Abstract
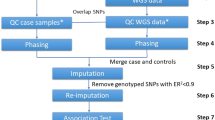
The discordance in results between independent genome-wide association studies (GWAS) indicates the potential for Type I and Type II errors. To identify the causes of variability underlying lack of reproducibility, here we present the results of a repeatability experiment on GWAS on a cohort of 1991 coronary artery disease individuals and 1500 controls (National Blood Service) provided by the Wellcome Trust Case Control Consortium. As part of the MicroArray Quality Control project, we identified quality control (QC) and association analysis steps with a major impact on the identification of candidate markers for possible classifiers. Different experimental conditions were used with the CHIAMO calling algorithm to assess the effects of batch size and case–control composition on downstream association analysis. Results showed that both composition and size create discordant single-nucleotide polymorphism (SNP) results for QC and statistical analysis and may contribute to the lack of reproducibility in GWAS. An interactive effect of batch size and composition contributes to discordant results in significantly associated loci. About 800 significant SNPs (Cochran–Armitage trend test, P<5.0 × 10−7) were found for batches of 2000 samples with separated cases and controls, whereas only 14 significant markers were found with one batch of all samples.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 6 print issues and online access
$259.00 per year
only $43.17 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Sladek R, Rocheleau G, Rung J, Dina C, Shen L, Serre D et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature 2007; 445: 881–885.
The Wellcome Trust Case Control Consortium. Genome-wide association study of 14 000 cases of seven common diseases and 3000 shared controls. Nature 2007; 447: 661–678.
Thorleifsson G, Walters GB, Gudbjartsson DF, Steinthorsdottir V, Sulem P, Helgadottir A et al. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat Genet 2009; 41: 18–24.
Shete S, Hosking FJ, Robertson LB, Dobbins SE, Sanson M, Malmer B et al. Genome-wide association study identifies five susceptibility loci for glioma. Nat Genet 2009; 41: 899–906.
Ioannidis JPA, Allison DB, Ball CA, Coulibaly I, Cui X, Culhane AC et al. Repeatability of published microarray gene expression analyses. Nat Genet 2009; 41: 149–155.
Zeggini E, Weedon MN, Lindgren CM, Frayling TM, Elliott KS, Lango H et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science 2007; 316: 1336–1341.
Scott LJ, Mohlke KL, Bonnycastle LL, Willer CJ, Li Y, Duren WL et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science 2007; 316: 1341–1345.
Easton DF, Pooley KA, Dunning AM, Pharoah PDP, Thompson D, Ballinger DG et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature 2007; 447: 1087–1093.
Ioannidis JPA . Non-replication and inconsistency in the genome-wide association setting. Hum Hered 2007; 64: 203–213.
Ioannidis JPA, Patsopoulos NA, Evangelou E . Heterogeneity in meta-analyses of genome-wide association investigations. PLoS One 2007; 2: e841.
Kavvoura FK, McQueen MB, Khoury MJ, Tanzi RE, Bertram L, Ioannidis JPA . Evaluation of the potential excess of statistically significant findings in published genetic association studies: application to Alzheimer's disease. Am J Epidemiol 2008; 168: 855–865.
Zeggini E, Ioannidis JPA . Meta-analysis in genome-wide association studies. Pharmacogenomics 2009; 10: 191–201.
Kingsmore SF, Lindquist IE, Mudge J, Gessler DD, Beavis WD . Genome-wide association studies: progress and potential for drug discovery and development. Nat Rev Drug Discov 2008; 7: 221–230.
Ioannidis JPA, Trikalinos TA, Khoury MJ . Implications of small effect sizes of individual genetic variants on the design and interpretation of genetic association studies of complex diseases. Am J Epidemiol 2006; 164: 609–614.
Kraft P, Zeggini E, Ioannidis JPA . Replication in genome-wide association studies. Stat Sci 2009; 24: 561–573.
Donnelly P . Progress and challenges in genome-wide association studies in humans. Nature 2008; 456: 728–731.
Saxena R, Voight BF, Lyssenko V, Burtt NP, de Bakker P, Chen H et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science 2007; 316: 1331–1336.
Gold B, Kirchhoff T, Stefanov S, Lautenberger JL, Viale A, Garber J et al. Genome-wide association study provides evidence for a breast cancer risk locus at 6q22.33. Proc Natl Acad Sci USA 2008; 105: 4340–4345.
Affymetrix White Paper Publication. BRLMM: an improved genotype calling method for the GeneChip Human Mapping 500K array set 2006.
Hua J, Craig DW, Brun M, Webster J, Zismann V, Tembe W et al. SNiPer-HD: improved genotype calling accuracy by an expectation-maximization algorithm for high-density SNP arrays. Bioinformatics 2007; 23: 57–63.
Carvalho B, Bengtsson H, Speed TP, Irizarry RA . Exploration, normalization, and genotype calls of high-density oligonucleotide SNP array data. Biostatistics 2007; 8: 485–499.
Hong H, Su Z, Ge W, Shi L, Perkins R, Fang H et al. Assessing batch effects of genotype calling algorithm BRLMM for the Affymetrix GeneChip Human Mapping 500K array set using 270 HapMap samples. BMC Bioinf 2008; 9: S17.
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D et al. PLINK: a toolset for whole-genome association and population-based linkage analysis. Am J Hum Genet 2007; 81: 559–575.
Armitage P . Tests for linear trends in proportions and frequencies. Biometrics 1971; 11: 375–386.
Lin S, Carvalho B, Cutler DJ, Arking DE, Chakravarti A, Irizarry RA . Validation and extension of an empirical Bayes method for SNP calling on Affymetrix microarrays. Genome Biol 2008; 9: R63.
MAQC Consortium. The MicroArray Quality Control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat Biotechnol 2006; 24: 1151–1161.
Acknowledgements
We acknowledge the contribution of S Paoli and R Flor for assistance in developing and administrating the FBK high-performance computing facility, on which most of the analyses were run, and of M Showe for useful comments on an earlier version of the paper. We truly thank F Goodsaid for scientific motivation and coordination of the MAQC Genome Wide Association working group and Dr George Wells from the University of Ottawa Heart Institute for providing access to the data.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Chierici, M., Miclaus, K., Vega, S. et al. An interactive effect of batch size and composition contributes to discordant results in GWAS with the CHIAMO genotyping algorithm. Pharmacogenomics J 10, 355–363 (2010). https://doi.org/10.1038/tpj.2010.47
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/tpj.2010.47
Keywords
This article is cited by
-
A genomic data archive from the Network for Pancreatic Organ donors with Diabetes
Scientific Data (2023)
-
M3-S: a genotype calling method incorporating information from samples with known genotypes
BMC Bioinformatics (2015)
-
Variability in GWAS analysis: the impact of genotype calling algorithm inconsistencies
The Pharmacogenomics Journal (2010)