucial for advancing genomic research. Considerable room remains for the improvement of HLA-B and especially HLA-DRB1 imputation methods, and no imputation method is as accurate as molecular genotyping. The application of large, ancestrally diverse HLA and SNP reference data sets and multiple imputation methods has the potential to make SNP-based HLA imputation methods a tractable option for determining HLA genotypes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 6 print issues and online access
$259.00 per year
only $43.17 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Welter D, MacArthur J, Morales J, Burdett T, Hall P, Junkins H et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res 2014; 42: D1001–D1006.
Martin AM, Nolan D, Gaudieri S, Almeida CA, Nolan R, James I et al. Predisposition to abacavir hypersensitivity conferred by HLA-B*5701 and a haplotypic Hsp70-Hom variant. Proc Natl Acad Sci USA 2004; 101: 4180–4185.
Mallal S, Nolan D, Witt C, Masel G, Martin AM, Moore C et al. Association between presence of HLA-B*5701, HLA-DR7, and HLA-DQ3 and hypersensitivity to HIV-1 reverse-transcriptase inhibitor abacavir. Lancet 2002; 359: 727–732.
Hung SI, Chung WH, Liou LB, Chu CC, Lin M, Huang HP et al. HLA-B*5801 allele as a genetic marker for severe cutaneous adverse reactions caused by allopurinol. Proc Natl Acad Sci USA 2005; 102: 4134–4139.
McCormack M, Alfirevic A, Bourgeois S, Farrell JJ, Kasperaviciute D, Carrington M et al. HLA-A*3101 and carbamazepine-induced hypersensitivity reactions in Europeans. N Engl J Med 2011; 364: 1134–1143.
Pavlos R, Mallal S, Phillips E . HLA and pharmacogenetics of drug hypersensitivity. Pharmacogenomics 2012; 13: 1285–1306.
Erlich H . HLA DNA typing: past, present, and future. Tissue Antigens 2012; 80: 1–11.
Jia X, Han B, Onengut-Gumuscu S, Chen WM, Concannon PJ, Rich SS et al. Imputing amino acid polymorphisms in human leukocyte antigens. PLoS ONE 2013; 8: e64683.
Hirschhorn JN, Daly MJ . Genome-wide association studies for common diseases and complex traits. Nat Rev Genet 2005; 6: 95–108.
McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, Ioannidis JP et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet 2008; 9: 356–369.
de Bakker PI, Raychaudhuri S . Interrogating the major histocompatibility complex with high-throughput genomics. Hum Mol Genet 2012; 21: R29–R36.
Traherne JA . Human MHC architecture and evolution: implications for disease association studies. Int J Immunogenet 2008; 35: 179–192.
Fernando MM, Stevens CR, Walsh EC, De Jager PL, Goyette P, Plenge RM et al. Defining the role of the MHC in autoimmunity: a review and pooled analysis. PLoS Genet 2008; 4: e1000024.
Moore JH, Asselbergs FW, Williams SM . Bioinformatics challenges for genome-wide association studies. Bioinformatics 2010; 26: 445–455.
de Bakker PI, McVean G, Sabeti PC, Miretti MM, Green T, Marchini J et al. A high-resolution HLA and SNP haplotype map for disease association studies in the extended human MHC. Nat Genet 2006; 38: 1166–1172.
Malkki M, Single R, Carrington M, Thomson G, Petersdorf E . MHC microsatellite diversity and linkage disequilibrium among common HLA-A, HLA-B, DRB1 haplotypes: implications for unrelated donor hematopoietic transplantation and disease association studies. Tissue Antigens 2005; 66: 114–124.
Zheng X, Shen J, Cox C, Wakefield JC, Ehm MG, Nelson MR et al. HIBAG-HLA genotype imputation with attribute bagging. Pharmacogenomics J 2013; 14: 192–200.
Li SS, Wang H, Smith A, Zhang B, Zhang XC, Schoch G et al. Predicting multiallelic genes using unphased and flanking single nucleotide polymorphisms. Genet Epidemiol 2011; 35: 85–92.
Dilthey A, Leslie S, Moutsianas L, Shen J, Cox C, Nelson MR et al. Multi-population classical HLA type imputation. PLoS Comput Biol 2013; 9: e1002877.
Cann HM, de Toma C, Cazes L, Legrand MF, Morel V, Piouffre L et al. A human genome diversity cell line panel. Science 2002; 296: 261–262.
Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE et al. An integrated map of genetic variation from 1,092 human genomes. Nature 2012; 491: 56–65.
Parkes M, Cortes A, van Heel DA, Brown MA . Genetic insights into common pathways and complex relationships among immune-mediated diseases. Nat Rev Genet 2013; 14: 661–673.
Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA et al. The variant call format and VCFtools. Bioinformatics 2011; 27: 2156–2158.
Erlich H, Bugawan T, Begovich AB, Scharf S, Griffith R, Saiki R et al. HLA-DR, DQ and DP typing using PCR amplification and immobilized probes. Eur J Immunogenet 1991; 18: 33–55.
Marsh SG, Albert ED, Bodmer WF, Bontrop RE, Dupont B, Erlich HA et al. Nomenclature for factors of the HLA system, 2010. Tissue Antigens 2010; 75: 291–455.
Mack SJ, S MA, Meyer D,. Single RM, Tsai Y, Erlich HA . Methods used in the generation and preparation of data for analysis in the 13th International Histocompatibility Work- shop vol 1. IHWG Press: Seattle, WA, USA, 2007.
Frangoul H, Crowe D . Cost saving associated with implementing a stepwise approach to HLA typing of related donors before hematopoietic SCT. Bone Marrow Transplant 2014; 49: 850–851.
Robinson J, Halliwell JA, McWilliam H, Lopez R, Parham P, Marsh SG . The IMGT/HLA database. Nucleic Acids Res 2013; 41: D1222–D1227.
Dilthey AT, Moutsianas L, Leslie S, McVean G . HLA*IMP—an integrated framework for imputing classical HLA alleles from SNP genotypes. Bioinformatics 2011; 27: 968–972.
Vlachopoulou E, Lahtela E, Wennerstrom A, Havulinna AS, Salo P, Perola M et al. Evaluation of HLA-DRB1 imputation using a Finnish dataset. Tissue Antigens 2014; 83: 350–355.
Wissemann WT, Hill-Burns EM, Zabetian CP, Factor SA, Patsopoulos N, Hoglund B et al. Association of Parkinson disease with structural and regulatory variants in the HLA region. Am J Hum Genet 2013; 93: 984–993.
Kuniholm MH, Xie X, Anastos K, Xue X, Reimers L, French AL et al. Human leucocyte antigen class I and II imputation in a multiracial population. Int J Immunogenet 2016; 43: 369–375.
Andersson G . Evolution of the human HLA-DR region. Front Biosci 1998; 3: d739–d745.
Gorski J . The HLA-DRw8 lineage was generated by a deletion in the DR B region followed by first domain diversification. J Immunol 1989; 142: 4041–4045.
Traherne JA, Horton R, Roberts AN, Miretti MM, Hurles ME, Stewart CA et al. Genetic analysis of completely sequenced disease-associated MHC haplotypes identifies shuffling of segments in recent human history. PLoS Genet 2006; 2: e9.
Zhang XC, Li SS, Wang H, Hansen JA, Zhao LP . Empirical evaluations of analytical issues arising from predicting HLA alleles using multiple SNPs. BMC Genet 2011; 12: 39.
Hsieh AR, Chang SW, Chen PL, Chu CC, Hsiao CL, Yang WS et al. Predicting HLA genotypes using unphased and flanking single-nucleotide polymorphisms in Han Chinese population. BMC Genomics 2014; 15: 81.
Leslie S, Donnelly P, McVean G . A statistical method for predicting classical HLA alleles from SNP data. Am J Hum Genet 2008; 82: 48–56.
Thorisson GA, Smith AV, Krishnan L, Stein LD . The International HapMap Project Web site. Genome Res 2005; 15: 1592–1593.
Single RM, Meyer D, Mack SJ, Lancaster A, Nelson MP, Fernández-Viña M et al. Haplotype Frequencies and Linkage Disequilibrium among classical HLA genes vol. 1. IHWG Press: Seattle, WA, USA, 2007.
Pillai NE, Okada Y, Saw WY, Ong RT, Wang X, Tantoso E et al. Predicting HLA alleles from high-resolution SNP data in three Southeast Asian populations. Hum Mol Genet 2014; 23: 4443–4451.
Khor SS, Yang W, Kawashima M, Kamitsuji S, Zheng X, Nishida N et al. High-accuracy imputation for HLA class I and II genes based on high-resolution SNP data of population-specific references. Pharmacogenomics J 2015; 15: 530–537.
Levin AM, Adrianto I, Datta I, Iannuzzi MC, Trudeau S, McKeigue P et al. Performance of HLA allele prediction methods in African Americans for class II genes HLA-DRB1, -DQB1, and -DPB1. BMC Genet 2014; 15: 72.
Sasazuki T, Inoko H, Morishima S, Morishima Y . Gene map of the HLA region, Graves' disease and Hashimoto Thyroiditis, and hematopoietic stem cell transplantation. Adv Immunol 2016; 129: 175–249.
Begovich AB, McClure GR, Suraj VC, Helmuth RC, Fildes N, Bugawan TL et al. Polymorphism, recombination, and linkage disequilibrium within the HLA class II region. J Immunol 1992; 148: 249–258.
Solberg OD, Mack SJ, Lancaster AK, Single RM, Tsai Y, Sanchez-Mazas A et al. Balancing selection and heterogeneity across the classical human leukocyte antigen loci: a meta-analytic review of 497 population studies. Hum Immunol 2008; 69: 443–464.
Klitz W, Hedrick P, Louis EJ . New reservoirs of HLA alleles: pools of rare variants enhance immune defense. Trends Genet 2012; 28: 480–486.
Mack SJ, Cano P, Hollenbach JA, He J, Hurley CK, Middleton D et al. Common and well-documented HLA alleles: 2012 update to the CWD catalogue. Tissue Antigens 2013; 81: 194–203.
Acknowledgements
We thank Janelle Noble, Marc Salit and P Scott Pine for helpful discussions and Abeer Madbouly for assistance with the PCA plots. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH, NIAID, NINDS, NMSS, ONR or United States Government. This work was supported by Office of Naval Research (ONR) grant N00014-08-1-1207 (KB, DP, PAG, JAH, AL, SJM, MM and VP), National Institutes of Health (NIH) grants U01AI067068 (JAH and SJM) and U19AI067152 (ARRA administrative supplement) (PAG) awarded by the National Institute of Allergy and Infectious Diseases (NIAID), R01GM109030 (JAH, SJM and DJP) and P01GM099568 (XZ) awarded by the National Institute of General Medical Sciences (NIGMS), RO1NS076492 (PAG), RO1NS046297 (PAG) and R01NS049477 (PAG) awarded by the National Institute of Neurological Disorders and Stroke (NINDS), and National Multiple Sclerosis Society (NMSS) grant RG 2899-D11 (PAG). PAG is a recipient of the Race to Erase MS Junior Investigator Award and the European Federation for Immunogenetics Julia Bodmer Award. This work was supported by the Australian National Health and Medical Research Council (NHMRC), Career Development Fellowship ID 1053756 (S.L.); and by the Victorian Life Sciences Computation Initiative (VLSCI) grant number VR0240 on its Peak Computing Facility at the University of Melbourne, an initiative of the Victorian Government, Australia (S.L.). Research at the Murdoch Childrens Research Institute was supported by the Victorian Government’s Operational Infrastructure Support Program. We thank President Barack H. Obama for his support and appreciation of American science and basic research.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
SL is a partner in Peptide Groove LLP. Peptide Groove has licensed HLA typing technology to Affymetrix Ltd.
Additional information
Supplementary Information accompanies the paper on the The Pharmacogenomics Journal website
Supplementary information
Rights and permissions
About this article
Cite this article
Pappas, D., Lizee, A., Paunic, V. et al. Significant variation between SNP-based HLA imputations in diverse populations: the last mile is the hardest. Pharmacogenomics J 18, 367–376 (2018). https://doi.org/10.1038/tpj.2017.7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/tpj.2017.7
This article is cited by
-
The lupus susceptibility allele DRB1*03:01 encodes a disease-driving epitope
Communications Biology (2022)
-
HLA-check: evaluating HLA data from SNP information
BMC Bioinformatics (2017)