Abstract
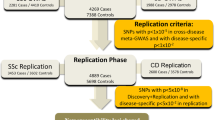
Up to 50% of multiple sclerosis (MS) patients do not respond to interferon-beta (IFN-β) treatment and determination of response requires lengthy clinical follow-up of up to 2 years. Response predictive genetic markers would significantly improve disease management. We aimed to identify IFN-β treatment response genetic marker(s) by performing a two-stage genome-wide association study (GWAS). The GWAS was carried out using data from 151 Australian MS patients from the ANZgene/WTCCC2 MS susceptibility GWAS (responder (R)=51, intermediate responders=24 and non-responders (NR)=76). Of the single-nucleotide polymorphisms (SNP) that were validated in an independent group of 479 IFN-β-treated MS patients from Australia, Spain and Italy (R=273 and NR=206), eight showed evidence of association with treatment response. Among the replicated associations, the strongest was observed for FHIT (Fragile Histidine Triad; combined P-value 6.74 × 10−6) and followed by variants in GAPVD1 (GTPase activating protein and VPS9 domains 1; combined P-value 5.83 × 10−5) and near ZNF697 (combined P-value 8.15 × 10−5).
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 6 print issues and online access
$259.00 per year
only $43.17 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Marziniak M, Meuth S . Current perspectives on interferon beta-1b for the treatment of multiple sclerosis. Adv Ther 2014; 31: 915–931.
Río J, Nos C, Tintoré M, Téllez N, Galán I, Pelayo R et al. Defining the response to interferon‐β in relapsing‐remitting multiple sclerosis patients. Ann Neurol 2006; 59: 344–352.
Roden D, Tyndale R . Pharmacogenomics at the tipping point: challenges and opportunities. Clin Pharmacol Toxicol 2011; 89: 323–327.
Byun E, Caillier SJ, Montalban X, Villoslada P, Fernández O, Brassat D et al. Genome-wide pharmacogenomic analysis of the response to interferon beta therapy in multiple sclerosis. JAMA Neurol 2008; 65: 337–344.
Comabella M, Craig DW, Morcillo-Suárez C, Río J, Navarro A, Fernández M et al. Genome-wide scan of 500 000 single-nucleotide polymorphisms among responders and nonresponders to interferon beta therapy in multiple sclerosis. JAMA Neurol 2009; 66: 972–978.
Esposito F, Sorosina M, Ottoboni L, Lim ET, Replogle JM, Raj T et al. A pharmacogenetic study implicates SLC9a9 in multiple sclerosis disease activity. Ann Neurol 2015; 78: 115–127.
The Australia and New Zealand Multiple Sclerosis Genetics Consortium (ANZgene) 2009. Genome-wide association study identifies new multiple sclerosis susceptibility loci on chromosomes 12 and 20. Nat Genet 2009; 41: 824–828.
International Multiple Sclerosis Genetics Consortium and Wellcome Trust. Case Control Consortium 2. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature 2011; 476: 214–219.
Pearce M, de Feo G . The MassARRAY® System is a rapid, cost-effective platform for the validation of data generated from GWAS and Next Gen Sequencing. Nat Methods 2010. doi:10.1038/an7787.
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Med Genet A 2007; 81: 559–575.
R Core Team. (2014) R: a language and environment for statistical computing. R Foundation for Statistical Computing: Vienna, Austria, 2012, ISBN 3-900051-07-0.
Li Q, Zheng G, Liang X, Yu K . Robust tests for single‐marker analysis in case‐control genetic association studies. Ann Hum Genet 2009; 73: 245–252.
Moldovan M, Langaas M . Exact conditional p-values from arbitrary ranking of a sample space: an application to genome-wide association studies. Adv Syst Sci Appl 2014; 14: 76–83.
McCullagh P . Proportional‐Odds Model. Encyclopedia of Biostatistics 2005; doi: 10.1002/0470011815.b2a10049.
Gamazon ER, Zhang W, Konkashbaev A, Duan S, Kistner EO, Nicolae DL et al. SCAN: SNP and copy number annotation. Bioinformatics 2010; 26: 259–262.
Hu Z, Chang Y-C, Wang Y, Huang C-L, Liu Y, Tian F et al. VisANT 4.0: Integrative network platform to connect genes, drugs, diseases and therapies. Nucleic Acids Res 2013; 41: W225–W231.
Cénit M, Blanco-Kelly F, de Las Heras V, Bartolomé M, De la Concha E, Urcelay E et al. Glypican 5 is an interferon-beta response gene: a replication study. Mult Scler 2009; 15: 913–917.
Mabb AM, Ehlers MD . Ubiquitination in postsynaptic function and plasticity. Annu Rev Cell Dev Biol 2010; 26: 179.
Mahurkar S, Moldovan M, Suppiah V, O’Doherty C . Identification of shared genes and pathways: a comparative study of multiple sclerosis susceptibility, severity and response to interferon Beta treatment. PloS One 2013; 8: e57655.
Sormani MP, De Stefano N . Defining and scoring response to IFN-β in multiple sclerosis. Nat Rev Neurol 2013; 9: 504–512.
Dobson R, Rudick RA, Turner B, Schmierer K, Giovannoni G . Assessing treatment response to interferon-β Is there a role for MRI? Neurology 2014; 82: 248–254.
Feng Z, Prentice R, Srivastava S . Research issues and strategies for genomic and proteomic biomarker discovery and validation: a statistical perspective. Pharmacogenomics 2004; 5: 709–719.
Zou G, Zhao H . The impacts of errors in individual genotyping and DNA pooling on association studies. Genet Epidemiol 2004; 26: 1–10.
Barrett JC, Cardon LR . Evaluating coverage of genome-wide association studies. Nat Genet 2006; 38: 659–662.
Tanaka Y, Nishida N, Sugiyama M, Kurosaki M, Matsuura K, Sakamoto N et al. Genome-wide association of IL28B with response to pegylated interferon-α and ribavirin therapy for chronic hepatitis C. Nat Genet 2009; 41: 1105–1109.
Rotstein D, Healy B, Malik M, Carruthers R, Musallam A et al. Differential effects of vitamin D in GA-versus IFN-Treated MS Patient. Neurology 2014; 82, S24. 005-S24. 005.
Capuano A, Dawson J, Graya G . Maximizing power in seroepidemiological studies through the use of the proportional odds model. Influenza Other Respir Viruses 2007; 3: 87–93.
Potamias G, Lakiotaki K, Katsila T, Lee MTM, Topouzis S, Cooper DN et al. Deciphering next-generation pharmacogenomics: an information technology perspective. Open Biol 2014; 4: 140071.
Butzkueven H, Chapman J, Cristiano E, Grand’Maison F, Hoffmann M, Izquierdo G et al. MSBase: an international, online registry and platform for collaborative outcomes research in multiple sclerosis. Mult Scler 2006; 12: 769–774.
Weiske J, Albring KF, Huber O . The tumor suppressor Fhit acts as a repressor of β-catenin transcriptional activity. Proc Natl Acad Sci USA 2007; 104: 20344–20349.
Nakagawa Y, Akao Y . Fhit protein inhibits cell growth by attenuating the signaling mediated by nuclear factor-κB in colon cancer cell lines. Exp Cell Res 2006; 312: 2433–2442.
The International Multiple Sclerosis Genetics Consortium 2007. Risk alleles for multiple sclerosis identified by a genomewide study. N Engl J Med 2007; 357: 851–862.
Giacalone G, Clarelli F, Osiceanu A, Guaschino C, Brambilla P, Sorosina M et al. Analysis of genes, pathways and networks involved in disease severity and age at onset in primary-progressive multiple sclerosis. Mult Scler 2015; 21: 1431–1442.
Arthur AT, Armati PJ, Bye C, Heard RN, Stewart GJ, Pollard JD et al. Genes implicated in multiple sclerosis pathogenesis from consilience of genotyping and expression profiles in relapse and remission. BMC Med Genet 2008; 9: 17.
Liggett T, Melnikov A, Tilwalli S, Yi Q, Chen H, Replogle C et al. Methylation patterns of cell-free plasma DNA in relapsing–remitting multiple sclerosis. J Neurol Sci 2010; 290: 16–21.
Lodhi IJ, Chiang S-H, Chang L, Vollenweider D, Watson RT, Inoue M et al. Gapex-5, a Rab31 guanine nucleotide exchange factor that regulates Glut4 trafficking in adipocytes. Cell Metab 2007; 5: 59–72.
Laity JH, Lee BM, Wright PE . Zinc finger proteins: new insights into structural and functional diversity. Curr Opin Struct Biol 2001; 11: 39–46.
Acknowledgements
The members of the ANZgene Consortium are: Alan Baxter (James Cook University, Townsville, Australia), Allan G Kermode (Sir Charles Gairdner Hospital, Perth, Australia), Bruce Taylor (Menzies Research Institute Tasmania, University of Tasmania, Hobart, Australia), David R Booth (ANZgene Consortium Chair) (Westmead Millennium Institute, University of Sydney, Sydney, Australia) david.booth@sydney.edu.au, Deborah Mason (Canterbury District Health Board, Christchurch, New Zealand), Graeme J Stewart (Westmead Millennium Institute, University of Sydney, Sydney, Australia), Helmut Butzkueven (University of Melbourne, Melbourne, Australia), Jac Charlesworth (Menzies Research Institute Tasmania, University of Tasmania, Hobart, Australia), James Wiley (Florey Institute of Neuroscience and Mental Health, University of Melbourne, Melbourne, Australia), Jeannette Lechner- Scott (Hunter Medical Research Institute, Newcastle, Australia), Judith Field (Florey Institute of Neuroscience and Mental Health, University of Melbourne, Melbourne, Australia), Lotti Tajouri (Bond University, Gold Coast, Australia), Lyn Griffiths (Griffith Institute of Health and Medical Research, Griffith University, Gold Coast, Australia), Mark Slee (School of Medicine, Flinders University of South Australia, Adelaide, Australia), Matthew A Brown (University of Queensland Diamantina Institute, Translational Research Institute, Brisbane, Australia), Pablo Moscato (Hunter Medical Research Institute, Newcastle, Australia), Rodney J Scott (Hunter Medical Research Institute, Newcastle, Australia), Simon Broadley (School of Medicine, Griffith University, Gold Coast, Australia), Steve Vucic (Westmead Millennium Institute, University of Sydney, Sydney, Australia), Trevor Kilpatrick (University of Melbourne, Melbourne, Australia), William M Carroll (Sir Charles Gairdner Hospital, Perth, Australia). We thank individuals with MS in Australia for supporting this research and all investigators of the study who have contributed to the recruitment of MS patients. This work was supported by a University of South Australia, Division of Health Sciences Research Development Grant.
Membership of the ANZgene Consortium
We would like to thank the Welcome Trust Case Control Consortium 2 for access to genotype data for a number of Australian patients included in the discovery stage of the current study. We also thank the Australian Genome Research Facility forassisting with genotyping of the samples in the validation study.
Author information
Authors and Affiliations
Consortia
Corresponding author
Ethics declarations
Competing interests
G Comi has received honoraria for consultancy and/or speaking activities in the past 12 months from Biogen, Novartis, Teva, Sanofi, Genzyme, Merck Serono, Bayer, Serono Symposia International Foundation, Roche, Almirall, Chugai, Receptos. V Martinelli has received honoraria and travel reimbursement for lectures at Meetings and Congresses from Teva Pharma, Bayer, Genzyme, Biogen Idec, Novartis and Merck Serono; he has received research support from Merck Serono and has served as a member in Advisory Board for Bayer, Genzyme, Biogen Idec, Novartis and Merck Serono.
Additional information
Supplementary Information accompanies the paper on the The Pharmacogenomics Journal website
PowerPoint slides
Rights and permissions
About this article
Cite this article
Mahurkar, S., Moldovan, M., Suppiah, V. et al. Response to interferon-beta treatment in multiple sclerosis patients: a genome-wide association study. Pharmacogenomics J 17, 312–318 (2017). https://doi.org/10.1038/tpj.2016.20
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/tpj.2016.20
This article is cited by
-
Pharmacogenetic Predictors of Response to Interferon Beta Therapy in Multiple Sclerosis
Molecular Neurobiology (2021)
-
The Challenge to Search for New Nervous System Disease Biomarker Candidates: the Opportunity to Use the Proteogenomics Approach
Journal of Molecular Neuroscience (2019)
-
A pharmacogenetic signature of high response to Copaxone in late-phase clinical-trial cohorts of multiple sclerosis
Genome Medicine (2017)
-
Neuroinflammation — using big data to inform clinical practice
Nature Reviews Neurology (2016)